Abstract
The activities of tigecycline alone and in combination with other antimicrobials are not well defined for carbapenem-intermediate or -resistant Acinetobacter baumannii (CIRA). Pharmacodynamic activity is even less well defined when clinically achievable serum concentrations are considered. Antimicrobial susceptibility testing of clinical CIRA isolates from 2001 to 2005 was performed by broth or agar dilution, as appropriate. Tigecycline concentrations were serially increased in time-kill studies with a representative of the most prevalent carbapenem-resistant clone (strain AA557; imipenem MIC, 64 mg/liter). The in vitro susceptibility of the strain was tested by time-kill studies in duplicate against the average free serum steady-state concentrations of tigecycline alone and in combination with various antimicrobials. Ninety-three CIRA isolates were tested and were found to have the following antimicrobial susceptibility profiles: tigecycline, MIC50 of 1 mg/liter and MIC90 of 2 mg/liter; minocycline, MIC50 of 0.5 mg/liter and MIC90 of 8 mg/liter; doxycycline, MIC50 of 2 mg/liter and MIC90 of ≥32 mg/liter; ampicillin-sulbactam, MIC50 of 48 mg/liter and MIC90 of 96 mg/liter; ciprofloxacin, MIC50 of ≥16 mg/liter and MIC90 of ≥16 mg/liter; rifampin, MIC50 of 4 mg/liter and MIC90 of 8 mg/liter; polymyxin B, MIC50 of 1 mg/liter and MIC90 of 1 mg/liter; amikacin, MIC50 of 32 mg/liter and MIC90 of ≥32 mg/liter; meropenem, MIC50 of 16 mg/liter and MIC90 of ≥128 mg/liter; and imipenem, MIC50 of 4 mg/liter and MIC90 of 64 mg/liter. Among the tetracyclines, the isolates were more susceptible to tigecycline than minocycline and doxycycline, according to FDA breakpoints (95%, 88%, and 71% of the isolates were susceptible to tigecycline, minocycline, and doxycycline, respectively). Concentration escalation studies with tigecycline revealed a maximal killing effect near the MIC, with no additional extent or rate of killing at concentrations 2× to 4× the MIC for tigecycline. Time-kill studies demonstrated indifference for tigecycline in combination with the antimicrobials tested. Polymyxin B, minocycline, and tigecycline are the most active antimicrobials in vitro against CIRA. Concentration escalation studies demonstrate that tigecycline may need to approach concentrations higher than those currently achieved in the bloodstream to adequately treat CIRA bloodstream infections. Future studies should evaluate these findings in vivo.
Bloodstream infections with Acinetobacter baumannii are occurring with increasing frequency, resulting in significant morbidity and mortality. The attributable mortality rate for infections ranges from 8 to 43% (11, 26), and A. baumannii continues to emerge as a health care-associated pathogen (35). Carbapenems, tetracyclines, and polymyxins are the most frequently active drugs against such strains (3, 12, 35). However, carbapenem-intermediate or -resistant A. baumannii (CIRA) strains are becoming increasingly prevalent (27), with few therapeutic options for the treatment of infections with this organism.
Among the tetracyclines, tigecycline is less active than minocycline against Acinetobacter spp. on a weight-per-weight basis (14); however, intravenous minocycline is not currently available in the United States. Therefore, tigecycline is likely the tetracycline of choice for the treatment of infections caused by CIRA. Prior investigations reveal that tigecycline is bacteriostatic against CIRA (24), but drug combinations with tigecycline at the achievable serum concentrations have not been sufficiently evaluated in vitro.
This study sought to determine the change in the activity of tigecycline against CIRA with increasing concentrations of drug. Additionally, we explored the activities of tigecycline in combination with other antimicrobial agents with activity against CIRA, including ampicillin-sulbactam, ciprofloxacin, levofloxacin, rifampin, polymyxin B, amikacin, meropenem, and imipenem. All combinations were modeled from average, steady-state, free, and serum concentrations. Finally, we describe the predictive value of the antimicrobial activities of doxycycline and minocycline for the antimicrobial activity of tigecycline.
MATERIALS AND METHODS
Bacterial strains.
