Abstract
Transposon Tn1403 from a clinical Pseudomonas strain is composed of three transposons, including Tn5393c. A related transposon Tn1404* from a plant-associated Pseudomonas strain lacks Tn5393 but includes a transposon carrying the tet(C) tetracycline resistance determinant. These compound transposons illustrate the role of preexisting transposons in generating clusters of antibiotic resistance genes.
Many clinical and environmental isolates of gram-negative bacteria that are resistant to a number of different antibiotics carry several antibiotic resistance genes clustered in large multiple-antibiotic resistance regions. The role of class 1 integrons and associated gene cassettes in forming such clusters is now well understood (3), but the role of other transposons and diversity-generating elements has received less attention. Recently, we determined the structure of a multiple-antibiotic resistance region that has six resistance genes, of which only one is in a gene cassette and one (sul1) is in the 3′-conserved segment (3′-CS) of the class 1 integron. Three further resistance genes are within or derived from known transposons, and one is associated with the novel insertion element CR1 (8, 9). Other examples of complex arrangements can be found in the recently reported sequences of several large plasmids or genomic islands. Where one transposon lies within another, they can move together as long as the transposition functions of the backbone transposon are not disrupted, and this piggybacking route is used by class 1 integrons, most of which are defective transposons that need transposition proteins to be supplied in trans.
Tn1403 was recovered from a multiply antibiotic resistant clinical strain of Pseudomonas aeruginosa isolated in the United States in 1973-1974 (5, 13), and it confers resistance to carbenicillin, chloramphenicol, streptomycin, and spectinomycin (5). Genes conferring resistance to these antibiotics are found in an array of three gene cassettes, blaP1-cmlA1-aadA1, located within an In4-type class 1 integron, In28, located within Tn1403 (10). In28 has lost the sul1 gene normally found in the 3′-CS, explaining the susceptibility to sulfonamides. However, the region to the right of In28 has been reported to contain a further aphC-type gene conferring resistance to streptomycin (13). Here, we have sequenced the remainder of Tn1403 and identified within it a further transposon, Tn5393c, which carries the strA-strB gene. The structure of Tn1403 was also compared to those of other class II transposons.
Structure of Tn1403.
Tn1403, which had been transposed into the IncW plasmid R388 (5), was obtained from G. Jacoby. The region to the right of In28 was sequenced using cloned EcoRI and HindIII fragments (10) as substrates. The region to the left of In28, which contains no EcoRI or HindIII sites, and the region to the right of the EcoRI site in the right-hand 38-bp inverted repeat (IR) were sequenced directly using R388::Tn1403 as the substrate or using PCR-amplified fragments purified using a QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA) according to the manufacturer's protocols. The sequence was determined on both strands. Automated sequencing, assembly, and analysis of sequences were carried out as described previously (9).
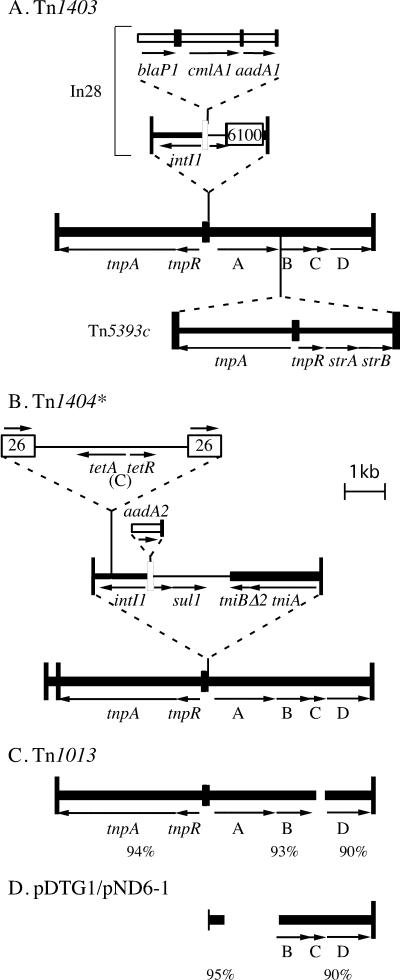
Tn1403 was found to be 19.63 kb long and is bounded by 38-bp IR and flanked by a 4-bp duplication of the target sequence within the trwC gene of R388. It is made up of three distinct entities (Fig. 1A), the transposon backbone of 7,884 bp, In28 (6.27 kb), which was sequenced previously (10), and a 5,470-bp version of Tn5393, known as Tn5393c, which lacks the IS found in Tn5393. Tn5393c is located in the right arm of the backbone (Fig. 1A). Tn5393 is a 6,705-bp transposon made up of a transposition module and a module carrying the strA and strB genes with an insertion sequence, IS1133, located between the modules (1). The Tn5393 relative in Tn1403 lacks the IS1133 element but is otherwise over 99.5% identical to the original sequence (GenBank accession no. M95402) and to Tn5393c (GenBank accession no. AF262622) (4).
