Abstract
An NAD(P)H-nicotine blue (quinone) oxidoreductase was discovered as a member of the nicotine catabolic pathway of Arthrobacter nicotinovorans. Transcriptional analysis and electromobility shift assays showed that the enzyme gene was expressed in a nicotine-dependent manner under the control of the transcriptional activator PmfR and thus was part of the nicotine regulon of A. nicotinovorans. The flavin mononucleotide-containing enzyme uses NADH and, with lower efficiency, NADPH to reduce, by a two-electron transfer, nicotine blue to the nicotine blue leuco form (hydroquinone). Besides nicotine blue, several other quinones were reduced by the enzyme. The NAD(P)H-nicotine blue oxidoreductase may prevent intracellular one-electron reductions of nicotine blue which may lead to semiquinone radicals and potentially toxic reactive oxygen species.
The unraveling of the details of nicotine catabolism in the model organism Arthrobacter nicotinovorans stays in the center of our research efforts. The final goal is to obtain a comprehensive picture of the biochemical steps implicated in this process (7) and to find ways for their ultimate use in biotechnological applications.
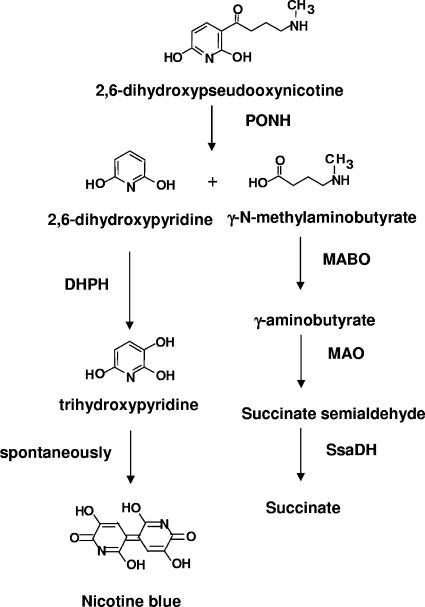
When grown on nicotine, A. nicotinovorans cultures turn dark blue due to the formation of a pigment known as nicotine blue (NB). Chemically, NB was shown to be a 4,5,4′,5′-tetrahydroxy-3,3′-diazadiphenoquinone-(2,2′) formed by the autocatalytic condensation of two molecules of trihydroxypyridine (THP) (18, 20). The steps in nicotine catabolism leading to THP (Fig. 1) start with the cleaving of 2,6-dihydroxypseudooxynicotine into 2,6-dihydroxypyridine (DHP) and γ-N-methylaminobutyrate by the enzyme 2,6-dihydroxypseudooxynicotine hydrolase (PONH) (26). γ-N-methylaminobutyrate is converted to the citric acid cycle intermediate succinate by the enzymes γ-N-methylaminobutyrate oxidase (MABO) (11), monoamine oxidase, and succinate semialdehyde dehydrogenase (13). DHP is hydroxylated to 5-hydroxy-2,6-(1H,3H)-pyridinedione by the flavoenzyme DHP hydroxylase (4). The enzyme reaction can be detected in vitro by monitoring at 578 nm the spontaneous formation of nicotine blue from THP.
FIG. 1.
Steps in nicotine catabolism leading from 2,6-dihydroxypseudooxynicotine to nicotine blue and succinate. The enzymes are PONH, DHPH, MABO, monoamine oxidase (MAO), and succinate semialdehyde dehydrogenase (SsaDH).
