Abstract
Revealing the cellular identity of organisms behind environmental eukaryote rRNA gene sequences is a major objective in microbial diversity research. We sampled an estuarine oxygen-depleted microbial mat in southwestern Norway and retrieved an 18S rRNA gene signature that branches in the MAST-12 clade, an environmental marine stramenopile clade. Detailed phylogenetic analyses revealed that MAST-12 branches among the heterotrophic stramenopiles as a sister of the free-living Bicosoecida and the parasitic genus Blastocystis. Specific sequence signatures confirmed a relationship to these two groups while excluding direct assignment. We designed a specific oligonucleotide probe for the target sequence and detected the corresponding organism in incubation samples using fluorescence in situ hybridization (FISH). Using the combined FISH-scanning electron microscopy approach (T. Stoeck, W. H. Fowle, and S. S. Epstein, Appl. Environ. Microbiol. 69:6856-6863, 2003), we determined the morphotype of the target organism among the very diverse possible morphologies of the heterotrophic stramenopiles. The unpigmented cell is spherical and about 5 μm in diameter and possesses a short flagellum and a long flagellum, both emanating anteriorly. The long flagellum bears mastigonemes in a characteristic arrangement, and its length (30 μm) distinguishes the target organism from other recognized heterotrophic stramenopiles. The short flagellum is naked and often directed posteriorly. The organism possesses neither a lorica nor a stalk. The morphological characteristics that we discovered should help isolate a representative of a novel stramenopile group, possibly at a high taxonomic level, in order to study its ultrastructure, physiological capabilities, and ecological role in the environment.
In the past two decades, the use of molecular tools has resulted in a new era in microbial diversity research. Analysis of the small subunit of the rRNA gene, amplified from genomic environmental DNA, has led to the discovery of numerous novel phylogenetic lineages up to the highest taxonomic levels in a wide variety of habitats (3, 11, 15, 21, 39). Such studies have dramatically changed our understanding of microbial diversity (27, 34) and have fueled ongoing surveys of global molecular environmental diversity across all three domains of life (Archaea, Bacteria, and Eukarya). For each domain, the ubiquitous presence of completely novel lineages without cultured representatives has been established (14, 26, 34). Unfortunately, the discovery of molecular signatures in environmental samples provides very little information beyond the existence of the organisms and their distribution in nature. The organisms themselves remain bewildering, and their basic biology is unknown. However, because of the great taxonomic, morphological, and ecological diversity of protists, it is essential to directly study the organisms behind novel uncultured microbial eukaryote sequences in order to confirm their uniqueness, to assess their relatedness to known protists, and to determine certain aspects of their lifestyles.
For example, in the domain Eukarya, the stramenopiles are a major phylum that dominates 18S rRNA gene sequence clone libraries retrieved from aquatic environmental samples (5, 24, 37, 44). The stramenopiles are a very diverse group of organisms that form one of eight major phylogenetic groups of eukaryotes (4). Their evolutionary diversification has produced a remarkable panoply of body forms and nutritional modes. This group includes unicellular and multicellular algae, fungus-like cells, and parasitic and free-living flagellates that may have an autotrophic, heterotrophic, mixotrophic, or osmotrophic lifestyle (7, 28, 33). Molecular characteristics like rRNA gene sequences and the presence of two flagella, one smooth and the other with tripartite rigid tubular hairs (mastigonemes), unite these diverse tubulocristate protists (and protists derived from these organisms) in a monophyletic clade (10).
A detailed phylogenetic analysis of uncultured marine stramenopiles (MAST) resulted in identification of 12 independent environmental sequence clades (23). An additional clade (the MAST-13 clade) has been reported only recently (44). Because of the great morphological and ecological diversity of the stramenopiles, determining the identities of the organisms in these uncultured MAST lineages has been considered a major goal in microbial diversity research (23).
Recently, Stoeck et al. (38) developed a culture-independent approach that involves a specific oligonucleotide probe, the use of this probe with environmental samples, light microscopic visualization of the target organisms via fluorescence in situ hybridization (FISH), and subsequent scanning electron microscopy (SEM) of the same individual cells. We successfully used this technique with an oxygen-depleted incubation culture obtained from an estuarine microbial mat in order to determine the cellular identity and lifestyle of a novel phylogenetic lineage branching within the uncultured MAST-12 clade.
MATERIALS AND METHODS
Terminology.
The classification of stramenopiles varies considerably, and we used the classification system suggested by Adl et al. (1).
Sampling site and sample collection.
Samples were collected in September 2005 from a microbial mat in a small estuarine system (Rivulet Åsen that connects Lake Hanagervatnet to a sandy bay) near the town of Farsund in southwestern Norway (58°04′N, 06°44′E). The microbial mat was located in a ca. 50-cm-deep pool away from the main currents. At the time of sampling, the ambient water in the pool was oxygen depleted (2.9 mg O2 liter−1) and the water temperature and salinity were 12°C and 4‰, respectively. Samples were taken at low tide by carefully scraping off the microbial mat with a palette knife. Undisturbed mat samples were transferred into 650-ml T-175 cell culture flasks (Sarstedt, Nürnberg, Germany). Ambient sterile filtered water was added to the samples using various headspace volumes (50% to 0%). During transport to the laboratory, samples were kept in a cooling box whose temperature was adjusted to 10 to 15°C. In the laboratory, the samples were stored for 4 months in the dark (to select for heterotrophic or mixotrophic organisms) at 12°C without any additives. The oxygen content in the flasks after 4 months of undisturbed incubation was 1.7 mg liter−1 in the surface layer and 0.6 mg liter−1 in the bottom water.