Isolates of A. baumannii complex were obtained from clinical specimens submitted to the clinical microbiology laboratory at Northwestern Memorial Hospital (Chicago, IL). One isolate per patient per calendar year was obtained, in accordance with the criteria of the CLSI (formerly the NCCLS) (21). Clinical specimens were collected from January 2001 to December 2005. Species identification was performed with the Vitek II system (bioMérieux, Balmes-les-Grottes, France) or manually with biochemicals, when necessary (4). Isolates with an imipenem or meropenem MIC of ≥8 mg/liter with the Vitek II system were included in the study. Isolates were stored in Mueller-Hinton broth with 15% glycerol and frozen at −80°C. Isolates were subcultured a minimum of three times prior to experimentation.
Molecular analysis.
Pulsed-field gel electrophoresis (PFGE) was performed with all isolates. The SmaI and ApaI restriction enzymes were used as described previously (13). The similarity between isolates was determined by visual comparison of the DNA banding patterns by using the criteria of Tenover et al. (31), and isolates with a difference of greater than six bands were considered genetically distinct.
Antimicrobial agents.
Laboratory-grade standard powders of the following antimicrobials were obtained and reconstituted according to the manufacturers' instructions: tigecycline (Wyeth Research, Pearl River, NY); ciprofloxacin, doxycycline, rifampin, polymyxin B, amikacin, and minocycline (Sigma Chemical Company, St. Louis, MO); meropenem (AstraZeneca, Waltham, MA); imipenem (Merck & Company, Rathway, NJ); ampicillin-sulbactam (Pfizer, Groton, CT); and levofloxacin (Johnson and Johnson, Springhouse, PA). Stock solutions of tigecycline were freshly prepared on the day of the experiments to avoid known degradation problems (5). All other antibiotics were freshly prepared or were prepared and frozen at −20°C and given a 6-month expiration date. All antimicrobials were required to pass quality control standards on the day of the experiment (8, 20). Ampicillin-sulbactam was tested as a combined product in a ratio of 2:1.
MIC determination.
Individual CIRA isolates were cultured on Trypticase soy agar with sheep blood (Remel, Lenexa, KS) and incubated in an incubator at 35°C. Four to six colonies were selected and inoculated in 1 ml of brain heart infusion broth (Remel) in sterile glass tubes. The broths were incubated for 2 h and then diluted with broth to a 0.5 McFarland standard.
Agar dilution.
All antibiotics with the exception of tigecycline were tested by agar dilution for MIC determination. Mueller-Hinton agar (Remel) was prepared according to the manufacturer's specifications, autoclaved, and supplemented with 25 mg of calcium/liter and 12.5 mg of magnesium/liter. Autoclaved replicators designed to deliver 1 to 2 μl (∼104 CFU) were used to inoculate antibiotic-containing agar. Agar plates were incubated at 35°C for 18 to 24 h. Growth greater than a single colony or a slight haze was required for substantiation of growth.
Broth microdilution.
Tigecycline MIC determinations were performed by broth microdilution. Mueller-Hinton broth (Becton Dickinson, Sparks, MD) was freshly prepared according to the manufacturer's specifications and autoclaved. The Mueller-Hinton broth was supplemented with 25 mg of calcium/liter and 12.5 mg of magnesium/liter (CAMHB). Approximately 5 × 105 CFU/ml was mixed with serial drug dilutions and a control solution to obtain a volume of 100 μl in sterile broth microdilution trays. The trays were incubated in ambient air at 35°C for 18 to 24 h.
Susceptibility interpretation.
MICs were interpreted with category designations according to the criteria of the CLSI (8). Breakpoints for tigecycline susceptibility were interpreted according to the FDA breakpoint of 2 mg/liter (Tygacil package insert; Wyeth Pharmaceuticals Inc., Philadelphia, PA). Positive predictive values for doxycycline and minocycline susceptibility were calculated for prediction of tigecycline susceptibility.
Time-kill studies.
Concentration escalation time-kill studies were completed in accordance with the guidelines of the CLSI (20). One bloodstream isolate representative of our most common genotype of CIRA (isolate AA557; imipenem MIC, 64 mg/liter) was used for all time-kill experiments. Glass tubes were autoclaved and filled with freshly prepared CAMHB to a volume of 10 ml with no antibiotic (growth control) and tigecycline at 0.5× MIC, 1× MIC, 2× MIC, or 4× MIC. In addition, a calculated average serum concentration at steady state was tested. All experiments were performed in duplicate.
Several colonies (4 to 6 CFU) were selected and suspended in 1 ml of CAMHB. The colonies were allowed to grow to mid-logarithmic growth (4 h). Suspensions were adjusted to a 0.5 McFarland standard (∼1 × 108 CFU/ml) in 1 ml of CAMHB. Then, 0.5 ml was withdrawn and mixed with CAMHB to equal a volume of 1 ml. One hundred microliters of this suspension was removed and diluted in the premade antibiotic and growth control solutions to equal a resultant concentration of ∼5 × 105 CFU/ml.