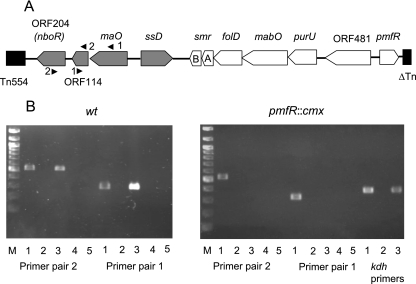
The gene for MABO forms one operon with the genes for the enzymes formyltetrahydrofolate deformylase (purU) and methylentetrahydrofolate dehydrogenase-cyclohydrolase (folD), which are required for the metabolism of methylenetetrahydrofolate formed during the oxidative demethylation of γ-N-methylaminobutyrate (11, 13) and the genes for a small, two-component multidrug resistance pump (SmrAB) (P. Ganas and R. Brandsch, unpublished data). The expression of this operon is regulated by the transcriptional activator PmfR (12). The genes for the monoamine oxidase (maO) and succinate semialdehyde dehydrogenase (ssD) are divergently transcribed (13). The gene cluster is located on the catabolic megaplasmid pAO1 (19) of A. nicotinovorans, flanked by a transposon similar to Tn554 and the truncated gene for a transposase (Fig. 2). The protein derived from orf481 exhibits a high degree of similarity to amino acid permeases but has not yet been functionally characterized. The succinate semialdehyde and monoamine oxidase encoded by the ssD and maO genes, respectively, have been analyzed in some detail (13) and are active in the deamination and oxidation of γ-N-aminobutyrate to succinate.
FIG. 2.
Transcriptional analysis by RT-PCR of the maO, ORF114, and nboR genes. (A) Schematic representation of the gene cluster implicated in γ-N-methylaminobutyrate catabolism. Genes analyzed in this study are shaded in gray. Positions and numbers of primer pairs used in RT-PCR (Table 1) are indicated. A, smrA; B, smrB. (B) Lanes 1, PCR with wild-type strain (wt) or pmfR::cmx strain A. nicotinovorans cells harboring pAO1; lanes 2 and 3, PCR with RNA and cDNA, respectively, of nicotine-induced bacteria; lanes 4 and 5, PCR with RNA and cDNA, respectively, of non-nicotine-induced bacteria. Control PCRs with kdh primers (Table 1) were performed. M, molecular size markers. Expected sizes of PCR DNA fragments from wild-type cells: primer pair 2, 540 bp; primer pair 1, 310 bp. Expected size of DNA fragment from cells with the disrupted pmfR gene, 360 bp.
Here we show that the gene for the pAO1 ORF204 forms one operon with the maO gene and that this operon is under the control of PmfR. The ORF204 protein was identified as a flavin mononucleotide (FMN)-dependent NAD(P)H-nicotine blue oxidoreductase (NBOR). Given the large amounts of nicotine blue produced, the presence of such an enzyme among the enzymes of nicotine catabolism may have its rationale in the need to prevent the intracellular formation of nicotine blue semiquinone radicals, which by redox cycling would lead to the formation of toxic reactive oxygen species (ROS).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli XL-1 Blue was employed both as a host for plasmids and as an expression strain and was grown in Luria-Bertani medium supplemented with the appropriate antibiotics at 37°C. A. nicotinovorans and A. nicotinovorans pmfR::cmx (12) were grown at 30°C in citrate medium (8).
Chemicals and biochemicals.
Endonuclease restriction enzymes were purchased from New England Biolabs (Frankfurt, Germany), PfuUltra DNA polymerase and T4 reverse transcriptase were purchased from Stratagene (Amsterdam, The Netherlands), the rapid DNA ligation kit was purchased from Roche Applied Science (Mannheim, Germany), and Ni-chelating Sepharose was purchased from Amersham Biosciences (Freiburg, Germany). 5-Diethoxyphosphoryl-5-methyl-1-pyrroline-N-oxide (DEPMPO) was from Calbiochem (Schwalbach, Germany). All other chemicals were obtained from Sigma (Steinheim, Germany) unless otherwise indicated and were of the highest purity available.
Reverse transcription-PCR (RT-PCR).
Total RNA was isolated from A. nicotinovorans and A. nicotinovorans pmfR::cmx (12) cultures grown in the presence or absence of nicotine with the help of the RNeasy kit (QIAGEN, Hilden, Germany) and reverse transcribed with T4 reverse transcriptase, and the respective cDNA fragments were applied as templates in PCRs with gene-specific primers (Table 1) as described previously (11, 27). PCRs with pAO1-carrying bacteria were performed as positive controls, and PCR with untranscribed RNA was performed as a negative control.