DNA extraction and amplification.
After 4 months of undisturbed incubation, we extracted DNA from the samples in order to investigate the phylogenetic diversity by constructing 18S rRNA gene clone libraries. For genomic DNA extraction, we used a DNEasy tissue kit (QIAGEN, Hildesheim, Germany) according to the manufacturer's instructions, with slight modifications. First, 6.5 ml of sample water was gently filtered onto a 0.45-μm-pore-size Durapore filter (diameter, 25 mm; Millipore, Schwalbach, Germany). The filter was then transferred into 180 μl of the kit's ATL buffer containing 20 μl proteinase K (20 mg ml−1). After this, we added 200 μl AL buffer and incubated the sample for 45 min at 70°C. For subsequent procedures we did not deviate from the manufacturer's instructions. The integrity and concentration of the extracted nucleic acids were evaluated photometrically with a Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technologies, Wilmington, DE) at a wavelength of 260 nm. As the DNA concentrations after extraction were too low for multiple PCRs, we amplified the genomic environmental DNA using a QIAGEN Repli-g mini-kit according to the manufacturer's instructions.
For PCR amplification of the 18S rRNA gene we used the universal eukaryotic primers EukA (5′-AACCTGGTTGATCCTGCCAGT-3′) and EukB (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) (25), followed by a seminested reaction with primers Euk82F (5′-GAA[AGT]CTG[CT]G AA[CT]GGCTC-3′) (21) and EukB covering the nearly complete 18S rRNA gene fragment. The PCR and clone library construction were performed as previously described in detail by Stoeck et al. (39). We performed an HaeIII restriction analysis (37) with 128 randomly selected positively screened (M13 PCR) clones and identified 57 different restriction patterns. At least one representative clone for each pattern was sequenced bidirectionally (M13 sequencing primers) by MWG-Biotech (Ebersheim, Germany). Environmental 18S rRNA gene sequences initially were compared to sequences in the GenBank database using gapped BLAST analysis (2) to determine their approximate phylogenetic affiliations.
We identified one sequence (KKTS_D3) that was closely related to uncultured marine stramenopile sequences. We subsequently performed a detailed phylogenetic analysis of stramenopile sequences to evaluate the exact position of KKTS_D3. We constructed evolutionary distance, maximum likelihood (ML), and Bayesian inference (BI) trees with KKTS_D3 and stramenopile sequences representing all major heterokont taxonomic groups, including uncultured environmental sequence clades. as described previously (5). To create evolutionary distance and ML trees, we used the program Modeltest to choose the model of DNA substitution that best fit our data sets from among 56 possible models (32). In the case of the BI analysis, the best substitution model was calculated using MrBayes (35).
Probe design and evaluation.
For initial detection of the organism behind the novel stramenopile sequence KKTS_D3 by FISH, we designed two oligonucleotide probes using the PROBE design tool of the ARB software package (22), as described by Pernthaler et al. (30) and Hugenholtz et al. (17). The PROBE design tool detected two oligonucleotide probes that exhibited no mismatch with the KKTS_D3 stramenopile sequence but exhibited mismatches with the next most similar sequences in the ARB database and that at the same time were likely to be in an appropriately accessible area of the 18S rRNA secondary structure. The first probe, MAST-12Nor1 (5′-CGUCCGAAAACGAAUUUG-3′), hybridized with positions 193 to 211 of the Saccharomyces cerevisiae secondary structure with class III accessibility (relative fluorescence of Cy3-labeled probes, 0.6 to 0.41) (6). This probe exhibited at least two mismatches with the next most similar sequences in the ARB database (17 autotrophic Phaeophyceae sequences). The second probe, MAST-12Nor2 (5′-UACAGUGCCAAUGGAGAC-3′; positions 845 to 863; class III accessibility), exhibited one mismatch with the most similar sequences in the ARB database, all of which are sequences of uncultured MAST-12 clones (BAQD200, BAQD16, BAQD14, BAQD21, BAQD224, BAQA21, and BAQA72). The next most similar sequences exhibited three mismatches with the MAST-12Nor2 probe (sequences of two autotrophic pelagophytes, one autotrophic diatom, one parasitic apicomplexa, one cercozoa, and one fungus). Both probes were labeled with the fluorescent dye Cy3 at the 5′ end (Operon Biotechnologies, Cologne, Germany).
As both probes targeted only environmental sequences with at least two and three mismatches with the next most similar known sequences of named protists, it was not possible to test the probes with cultured isolates that exhibited either zero or one mismatch with the oligonucleotides. Therefore, we tested the probes using an alternative method, as suggested by Pernthaler et al. (30). In brief, we performed FISH directly with environmental samples as described in detail previously (38). The relative abundance of the cells detected and the average cell brightness were determined using different formamide concentrations (0, 10, 20, 30, 40, and 50%) by counting and image analysis (ImageJ [http://rsb.info.nih.gov/ij/index.html]). When the stringency was too low, cell counts may have been higher because nontarget populations were also detected. However, if the stringency was too high, the mean cell brightness of the target population rapidly declined. We used no-probe and nonsense probe (Cy3-labeled complement to target probes) samples as controls.
FISH-SEM.