Serial samples were obtained for colony quantification. The plates at time zero functioned as the purity plates. Aliquots were removed at time zero and 2, 4, 8, 24, and 48 h. Antibiotic carryover was minimized through serial dilution with normal saline (10−1 to 10−8). Aliquots of 25 μl were plated on Trypticase soy agar with sheep blood (Remel) in duplicate. Bacterial counts (expressed as log10 CFU/ml) were completed after incubation from 18 to 24 h at 35°C in ambient air. The lower limit of detection was 40 CFU/ml. Colony counts were averaged between the duplicate plates for each experiment. This resulted in an average of four quantified growth plates per time point for each drug concentration.
Synergy studies with time-kill.
To determine the achievable serum concentrations based on FDA-approved labeling, the free drug area under the curve for 24 h (fAUC24) or the free drug maximum concentrations (fCmax) and free drug minimum concentrations (fCmin) were obtained from FDA-approved package inserts or the literature for all drugs under steady-state conditions. Steady-state concentrations were estimated by use of the following equations: fAUC24/24 h or (fCmax + fCmin)/2, where fCmin represents the trough concentration at the end of a dosing interval (i.e., the lowest value prior to normal redosing of the antibiotic).
The following concentrations were used for the time-kill experiments: rifampin, 2 mg/liter (rifampin package insert; Bedford Laboratories, Bedford, OH); meropenem, 8 mg/liter (9); imipenem, 4 mg/liter (30); ampicillin-sulbactam in a 2:1 ratio, 48 mg/liter (28); ciprofloxacin, 1 mg/liter (Cipro package insert; Bayer Pharmaceuticals West Haven, CT); levofloxacin, 4 mg/liter (Levaquin package insert; Ortho-McNeal Pharmaceutical, Raritan, NJ); amikacin, 32 mg/liter (amikacin package insert; Sicor Pharmaceuticals, Irvine, CA); polymyxin B, 2 mg/liter; and tigecycline, 0.125 mg/liter (16).
Pharmacodynamic endpoints.
Bactericidal activity (99.9% killing) was defined as a 3-log10-CFU/ml reduction in the colony count from the initial inoculum (1, 20). Enhancement of activity was defined as an increase in killing of 2 log10 CFU/ml by a combination of antimicrobials compared with the killing by the most active single antimicrobial agent (1). Improvement was defined as a <2-log10 increase in killing in comparison to that by the most active single agent, while combinations that resulted in 1-log10 bacterial growth in comparison to that achieved with tigecycline alone were considered antagonistic (1). Other changes in colony counts were considered indifference. Colony counts were determined between 18 and 24 h.
RESULTS
A total of 93 CIRA isolates were identified by the Vitek II system and tested for their antimicrobial susceptibilities (Table 1). PFGE analysis revealed that 27% (25/93) of the isolates were genetically distinct. The lungs and blood were the most common sources of the isolates (50% and 20%, respectively). MIC50 and MIC90 values are presented in Table 1. Phenotypic imipenem resistance (MIC ≥ 8 mg/liter) was identified in 28 isolates by agar dilution. Only seven of these isolates were genotypically distinct. During the time period from 2001 to 2004, only four isolates displayed imipenem MICs ≥8 mg/liter. PFGE analysis revealed that all of these isolates were genotypically distinct. The remaining 24 isolates were obtained in 2005, and PFGE analysis revealed that a single clone of A. baumannii was responsible for 88% (21/24) of the A. baumannii isolates with resistance to imipenem.
TABLE 1.