FIG. 1.
Structure of Tn1403 and related transposons. A. The structure of Tn1403 is derived from the previously determined sequence of In28 (10) and the sequence determined here. B. The structure of Tn1404 was deduced by joining the partial sequences available in GenBank accession numbers AF157797 to AF157801 using available sequences as follows. AF157801 was linked to AF157800 using the sequence of the transposition module of Tn1403 reported here. AF157800 was linked to AF157799 using the sequence of the pSC101 tet(C) region in X01654, and the gap in the 5′-CS between AF157799 and AF157798 was filled from AF313472. The end of the aadA2 gene cassette was from U12338, and the 3′-CS and tni region were from AJ223604, which has an In0-like segment that does not include IS1326. C. Tn1013, from GenBank accession no. AM261760, is between 90 and 94% identical to Tn1403 with a deletion of 78 bp in orfC. D. Other regions related to the Tn1403 backbone. pDTG1 and pND6-1, which are identical to one another in this region, are from GenBank accession numbers AY208917 and AY491307. High identity with Tn1403 extends from between resI and resII (bp 179 in Fig. 2) to the outer end of Tn1403, indicating that they may be derived from a transposon related to Tn1403. Tall vertical bars represent the IR, and medium vertical bars represent the res sites of transposons. Open vertical bars represent the attI site of class 1 integrons, and cassettes are shown as an open box with a short vertical bar (representing the 59-be) at one end. ISs are shown as open boxes with the IS number within. Genes are indicated by arrows with gene names below or A, B, C, and D indicating orfA to orfD.
The Tn1403 backbone transposon.
The 3.74-kb region to the left of In28 was found to contain IRtnp, the tnpA and tnpR genes, encoding the TnpA transposase and TnpR resolvase, and most of the res site. Because In28 is located within resI (10, 13), part of the resI subsite lies to the right of In28. The closest relatives of this tnp module (94% or 93% identical over the full length) are found in Tn1013 (GenBank accession no. AM261760), which is related to the Tn1403 backbone along its full length (Fig. 1C), and Tn4656, a catabolic transposon encoding xylene and toluene degradation determinants (GenBank accession no. AB062597) (12). Other close relatives are listed in Table 1. However, the sequence of short regions (GenBank accession no. AF157801 and AF157800) from either end of the transposition module of a transposon called Tn1404 (11), but here named Tn1404* to avoid confusion with the previously known Tn1404 (7), are identical to the Tn1403 sequence, indicating that Tn1404* contains the same transposition module. Potentially characteristic differences defining two evolutionary groups are found in the short region between the initiation codon of the tnpR gene and the res site (Fig. 2). Tn1403, Tn4656, and Tn1013 have related sequences, and Tn501, Tn4653, and Tn1721 have a sequence that is significantly diverged from the Tn1403 segment. The equivalent regions from the more distantly related class II transposons Tn21 and Tn1696 are shorter.
TABLE 1.
Relationships between transposon transposition modulesa
| Transposon | % Identity with:
|
||||||
|---|---|---|---|---|---|---|---|
| Tn1013 | Tn4656 | Tn4653 | Tn501 | Tn1721 | Tn21 | Tn1696 | |
| Tn1403 | 94.6 | 93.2 | 91.9 | 90.6 | 90.5 | 73.3 | 73.2 |
| Tn1013 | 92.7 | 91.9 | 91.3 | 91.3 | 73.2 | 72.8 | |
| Tn4656 | 92.1 | 90.8 | 90.5 | 73.1 | 72.9 | ||
| Tn4653 | 94.1 | 93.3 | 73.6 | 73.0 | |||
| Tn501 | 95.6 | 74.2 | 73.9 | ||||
| Tn1721 | 73.6 | 73.2 | |||||
| Tn21 | 81.5 | ||||||
The numbers represent the percent nucleotide identities between the transposition modules. For the GenBank accession numbers, see the legend to Fig. 2.