FISH-SEM was performed as described in detail by Stoeck et al. (38). In short, 5 ml of preserved sample water (50% [final concentration] Bouin's fixative and 0.1% [final concentration] glutaraldehyde) was filtered onto a Corning Costar filter transwell (polycarbonate; diameter, 25 mm; pore size, 0.4 μm; Corning Inc. Corning, NY). The cells collected on the filter were washed by gradually removing the fixative with 1× phosphate-buffered saline using three to five cycles until the yellowish color of the fixative disappeared. The washing buffer was then gradually replaced with a standard hybridization buffer containing formamide at a concentration of 20%, which was identified as the optimal concentration for the highest-stringency conditions. Leaving approximately 300 μl of hybridization buffer on the filter, we transferred the well to a six-well transwell tissue culture tray, and 50 μl of probe solution (30 ng μl−1 in molecular-grade distilled water) was added. The wells were then placed into incubation chambers and incubated for 2 h at 46°C, which was followed by 10 min of washing with preheated washing buffer at 48°C. After this, the cells were washed with distilled water, counterstained with DAPI (4′,6′-diamidino-2-phenylindole), and dehydrated first with an ethanol series and then with a hexamethyldisilazane series. In the final hexamethyldisilazane evaporation step the preparation was incubated overnight.
For fluorescence microscopy the filters were cut out from the transwell and mounted on an adhesive silicon spacer (Whatman Schleicher and Schuell, Middlesex, United Kingdom) attached to a glass microscope slide. The filters were scanned with appropriate epifluorescence illumination using a Zeiss Axiophot II with a ×40 (dry) Neofluar objective and DAPI- and Cy3-specific filter sets. We checked for chlorophyll autofluorescence using a Zeiss 01 filter set. Once a positively Cy-3-stained target cell was located, the appropriate filter area was photographed, cut out, mounted on aluminum stubs for SEM, and sputter coated with 10 to 15 nm of gold using a cooled Cressington 108auto sputter coater (Cressington, Watford, United Kingdom). SEM was performed with a Zeiss DSM 940. The target cell was located at a low SEM magnification, using the photographs obtained with the epifluorescence microscope.
SEM.
As some details of the target cell were not clearly visible using the FISH-SEM approach, we also performed a standard SEM analysis once we identified the target cell's morphotype. The standard SEM protocol described by Stoeck et al. (40) was used.
Nucleotide sequence accession number.
The sequence of clone KKTS_D3 retrieved from a microbial mat sample from Norwegian coastal water has been deposited in the GenBank database under accession number EF219381.
RESULTS
Phylogeny.
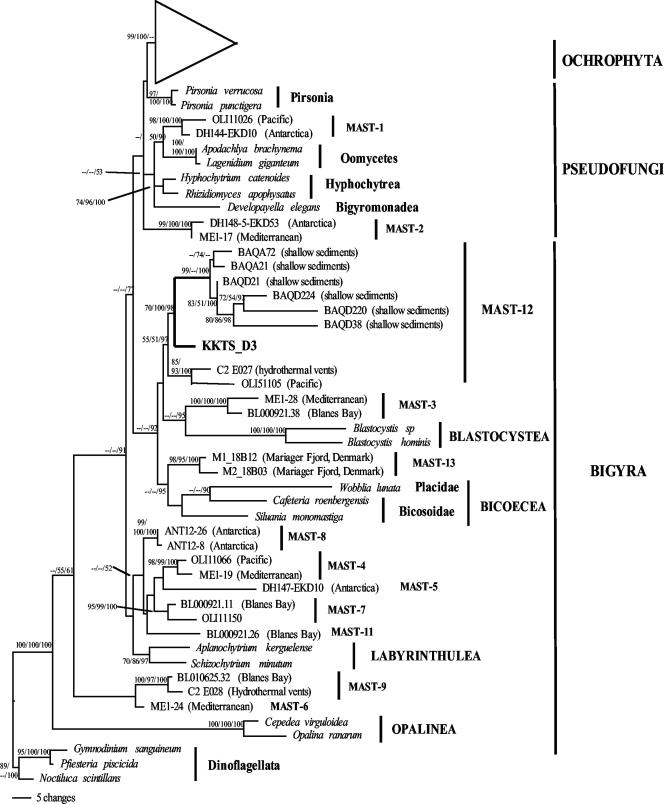
We analyzed 128 clones and detected 57 different protist phylotypes (based on 97% sequence similarity), which showed affiliations to nearly all major eukaryote lineages (unpublished data). One of the clones, KKTS_D3, exhibited 93.14% sequence similarity to an uncultured marine stramenopile sequence (BAQA72) as its closest database match. Because uncultured marine stramenopiles are prime targets for cellular identification, we performed a detailed analysis to determine the exact phylogenetic position of KKTS_D3. All three analyses (ML, evolutionary distance, and BI) identified KKTS_D3 as a member of the MAST-12 clade (ML distance, 70%; evolutionary distance, 100%), and posterior probability analysis (98%) supported this conclusion (Fig. 1). The bootstrap support for the complete MAST-12 clade, however, contrasts with the posterior probability support (97%) and was relatively low (55% and 51%). Eliminating KKTS_D3 from the phylogenetic analyses did not significantly increase the bootstrap support for MAST-12 (57% and 53%) (results not shown). With the exception of one sequence at the base of the group (OLI51105), all sequences in the MAST-12 clade originated from anoxic sediments (shallow coastal waters and a deep-sea hydrothermal vent). The MAST-12 clade branches within the heterotrophic stramenopiles and exhibits weakly supported phylogenetic affinity with Blastocystis (Oplinata) and the Bicosoecida. All sequences in this clade except KKTS_D3 originated from marine samples.