In vitro susceptibilities for 93 CIRA isolates
| Drug | MIC (mg/liter)
|
No. (%) susceptible | ||
|---|---|---|---|---|
| 50% | 90% | Breakpoint | ||
| Polymyxin B | 1 | 1 | 2 | 93 (100) |
| Minocycline | 0.5 | 8 | 4 | 77 (83) |
| Doxycycline | 2 | >32 | 4 | 52 (56) |
| Tigecycline | 1 | 2 | 2 | 88 (95) |
| Rifampin | 4 | 8 | None | NA |
| Meropenem | 16 | >128 | 4 | 4 (4) |
| Imipenem | 4 | 64 | 4 | 19 (20) |
| Ampicillin-sulbactam | 48 | 96 | 12 | 19 (20) |
| Ciprofloxacin | >16 | >16 | 1 | 0 (0) |
| Amikacin | 32 | >32 | 16 | 36 (38) |
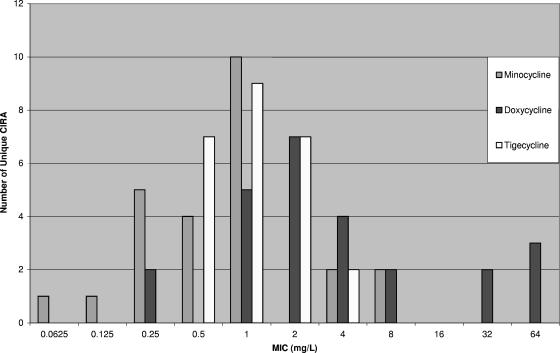
Among the tetracyclines, susceptibility to tigecycline was greater than that to minocycline and doxycycline (95%, 88%, and 71% of the isolates were susceptible to tigecycline, minocycline, and doxycycline, respectively). The positive predictive value of minocycline and doxycycline susceptibility for tigecycline susceptibility was high (98% for both drugs). MIC results and positive predictive values did not change appreciably when only the unique isolates (n = 25) were considered. Both tigecycline and minocycline remained very active, with identical isolate susceptibilities to both drugs (92%), and doxycycline was, again, less active (72%). The positive predictive value of minocycline and doxycycline susceptibility for tigecycline susceptibility remained high (96% and 100%, respectively). The MICs for tigecycline and minocycline were unimodally distributed, with only 8% and 16% of the unique isolates having MICs greater than the breakpoint; however, minocycline displayed greater activity than tigecycline on a weight-per-weight basis (Fig. 1).
FIG. 1.
Distribution of MICs of the tetracyclines (n = 25).
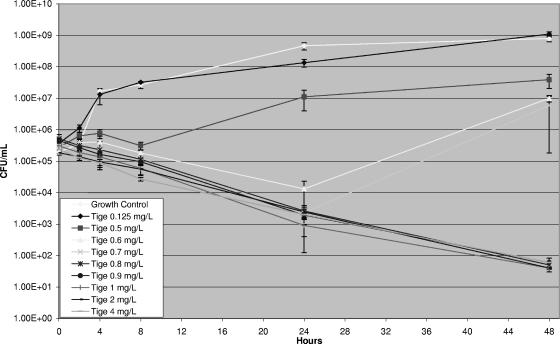
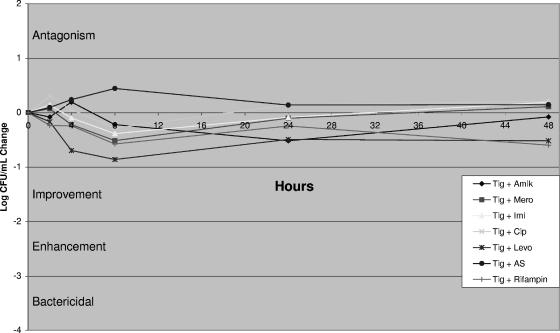
Concentration escalation studies with tigecycline revealed a maximal killing effect near the MIC (Fig. 2) without an increased rate or extent of killing at multiples of the MIC (data not shown). Time-kill studies demonstrated indifference for tigecycline in combination with the antimicrobials tested (Fig. 3). All combinations resulted in less than a 1-log-CFU net change in killing compared to the killing achieved with the most active agent alone.
FIG. 2.
Time-kill curve with escalating concentrations of tigecycline (MIC = 1 mg/liter).
FIG. 3.
Quantitative change in CFU/ml and qualitative interpretation. Changes in colony counts ±1 × 101 were interpreted as indifference. The concentrations used were as follows: tigecycline (Tig)-amikacin (Amik), 0.125 and 32 mg/liter, respectively; tigecycline and meropenem (Mero), 0.125 and 8 mg/liter, respectively; tigecycline and imipenem (Imi), 0.125 and 4 mg/liter, respectively; tigecycline and ciprofloxacin (Cip), 0.125 and 1 mg/liter, respectively; tigecycline and levofloxacin (Levo), 0.125 and 4 mg/liter, respectively; tigecycline and ampicillin-sulbactam (AS), 0.125 and 48 mg/liter, respectively; and tigecycline and rifampin, 0.125 and 2 mg/liter, respectively.