FIG. 2.
Comparison of the region between tnpR and res in class II transposons. Sequences are from this study for Tn1403 and from GenBank accession no. AB062597 for Tn4656, AM261760 for Tn1013, X61367 for Tn1721, Z00027 for Tn501, AJ344068 for Tn4653, AF071413 for Tn21, U12338 for Tn1696, and X90708 for Tn4378. The start codons and potential alternate start codons for the tnpR gene are underlined, and the crossover position within resI is in bold. Positions of bases duplicated by insertion of a class 1 integron or Tn5053 are boxed.
The 9,628-bp region to the right of In28 in Tn1403, with the Tn5393 and one copy of the flanking duplication removed, differed at only four positions from the sequence of the right arm of Tn1404* (GenBank accession number AF157797). The products of orfA and orfC were previously identified as homologues of a sulfate permease and a DksA-like DnaK suppressor protein, respectively (11). The OrfB protein is related to a series of proteins annotated as universal stress proteins (UspA). Close relatives of this DNA sequence were found in Tn1013 and in plasmids pND6-1 and pDTG1 (GenBank accession numbers AY208917 and AF491307) (2, 6), which are identical to one another in this region. However, in pDTG1 and pND6-1 the sulfate permease gene (orfA) is missing (Fig. 1D).
Relationship to Tn1404*.
The closest relative of Tn1403 is the partially sequenced transposon Tn1404* isolated from a tetracycline-resistant Pseudomonas strain from a Michigan apple orchard (11). The structure of Tn1404* (Fig. 1B) was deduced from the available sequence mapping (11) using sequence determined here and standard sequences to fill the gaps. Tn1404* contains a class 1 integron in the same location as In28 but it has an In5-type backbone lacking IS1326, in contrast to the In4-type backbone of In28. The cassette arrays also differ. Tn1404* lacks Tn5393c but contains a transposon consisting of the tet(C) tetracycline resistance determinant flanked by two copies of IS26 (11). This transposon lies within the intI1 gene of the integron. These two compound transposons illustrate the role of preexisting transposons in generating different clusters of antibiotic resistance genes within the same backbone, adding to the diversity generated by variability in integron-associated gene cassettes.
Nucleotide sequence accession number.
The 12.9 kb of sequence reported in this paper has been added to GenBank accession no. AF313472 to give AF313472.2.
Acknowledgments
This work was supported by NHMRC grants 192108 and 352352.
Footnotes
Published ahead of print on 29 January 2007.
REFERENCES
- 1.Chiou, C.-S., and A. L. Jones. 1993. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J. Bacteriol. 175732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dennis, J. J., and G. J. Zylstra. 2004. Complete sequence and genetic organization of pDTG1, the 83 kilobase napthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 341753-768. [DOI] [PubMed] [Google Scholar]
- 3.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1109-119. [DOI] [PubMed] [Google Scholar]
- 4.L'Abée-Lund, T. M., and H. Sørum. 2000. Functional Tn5393-like transposon in the R plasmid pRAS2 from the fish pathogen Aeromonas salmonicida subspecies salmonicida isolated in Norway. Appl. Environ. Microbiol. 665533-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lévesque, R. C., and G. A. Jacoby. 1988. Molecular structure and interrelationships of multiresistance β-lactamase transposons. Plasmid 1921-29. [DOI] [PubMed] [Google Scholar]
- 6.Li, W., J. Shi, X. Wang, Y. Han, W. Tong, L. Ma, B. Liu, and B. Cai. 2004. Complete nucleotide sequence and organization of the napthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 336231-240. [DOI] [PubMed] [Google Scholar]
- 7.Partridge, S. R., C. M. Collis, and R. M. Hall. 2002. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R151. Antimicrob. Agents Chemother. 462400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Partridge, S. R., and R. M. Hall. 2004. Complex multiple antibiotic and mercury resistance region derived from the r-det of NR1 (R100). Antimicrob. Agents Chemother. 484250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 453014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnabel, E. L., and A. L. Jones. 1999. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 654898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuda, M., and H. Genka. 2001. Identification and characterization of Tn4656, a novel class II transposon carrying a set of toluene-degrading genes from TOL plasmid pWW53. J. Bacteriol. 1836215-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vézina, G., and R. C. Levesque. 1991. Molecular characterization of the class II multiresistance transposable element Tn1403 from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]