FIG. 1.
Bayesian inference tree for stramenopile 18S rRNA gene sequences, showing the phylogenetic position of clone KKTS_D3 retrieved from an enrichment sample of an oxygen-depleted microbial mat from a small Norwegian estuary. This clone branches within the environment-specific MAST-12 clade. The tree is based on 1,600,000 generations with two runs (each run with three hot chains and one cold chain) using a general time-reversible model. The gamma shape parameter, the rate of invariable sites, the rate matrix, and the base frequencies of the substitution model were estimated by MrBayes (35). The sample frequency was set at 1,000, resulting in 1,601 trees. The numbers at the nodes are ML bootstrap/evolutionary distance bootstrap/Bayesian posterior probability values. For ML and evolutionary distance trees, we used the TrN substitution model with a gamma shape parameter of 0.6482, a rate of invariable sites of 0.3339, and base frequencies and a rate matrix suggested by Modeltest (32). The tree is based on 906 unambiguously aligned positions.
An analysis of specific heterokont sequence signatures confirmed the assignment to the stramenopiles (U at nucleotide position 1743) and eliminated the possibility of an affiliation with the phylum Ochrophyta (G at position 1137) (Table 1). The possibility of direct assignment to either the Bicosoecida or the Opalinata (including Blastocystis) could be eliminated based on the presence of stramenopile outgroup signatures in KKTS_D3 at positions 982 and 1019. Nevertheless, a relationship to the latter groups was confirmed by the presence of a G at position 996 and a C at position 1008, which are found exclusively in the Opalinata and Bicosoecida. Surprisingly, we identified a U at position 1649 in KKTS_D3, whereas all other known stramenopiles (including other MAST-12 sequences) have an A at this position. We verified this nucleotide by a second independent sequencing analysis of another clone with the same restriction pattern in order to exclude the possibility that there was a sequencing artifact. Also, the possibility of PCR artifacts could be eliminated as we obtained the same result with an independent 18S rRNA gene clone library from another incubation sample obtained in the same sampling area (GenBank accession number EF219382; clone KKTS_E5).
TABLE 1.
Comparison of the KKTS_D3 18S rRNA sequence with specific selected stramenopile sequence signaturesa
| Nucleotide positionb | Taxon | Nucleotide | Nucleotide in KKTS_D3 (nucleotide in other MAST-12 sequences) |
|---|---|---|---|
| 1743 | Stramenopiles | U | U (U) |
| 996 | Stramenopiles | U (except Opalinata and Bicosoecida [G]) | G (G) |
| 1008 | Stramenopiles | A (except Opalinata and Bicosoecida [G]) | C (C) |
| 1649 | Stramenopiles | A | U (A) |
| 982 | Stramenopiles | C | T (T)c |
| 1019 | Stramenopiles | G | U (U)d |
| 1137 | Ochrophytae | A | G (G)f |
Stramenopile sequence signatures are described in reference 10.
Position in the Skeletonema costatum reference sequence (GenBank accession no. M54988, X52006, and X85395).
T is the common nucleotide in the heterokont outgroup.
U is the common nucleotide in the heterokont outgroup.
Terminology used in reference 10.
G is the common nucleotide in the heterokont outgroup.
Probe evaluation and FISH.
The use of probe MAST-12Nor1 (5′-CGUCCGAAAACGAAUUUG-3′) did not result in any hybridization even with 0% formamide. Thus, we did not use this probe in subsequent experiments. However, the second probe (MAST-12Nor2 [5′-UACAGUGCCAAUGGAGAC-3′]) was tested successfully. We observed intense fluorescence signals only with formamide concentrations of 0 and 10%. With 20% formamide, the signal intensity (relative fluorescence) decreased significantly, and no signal was detectable at formamide concentrations of more than 20% (results not shown). The mean cell counts for fluorescence-labeled cells were very similar (ca. 18 cells/ml) at formamide concentrations of 0% and 10%. Thus, in subsequent experiments we used 10% formamide in the hybridization buffer.
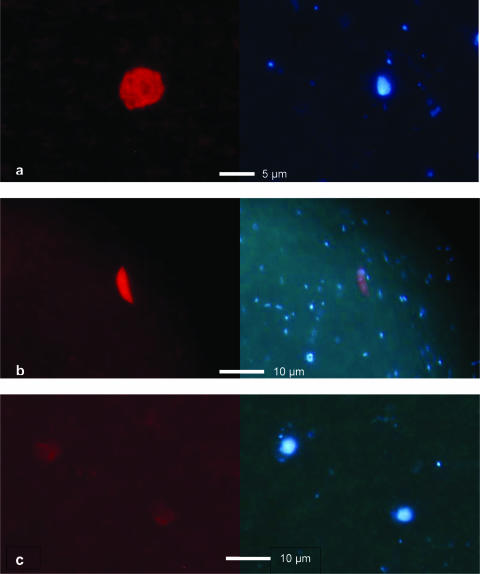
Fluorescence microscopy revealed spherically shaped cells that had a diameter of about 4 to 6 μm and a clearly blue-stained round nucleus (Fig. 2a). The target cells did not exhibit a reddish autofluorescence signal upon UV excitation like other chlorophyll-containing cells on the same filter (Fig. 2b). Additionally, we counterchecked for red fluorescence of target cells in the Cy3 channel by using probe MAST-12Nor2 with fluorescein isothiocyanate as a fluorescent dye. We observed clear signals with the green emission fluorescein isothiocyanate-specific filter set but not with the Cy3-specific filter set (results not shown). Unlabeled nontarget cells were easily distinguished from labeled target cells (Fig. 2c).