Polymyxin B at 2 mg/liter in combination with tigecycline at 0.125 mg/liter initially appeared to result in antagonism, but subsequent testing revealed the variable action of polymyxin B at 2 mg/liter as a single agent (Table 2). Polymyxin B at 2 mg/liter was inconsistent for the continued suppression of bacterial growth at 24 h.
TABLE 2.
Pharmacodynamic effects of polymyxin B and tigecycline plus polymyxin B against CIRA
| Expt no. | Polymyxin B (2 mg/liter)
|
Tigecycline (0.125 mg/liter) + polymyxin B (2 mg/liter)
|
Interpretation | ||||
|---|---|---|---|---|---|---|---|
| Log10 CFU/ml at time zero | Log10 CFU/ml at time 24 h | Change in log10 CFU/ml from time zero to 24 h | Log10 CFU/ml at time zero | Log10 CFU/ml at time 24 h | Change in log10 CFU/ml from time zero to 24 h | ||
| 1 | 5.52 | 3.95 | −1.57 | NAa | NA | NA | NA |
| 2 | 5.20 | 1.60 | −3.6 | 5.44 | 3.99 | −1.45 | Antagonism |
| 3 | 5.20 | 4.64 | −0.56 | 5.01 | 1.60 | −3.41 | Enhancement |
| 4 | 5.30 | 6.34 | +1.04 | 5.26 | 1.60 | −3.65 | Bactericidal |
| 5 | 5.36 | 2.20 | −3.16 | 5.44 | 5.76 | +0.32 | Antagonism |
NA, not applicable.
DISCUSSION
Practitioners treating patients infected with multidrug-resistant Acinetobacter spp. often have limited antimicrobial options. Many patients receive combination antimicrobial therapy, and the pharmacodynamic interaction between the therapies is ill defined. Tigecycline, a glycylcycline, has recently gained FDA approval, but few studies have evaluated its activity in combination with other antimicrobials against CIRA. Moreover, investigators have concerns about using tigecycline for bloodstream infections because of low serum concentrations (15). This study sought to evaluate the effect of unbound tigecycline at clinically achievable serum concentrations against CIRA.
The emergence of dominant clones of A. baumannii (such as the single clone that represented 88% of our A. baumannii isolates with imipenem MICs ≥8 mg/liter) has been reported throughout the world (22, 32, 34), and its prevalence continues to increase. At Northwestern Memorial Hospital, CIRA accounted for 55% of all A. baumannii isolates, and only 41% of the CIRA isolates were genetically distinct. Therefore, we conducted our time-kill studies with a strain representative of the strains responsible for this outbreak.
Two in vitro studies have previously evaluated the comparative activities of tigecycline alone or in combination with other antimicrobials against A. baumannii (24, 25). In the first analysis, tigecycline was evaluated with a variety of antimicrobials and pathogens by checkerboard analysis (25). Among the A. baumannii isolates studied, time-kill studies verified a single synergistic result for tigecycline plus amikacin. It should be noted that tigecycline was tested at a concentration of 2 mg/liter, a concentration not achievable as free drug in human serum with current dosing strategies. In addition, it is not clear if CIRA isolates were tested. In the second study, tigecycline was assessed as monotherapy against CIRA and achieved nearly bactericidal activity against two separate strains (2.99- and 2.84 log10-CFU/ml decreases). However, the study did not evaluate the effect of combination therapy (24).
We demonstrate that tigecycline achieved maximal killing near the MIC for the isolate tested. Additionally, tigecycline appears to exhibit predictable kinetics in humans. In phase I trials, dose-escalation studies evaluated single doses between 12.5 mg and 300 mg (18). Linear regression of the AUC from time zero to infinity demonstrates that the kinetics remain linear within these specified doses (r2 = 0.99). It should be noted, however, that the safety of this dosage range has not been extensively evaluated, but the most prominent adverse reaction to tigecycline is gastrointestinal intolerance (2, 10). Yet sufficient numbers of subjects have not received increased dosages to imply safety. Mathematical modeling could be employed to predict the dosage of tigecycline needed to optimize therapy for CIRA bloodstream infections. It should also be noted that protein binding may increase with increasing tigecycline concentrations, as shown by in vitro protein binding studies. Yet these studies have shown absolute changes of protein binding of only 6 to 16% with various concentrations (18). Therefore, it is reasonable to suggest that increased dosages of tigecycline might improve outcomes in patients with CIRA bloodstream infections. Since tigecycline is known to have significant postantibiotic effects with other organisms (33), the optimal scheduling of the drug for the treatment of CIRA bloodstream infections is unclear.