FIG. 2.
FISH of a microbial consortium retrieved from a microbial mat sample using the KKTS_D3-specific probe MAST-12Nor2 labeled with the fluorochrome Cy3 (left panels) and DAPI stain to visualize the nuclei (right panels). (a) Positively labeled target cell. No red chlorophyll autofluorescence was detectable with UV excitation (right panel), indicating that the target cell was unpigmented (compare with panel b). (b) Chlorophyll-containing (photoautotroph) algae in a negative control without a probe, showing autofluorescence of chlorophyll with the Cy3-specific filter set. However, chlorophyll also showed red autofluorescence with the DAPI-specific filter set, which allowed us to determine if the signal obtained with the Cy3 filter could be ascribed to Cy3 labeling or a chlorophyll signal. (c) Unlabeled nontarget cells from the same filter as the cell in panel a. The exposure time used for the Cy3 image was nearly three times the exposure time used in panel a to visualize the cells. The fluorescence intensity of the nontarget cells hybridized with MAST-12Nor2 is the same order of magnitude as the fluorescence intensity of cells in negative controls (without a probe) and in hybridizations with a nonsense probe.
Combined FISH and SEM analyses.
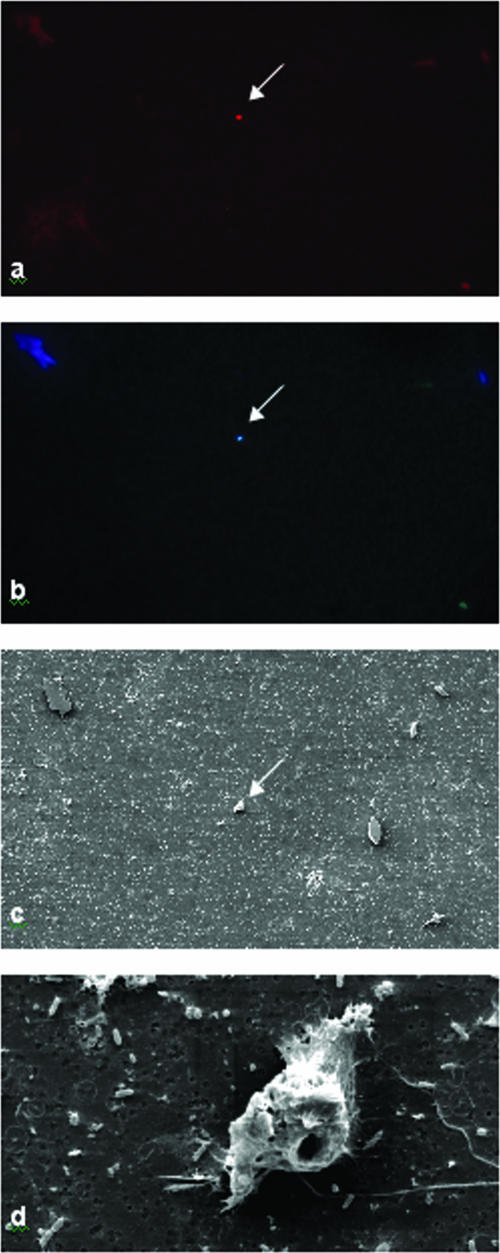
Despite the small target cell size, we succeeded in visualizing fluorescently labeled cells and observing the same cells with the scanning electron microscope (Fig. 3). However, the SEM quality of individual FISH-labeled cells did not entirely meet our expectations based on previously described experiences (38) with different protists. As analysis of other organisms in the same sample (e.g., diverse ciliates, flagellates, and diatoms) resulted in high-quality SEM pictures (available upon request) and because we used several alternative fixatives, we did not attribute the partial damage of the target cells to the preparation technique itself; instead, we attributed it to the target cell's ultrastructural properties. This hypothesis is supported by the hole at the bottom (posterior) of the cell that was exclusively and regularly observed with FISH-labeled target cells. FISH-SEM analyses confirmed the size and the shape of the target cells determined in the FISH analysis.
FIG. 3.
Images obtained at different stages of the combined FISH-SEM protocol to obtain information about the morphology of the target organism behind environmental sequence KKTS_D3. (a) FISH staining in the red channel (Cy3) using probe MAST-12Nor2. (b) DAPI staining in the UV channel to visualize the nucleus of the target cell and to confirm that the Cy3-stained structure was a eukaryotic cell. (c) Same filter area as in panels a and b examined by SEM. (d) Higher magnification of a target cell, revealing the first clues about its morphology.
Despite the fact that FISH-SEM analysis did not provide detailed insights regarding the cell's morphotype, the unique combination of information that we obtained from a series of SEM pictures (especially the characteristic aperture at the posterior of the cell combined with the cell size, cell shape, flagella, and abundance in the sample) was sufficient to unmistakably identify this organism in subsequent SEM analysis of the mixed microbial mat protist community without prior labeling of the target species.
SEM analyses.
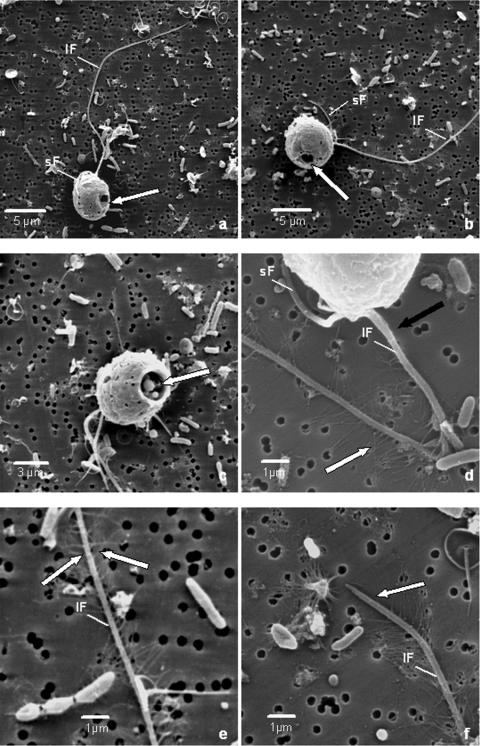
SEM without preceding FISH analysis revealed additional cellular properties of the target cells. Each cell is characterized by two flagella located opposite the cell's opening (Fig. 4a, b, and c), one short flagellum that is about one-half the size of the cell and one conspicuously long flagellum (up to 30 μm long). The latter flagellum tapers in the last 2 to 3 μm of the tip (Fig. 4a). The short flagellum is smooth without any flagellar hairs (mastigonemes) (Fig. 4d), but the long flagellum has two opposed identical pairwise rows of tubular mastigonemes (Fig. 4d, e, and f). High-magnification SEM of the mastigonemes indicated that they are tripartite; however, an ultrastructural analysis using pure cultures of the target cells is needed to confirm this result. The base of the long flagellum, as well as its tip, does not have mastigonemes (Fig. 4d and f). Often, the short flagellum is directed posteriorly and sometimes even appears to be attached to the cell's outer cortex (Fig. 4a). The long flagellum is usually directed anteriorly (Fig. 4a and b). We did not observe flagellar scales, a flagellar wing, swelling at the base of the flagellum, or a stalk. None of the cells studied had anybody scales.
FIG. 4.
SEM images of a target cell. (a and b) Overview of the target cell's general morphotype. The round regularly shaped opening is visible at the posterior of the cell, indicated by an arrow. The short flagellum (sF) is directed posteriorly and seems to be attached to the cell's surface in panel a. The long flagellum (lF) is directed anteriorly and tapers towards its end. (c) Opening at the posterior of the cell (arrow). (d) Arrangement of flagellar hairs (mastigonemes) on the long flagellum, indicated by a white arrow. The base of the flagellum does not have flagellar hairs (black arrow). (e) Detailed view of the arrangement of flagellar hairs (arrow) on the long flagellum. (f) Detailed view of the tapering tip of the long flagellum. The last 1 to 2 μm of the flagellum does not have flagellar hairs (arrow).
DISCUSSION
Identifying the organisms behind novel environmental lineages has been one of several major targets in microbial biodiversity research (9, 23, 38, 41). This is especially important in the case of evolutionarily interesting lineages at higher taxonomic levels and in the case of lineages that occur frequently in different environmental systems and whose morphological diversity and ecological function are unknown. The uncultured MAST clades identified recently (23, 44) meet all of these criteria and therefore deserve further study. Due to the information obtained as a result of the research that we conducted, we were able to determine the morphotype for the 18S rRNA gene sequence that branches within the MAST-12 clade which we retrieved from a Norwegian estuarine microbial mat sample.
Our phylogenetic reconstruction of stramenopiles using 18S rRNA gene signatures is consistent with previous analyses. The previous analyses confirmed the monophyly of phototrophic stramenopiles, resulting in controversies regarding the evolution of stramenopiles (20, 23, 43) and the relationships among basal heterotrophic groups (10, 23). The latter groups branch as separate lineages before the phototrophic radiation and include the Opalinea, the Labyrinthulomycetes, the Bicosoecida, Blastocystis, Developayella, the Hyphochytriales, the Peronosporomycetes, and Pirsonia. We also agree that all 13 novel uncultured marine stramenopile clades should be placed in these heterotrophic basal groups (23, 44). The data indicate that there has been severe undersampling of these organisms compared to their autotrophic relatives. However, some of the MAST clades may actually represent known flagellates having uncertain phylogenetic affinities that have not been sequenced yet. For example, the taxonomically unaffiliated zooflagellates Bordnamonas (19), Glissandra (29), and Kiitoksia (42) may be unrecognized heterotrophic stramenopiles (10). Therefore, it may be useful to determine the 18S rRNA gene sequences of such organisms because in phylogenetic analyses some of them may turn out to be members of a MAST clade.