Several limitations exist for our study. The current model examining combinations of antimicrobials used fixed drug concentrations and does not definitively simulate the in vivo concentrations of the drug over time. The combinations of antimicrobials used in this study were tested with the average free serum steady-state concentrations of tigecycline alone and in combination with various antimicrobials, with the exception of polymyxin B. There are no published papers detailing the achievable serum concentrations of polymyxin B. Hence, the CLSI breakpoint was modeled (8). Tigecycline was tested at concentrations consistent with an intravenous dosage of 100 mg given every 12 h. While this is higher than the current FDA-approved dosage, the lack of a favorable response may indicate that even higher concentrations need to be tested. Ampicillin-sulbactam was tested as a combination, as it is commercially available only as a combination product in the United States. Finally, we noted the regrowth of A. baumannii with concentrations of tigecycline between 0.5 mg/liter and 0.7 mg/liter after initial killing. As we did not measure the final tigecycline concentrations, we are unable to determine if regrowth was secondary to antibiotic degradation; however, it is reassuring that this effect did not continue when the concentrations approached the MIC (MIC = 1 mg/liter).
The variable response that we noted for polymyxin B is paradoxical. Essentially, after 8 h the bacteria either resumed logarithmic growth or remained present at very low numbers. Similar results have been observed with Pseudomonas aeruginosa (29). One possibility is that the drug degraded and the isolate was able to proliferate. This is well known for tigecycline (5), but polymyxin B is generally stable at neutral or acidic pHs within room temperature ranges (23). The pH of our CAMHB was verified as neutral after 24 h of bacterial growth (data not shown). Thus, it is unlikely that polymyxin B hydrolysis occurred in our experiment, even though the drug concentrations were never verified. In order to test if tigecycline was antagonistic with polymyxin B, we increased the polymyxin B concentration to 8× to 16× the MIC. Thus, we were able to remove the inconsistency associated with polymyxin B. No differences were observed when polymyxin B was combined with tigecycline; therefore, tigecycline is not likely antagonistic with polymyxin B.
This is the largest published study to date to directly compare the activity of tigecycline against CIRA to those of minocycline and doxycycline. This remains important, as doxycycline and tigecycline are the only intravenous tetracyclines available for use in the United States, and the low bioavailability of oral tetracyclines probably precludes their use for the treatment of serious CIRA infections. Previously, tigecycline has been compared to doxycycline (3) and to minocycline in aggregate (12, 14, 17) and at the organism level (6). These studies revealed that tigecycline had fair activity against A. baumannii, with MICs ranging from 0.03 mg/liter to 16 mg/liter (MIC50, 0.5 mg/liter; MIC90, 2 mg/liter) (3, 6, 12, 14, 17, 19). Tigecycline should be more active than doxycycline and minocycline because of immunity from genetic determinants encoding for efflux (7). The tet(A) gene results in resistance to tetracycline but does not currently confer resistance to minocycline or tigecycline (7). The tet(B) gene results in resistance to tetracycline and minocycline but does not currently render tigecycline inactive (7). Despite these genetic observations, other authors have obtained findings similar to ours and have demonstrated that minocycline may be more active than tigecycline on a weight-per-weight basis (6, 14, 17). However, much of this reduced activity occurs close to the FDA breakpoint (MIC = 2 mg/liter) (Fig. 1) (14). Clearly, the value set for the breakpoint affects designations of susceptibility. If a breakpoint of 1 mg/liter is considered for tigecycline and minocycline, the susceptibilities of our isolates would decrease to 70% (n = 65/93) and 72% (n = 66/93), respectively.
Conclusions.
As antimicrobials and antimicrobial combinations with activities against A. baumannii are waning, polymyxin B, minocycline, and tigecycline are emerging as the most active agents for the treatment of these infections. Our data demonstrate no antagonism for tigecycline in combination with other antimicrobials possessing activities against gram-negative organisms modeled with average, serum, and steady-state concentrations. Concentration-escalation studies demonstrate that tigecycline achieved maximal killing near the MIC. If patients with CIRA bloodstream infections are treated with tigecycline, dose escalation may be needed to adequately treat these infections. Although the safety and tolerability associated with elevated dosages for tigecycline are not known, such approaches may improve the outcomes in patients with CIRA bloodstream infections. Future clinical studies should be performed to examine the role of tigecycline and increased tigecycline dosing strategies for the management of serious CIRA infections.