With the exception of one clone (OLI51105 [16]), all lineages in the MAST-12 clade originated from anoxic sediments like the Guaymas hydrothermal vent deep-sea basin (15) or intertidal sediments from the west coast of the United States (13). Likewise, the KKTS_D3 sequence was obtained from an organism that thrives under at least suboxic conditions. This supports the suggestion that MAST-12 is an environment-specific cluster that occurs in oxygen-depleted habitats (23). However, KKTS_D3 is the first sequence that was obtained from a freshwater sample. This fact does not necessarily suggest that MAST organisms are also distributed in freshwater environments, as the sampling area was a small intermittent estuary periodically influenced by marine water (fundamental ecological niche versus realized ecological niche). Nevertheless, thus far the 18S rRNA gene diversity of only a few freshwater habitats, particularly sediments, has been analyzed (36); therefore, based on our findings the possibility that “true” freshwater representatives of the MAST clades may be detected in future freshwater molecular diversity surveys cannot be eliminated.
Using the culture independent FISH-SEM approach, we microscopically visualized and characterized the organisms behind the MAST-12 sequence KKTS_D3. We designed two specific probes, one of which proved to be successful (MAST-12Nor2). We point out that use of this probe directly with environmental samples may require additional evaluation of its specificity. The MAST-12Nor2 probe exhibits three mismatches with the next closest known relatives that do not belong to the heterotrohic stramenopiles (two autotrophic Pelagophyceae, one autotrophic diatom, one parasitic apicomplexa, one cercozoa, and one fungus). These organisms are morphologically easily distinguishable from the stramenopile target organism. However, MAST12-Nor2 exhibits only a single mismatch with seven other environmental sequences (BAQD200, BAQD16, BAQD14, BAQD21, BAQD224, BAQA21, and BAQA72), all of which belong to the MAST-12 clade (13). Considering that significant binding of the probe occurs only at a low formamide concentration (10%), we cannot exclude the possibility that the probe also targets one of the organisms behind the BAQ sequences, six of which exhibit a maximum sequence difference of only 0.65% with each other. However, we can largely exclude the possibility that there was targeting of one of these BAQ organisms in the sample which we incubated, as we did not find any BAQ sequences in any of our samples (clone libraries).
Consistent with the molecular phylogeny (Fig. 1) and the specific sequence signatures (Table 1), the morphological details that we obtained from our FISH-SEM and SEM analyses allow assignment of KKTS_D3 to the heterotrophic stramenopiles. The target organism seems to be morphologically related to the Bicosoecida, as only in this group are aloricate naked species with a long, mastigoneme-bearing flagellum and a smooth shorter flagellum common (18, 33). However, even though it is possible that some previously described but unassigned flagellates with conspicuously long flagella (12, 31) could belong to the stramenopiles, no currently classified members of the Bicosoecida have a flagellum that is up to 30 μm long, like the organism described here. Thus, the long flagellum is an identifying characteristic of the novel organism detected. The possibility that the novel organism is related members of the genus Blastocystis can be largely eliminated because of the parasitic nature of the latter organisms and their distinctively different morphology (no flagella) (8).
Currently, we cannot explain the conspicuous aperture that was observed only at the posterior of the target cells (Fig. 3 and 4). The fact that such a structure has not been observed and recorded previously suggests that we might have been dealing with a preparation (fixation) artifact. On the other hand, finding a preparation artifact which appears with such regularity in the same symmetrical, spherical shape and at the same position despite the use of various preparation techniques would be unlikely. To solve the dilemma, pure cultures of the KKTS_D3 stramenopile flagellate must be obtained. Then ultrastructural analysis would be the next step in elucidating the nature of the aperture and comparing the organism to previously described (heterotrophic) stramenopiles. Using the FISH probe with natural samples should help us evaluate the abundance, dynamics, distribution, and quantitative role in the microbial web of this facultative anaerobe in natural systems.
Acknowledgments
We thank K. Hausmann, H. Arndt, J. Boenigk, R. A. Andersen, and H. Preisig for comments and advice on SEM photographs. We are grateful to M. Huckins for proofreading the manuscript. We also thank two anonymous reviewers who helped to improve the manuscript.
This study was supported by Deutsche Forschungsgemeinschaft grant STO 414/2-3 to T.S.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Adl, S. M., A. G. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, O. Moestrup, S. E. Mozley-Standridge, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaral Zettler, L. A., F. Gomez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Microbiology: eukaryotic diversity in Spain's River of Fire. Nature 417:137. [DOI] [PubMed] [Google Scholar]
- 4.Baldauf, S. L., D. Bhattacharya, J. Cockrill, P. Hugenholtz, J. Pawlowski, and G. B. Simpson. 2004. The tree of life: an overview, p. 43-75. In J. Cracraft and M. J. Donoghue (ed.), Assembling the tree of life. Oxford University Press, Oxford, United Kingdom.