Acknowledgments
We thank Farida Siddiqui for her help in completing the time-kill studies.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Allen, G. P., R. Cha, and M. J. Rybak. 2002. In vitro activities of quinupristin-dalfopristin and cefepime, alone and in combination with various antimicrobials, against multidrug-resistant staphylococci and enterococci in an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 462606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babinchak, T., E. Ellis-Grosse, N. Dartois, G. M. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline for the treatment of complicated intra-abdominal infections: analysis of pooled clinical trial data. Clin. Infect. Dis. 41(Suppl. 5)S354-S367. [DOI] [PubMed] [Google Scholar]
- 3.Bouchillon, S. K., D. J. Hoban, B. M. Johnson, T. M. Stevens, M. J. Dowzicky, D. H. Wu, and P. A. Bradford. 2005. In vitro evaluation of tigecycline and comparative agents in 3049 clinical isolates: 2001 to 2002. Diagn. Microbiol. Infect. Dis. 51291-295. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet, P., and P. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov., and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36228-240. [Google Scholar]
- 5.Bradford, P. A., P. J. Petersen, M. Young, C. H. Jones, M. Tischler, and J. O'Connell. 2005. Tigecycline MIC testing by broth dilution requires use of fresh medium or addition of the biocatalytic oxygen-reducing reagent oxyrase to standardize the test method. Antimicrob. Agents Chemother. 493903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, P. A., D. T. Weaver-Sands, and P. J. Petersen. 2005. In vitro activity of tigecycline against isolates from patients enrolled in phase 3 clinical trials of treatment for complicated skin and skin-structure infections and complicated intra-abdominal infections. Clin. Infect. Dis. 41(Suppl. 5)S315-S332. [DOI] [PubMed] [Google Scholar]
- 7.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Conte, J. E., Jr., J. A. Golden, M. G. Kelley, and E. Zurlinden. 2005. Intrapulmonary pharmacokinetics and pharmacodynamics of meropenem. Int J. Antimicrob. Agents 26449-456. [DOI] [PubMed] [Google Scholar]
- 10.Ellis-Grosse, E. J., T. Babinchak, N. Dartois, G. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline in the treatment of skin and skin-structure infections: results of 2 double-blind phase 3 comparison studies with vancomycin-aztreonam. Clin. Infect. Dis. 41(Suppl. 5)S341-S353. [DOI] [PubMed] [Google Scholar]
- 11.Falagas, M. E., P. Kopterides, and I. I. Siempos. 2006. Attributable mortality of Acinetobacter baumannii infection among critically ill patients. Clin. Infect. Dis. 43389-390. [DOI] [PubMed] [Google Scholar]
- 12.Gales, A. C., and R. N. Jones. 2000. Antimicrobial activity and spectrum of the new glycylcycline, GAR-936 tested against 1,203 recent clinical bacterial isolates. Diagn. Microbiol. Infect. Dis. 3619-36. [DOI] [PubMed] [Google Scholar]
- 13.Gouby, A., M. J. Carles-Nurit, N. Bouziges, G. Bourg, R. Mesnard, and P. J. Bouvet. 1992. Use of pulsed-field gel electrophoresis for investigation of hospital outbreaks of Acinetobacter baumannii. J. Clin. Microbiol. 301588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henwood, C. J., T. Gatward, M. Warner, D. James, M. W. Stockdale, R. P. Spence, K. J. Towner, D. M. Livermore, and N. Woodford. 2002. Antibiotic resistance among clinical isolates of Acinetobacter in the UK, and in vitro evaluation of tigecycline (GAR-936). J. Antimicrob. Chemother. 49479-487. [DOI] [PubMed] [Google Scholar]
- 15.Livermore, D. M. 2005. Tigecycline: what is it, and where should it be used? J. Antimicrob. Chemother. 56611-614. [DOI] [PubMed] [Google Scholar]
- 16.Meagher, A. K., P. G. Ambrose, T. H. Grasela, and E. J. Ellis-Grosse. 2005. Pharmacokinetic/pharmacodynamic profile for tigecycline—a new glycylcycline antimicrobial agent. Diagn. Microbiol. Infect. Dis. 52165-171. [DOI] [PubMed] [Google Scholar]