- 5.Behnke, A., J. Bunge, K. Barger, H. W. Breiner, V. Alla, and T. Stoeck. 2006. Microeukaryote community patterns along an O2/H2S gradient in a supersulfidic anoxic fjord (Framvaren, Norway). Appl. Environ. Microbiol. 72:3626-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens, S., C. Ruhland, J. Inacio, H. Huber, A. Fonseca, I. Spencer-Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackwell, W. H., and M. J. Powell. 2000. A review of group filiation of stramenopiles, additional approaches to the question. Evol. Theor. 12:49-88. [Google Scholar]
- 8.Boreham, P. F., and D. J. Stenzel. 1993. Blastocystis in humans and animals: morphology, biology, and epizootiology. Adv. Parasitol. 32:1-70. [DOI] [PubMed] [Google Scholar]
- 9.Caron, D. A., P. D. Countway, and M. V. Brown. 2004. The growing contributions of molecular biology and immunology to protistan ecology: molecular signatures as ecological tools. J. Eukaryot. Microbiol. 51:38-48. [DOI] [PubMed] [Google Scholar]
- 10.Cavalier-Smith, T., and E. E. Chao. 2006. Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J. Mol. Evol. 62:388-420. [DOI] [PubMed] [Google Scholar]
- 11.Countway, P. D., R. J. Gast, P. Savai, and D. A. Caron. 2005. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J. Eukaryot. Microbiol. 52:95-106. [DOI] [PubMed] [Google Scholar]
- 12.Dangeard, P. 1934. Mémoire sur l'Apistonema submarinum sp. nov. et considérations générales sur la structure des protozoaires et des protophytes. Botaniste 26:261-346. [Google Scholar]
- 13.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgcomb, V. P., D. T. Kysela, A. Teske, A. de Vera Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillou, L., S. Y. Moon-Van Der Staay, H. Claustre, F. Partensky, and D. Vaulot. 1999. Diversity and abundance of Bolidophyceae (Heterokonta) in two oceanic regions. Appl. Environ. Microbiol. 65:4528-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugenholtz, P., G. W. Tyson, and L. L. Blackall. 2001. Design and evaluation of 16S rrna-targeted oligonucleotide probes for fluorescence in situ hybridization, p. 29-42. In M. Aquino de Muro and R. Rapley (ed.), Gene probes: principles and protocols. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 18.Karpov, S. A. 2000. Flagellate phylogeny—an ultrastructural approach, p. 336-360. In B. S. C. Leadbeater and J. C. Green (ed.), The flagellates: unity, diversity and evolution. CRC Press, Taylor and Francis, New York, NY.
- 19.Larsen, J., and D. J. Patterson. 1990. Some flagellates (Protozoa) from tropical marine sediments. J. Nat. Hist. 24:801-937. [Google Scholar]
- 20.Leipe, D. D., S. M. Tong, C. L. Goggin, S. B. Slemenda, N. J. Pieniazek, and M. L. Sogin. 1996. 16S-like rDNA sequence from Developayella elegans, Labyrinthuloides haliotidis, and Proteromonas lacertae confirm that the stramenopiles are a primarily heterotrophic group. Eur. J. Protistol. 32:449-458. [Google Scholar]
- 21.Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Alio, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massana, R., J. Castresana, V. Balague, L. Guillou, K. Romari, A. Groisillier, K. Valentin, and C. Pedros-Alio. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70:3528-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massana, R., R. Terrado, I. Forn, C. Lovejoy, and C. Pedros-Alio. 2006. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ. Microbiol. 8:1515-1522. [DOI] [PubMed] [Google Scholar]
- 25.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 26.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 27.Moreira, D., and P. Lopez-Garcia. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 28.Patterson, D. J. 1999. The diversity of eukaryotes. Am. Nat. 154:S96-S124. [DOI] [PubMed] [Google Scholar]
- 29.Patterson, D. J., and A. G. Simpson. 1996. Heterotrophic flagellates from coastal marine and hypersaline sediments in Western Australia. Eur. J. Protistol. 32:423-448. [Google Scholar]
- 30.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 31.Perty, M. 1852. Zur Kenntnis kleinster Lebensformen nach Bau, Funktion, Systematik, mit Specialverzeichnis der in der Schweiz beobachteten. Jent & Reinert, Bern Switzerland.
- 32.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 33.Preisig, H. R., N. Vørs, and G. Hällfors. 1991. Diversity of heterotrophic heterokont flagellates. Syst. Assoc. Spec. Vol. 45:361-399. [Google Scholar]
- 34.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 35.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 36.Slapeta, J., D. Moreira, and P. Lopez-Garcia. 2005. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. Biol. Sci. 272:2073-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoeck, T., W. H. Fowle, and S. S. Epstein. 2003. Methodology of protistan discovery: from rRNA detection to quality scanning electron microscope images. Appl. Environ. Microbiol. 69:6856-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoeck, T., B. Hayward, G. T. Taylor, R. Varela, and S. S. Epstein. 2006. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157:31-43. [DOI] [PubMed] [Google Scholar]
- 40.Stoeck, T., M. V. J. Schwarz, J. Boenigk, M. Schweikert, S. von der Heyden, and A. Behnke. 2005. Cellular identity of an 18S rRNA gene sequence clade within the class Kinetoplastea: the novel genus Actuariola gen. nov. (Neobodonida) with description of the type species Actuariola framvarensis sp. nov. Int. J. Syst. Evol. Microbiol. 55:2623-2635. [DOI] [PubMed] [Google Scholar]
- 41.Stoeck, T., G. T. Taylor, and S. S. Epstein. 2003. Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea). Appl. Environ. Microbiol. 69:5656-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vørs, N. 1992. Heterotrophic amoebae, flagellates and heliozoa from the Tvärminne area, Gulf of Finland, in 1988-1990. Ophelia 36:1-109. [Google Scholar]
- 43.Yoon, H. S., J. D. Hackett, G. Pinto, and D. Bhattacharya. 2002. The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. USA 99:15507-15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuendorf, A., A. Behnke, J. Bunge, K. Barger, and T. Stoeck. 2006. Diversity estimates of microeukaryotes below the chemocline of the anoxic Mariager Fjord, Denmark. FEMS Microbiol. Ecol. 58:476-491. [DOI] [PubMed] [Google Scholar]