- 17.Milatovic, D., F. J. Schmitz, J. Verhoef, and A. C. Fluit. 2003. Activities of the glycylcycline tigecycline (GAR-936) against 1,924 recent European clinical bacterial isolates. Antimicrob. Agents Chemother. 47400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muralidharan, G., M. Micalizzi, J. Speth, D. Raible, and S. Troy. 2005. Pharmacokinetics of tigecycline after single and multiple doses in healthy subjects. Antimicrob. Agents Chemother. 49220-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathwani, D. 2005. Tigecycline: clinical evidence and formulary positioning. Int J. Antimicrob. Agents 25185-192. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 21.National Committee for Clinical Laboratory Standards. 2002. Analysis and presentation of cumulative antimicrobial susceptibility test data; approved guideline. NCCLS document M39-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 22.Nemec, A., L. Dijkshoorn, and T. J. van der Reijden. 2004. Long-term predominance of two pan-European clones among multi-resistant Acinetobacter baumannii strains in the Czech Republic. J. Med. Microbiol. 53147-153. [DOI] [PubMed] [Google Scholar]
- 23.Orwa, J. A., C. Govaerts, K. Gevers, E. Roets, A. Van Schepdael, and J. Hoogmartens. 2002. Study of the stability of polymyxins B(1), E(1) and E(2) in aqueous solution using liquid chromatography and mass spectrometry. J. Pharm. Biomed. Anal. 29203-212. [DOI] [PubMed] [Google Scholar]
- 24.Pachon-Ibanez, M. E., M. E. Jimenez-Mejias, C. Pichardo, A. C. Llanos, and J. Pachon. 2004. Activity of tigecycline (GAR-936) against Acinetobacter baumannii strains, including those resistant to imipenem. Antimicrob. Agents Chemother. 484479-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen, P. J., P. Labthavikul, C. H. Jones, and P. A. Bradford. 2006. In vitro antibacterial activities of tigecycline in combination with other antimicrobial agents determined by chequerboard and time-kill kinetic analysis. J. Antimicrob. Chemother. 57573-576. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Bano, J., A. Pascual, J. Galvez, M. A. Muniain, M. J. Rios, L. Martinez-Martinez, R. Perez-Cano, and E. J. Perea. 2003. Acinetobacter baumannii bacteremia: clinical and prognostic features. Enferm. Infecc. Microbiol. Clin. 21242-247. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 27.Ruiz, J., M. L. Nunez, J. Perez, E. Simarro, L. Martinez-Campos, and J. Gomez. 1999. Evolution of resistance among clinical isolates of Acinetobacter over a 6-year period. Eur. J. Clin. Microbiol. Infect. Dis. 18292-295. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz, J. I., L. E. Jauregui, K. A. Bachmann, M. E. Martin, and D. P. Reitberg. 1988. Multiple-dose pharmacokinetics of intravenously administered cefoperazone and sulbactam when given in combination to infected, seriously ill, elderly patients. Antimicrob. Agents Chemother. 32730-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tam, V. H., A. N. Schilling, G. Vo, S. Kabbara, A. L. Kwa, N. P. Wiederhold, and R. E. Lewis. 2005. Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 493624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tegeder, I., A. Schmidtko, L. Brautigam, A. Kirschbaum, G. Geisslinger, and J. Lotsch. 2002. Tissue distribution of imipenem in critically ill patients. Clin. Pharmacol. Ther. 71325-333. [DOI] [PubMed] [Google Scholar]
- 31.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dessel, H., L. Dijkshoorn, T. van der Reijden, N. Bakker, A. Paauw, P. van den Broek, J. Verhoef, and S. Brisse. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155105-112. [DOI] [PubMed] [Google Scholar]
- 33.van Ogtrop, M. L., D. Andes, T. J. Stamstad, B. Conklin, W. J. Weiss, W. A. Craig, and O. Vesga. 2000. In vivo pharmacodynamic activities of two glycylcyclines (GAR-936 and WAY 152,288) against various gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 44943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vila, J., J. Ruiz, M. Navia, B. Becerril, I. Garcia, S. Perea, I. Lopez-Hernandez, I. Alamo, F. Ballester, A. M. Planes, J. Martinez-Beltran, and T. J. de Anta. 1999. Spread of amikacin resistance in Acinetobacter baumannii strains isolated in Spain due to an epidemic strain. J. Clin. Microbiol. 37758-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wisplinghoff, H., M. B. Edmond, M. A. Pfaller, R. N. Jones, R. P. Wenzel, and H. Seifert. 2000. Nosocomial bloodstream infections caused by Acinetobacter species in United States hospitals: clinical features, molecular epidemiology, and antimicrobial susceptibility. Clin. Infect. Dis. 31690-697. [DOI] [PubMed] [Google Scholar]