TABLE 1.
Oligonucleotides used in RT-PCRs and cloning
| Primer or primer pair | Sequence | Description or use |
|---|---|---|
| Primer pair 1 | 5′-GCGAACCCGAATTTGTAGGC-3′ | Forward maO primer |
| 5′-CTCGTATCCCTGATGGACTG-3′ | Reverse orf114 primer | |
| Primer pair 2 | 5′-CTAATCCTTGAGAACGGCTG-3′ | Forward orf114 primer |
| 5′-GAGAACCCGGTTGATAGAGC-3′ | Reverse nboR primer | |
| ssD primer pair | 5′-CGTTGGAGATGGGCAAGCC-3′ | Forward ssD primer |
| 5′-GGGAATCTTTCAGGAGTGGC-3′ | Reverse ssD primer | |
| kdh primer pair | 5′-GGCAACAGACAGGCATGGATAGG-3′ | Forward kdh primer |
| 5′-GATCAACAGCAAGTACAGCAACC-3′ | Reverse kdh primer | |
| Forward primer | 5′-GAAAGGATTGGATCCATGGCAGCAAAATAC-3′ | Cloning of orf204 |
| Reverse primer | 5′-GTTTGCTGTCGACGCGCGT-3′ | Cloning of orf204 |
| 5′ maO primer pair | 5′-CACATCCGGAACTTTCCCT-3′ | Forward primer |
| 5′-CGAGTTTTGTAGCAGCTGC-3′ | Reverse 279-bp EMSA fragment | |
| 5′ ssD primer pair | 5′-GATCTCTGGAGAAGCAATGTTG-3′ | Forward primer |
| 5′-GTCATTTGTAGTCCTCAGAAG-3′ | Reverse 228-bp EMSA fragment |
EMSA.
DNA fragments were generated with primers listed in Table 1. The fragments were 5′ end labeled with [γ-32P]ATP by using the Ready-to-Go polynucleotide kinase kit according to the instructions of the supplier (Amersham Biosciences). Electrophoretic mobility shift assays (EMSA) of the labeled DNA fragments in the presence of purified PmfR were performed as described previously (12).
Cloning of the ORF204 gene (nboR).
pH6EX3 (5) was used as the expression vector for the cloning of the ORF204 gene. The gene was amplified by PCR with the primer pair indicated in Table 1 carrying BamHI and SalI restriction enzyme recognition sites, whole A. nicotinovorans bacteria carrying pAO1 as a template, and Pfu-Turbo high-fidelity DNA polymerase (Stratagene) as outlined in reference 11. The DNA fragments were BamHI-SalI digested and ligated into the pH6EX3 vector digested with the same restriction enzymes, resulting in pH6EX3nboR. The ligated DNA was used to transform E. coli XL-1 Blue competent bacteria. Recombinant plasmid DNA was isolated from transformed bacteria selected on Luria-Bertani plates with 50 μg/ml ampicillin, and positive clones were checked by sequencing.
Purification of the His-tagged oxidoreductase.
The growth of E. coli XL-1 Blue harboring pH6EX3nboR, the induction and overexpression of nboR, and the preparation of cleared bacterial lysates were carried out according to methods described in reference 12. The lysates were filtered through a 0.45-μm-pore-size filter and loaded onto 1-ml HisTrap high-performance columns equilibrated with 40 mM HEPES buffer (pH 7.5)-0.5 M NaCl which were connected to an ÁKTA basic 10 system (Amersham, Freiburg, Germany), and the His-tagged NBOR was purified using an imidazole gradient (80 to 500 mM) in HEPES buffer. Pure NBOR protein samples eluted at 400 mM imidazole. The purified samples were further loaded onto HiPrep desalting columns (Amersham) previously equilibrated with 40 mM HEPES buffer (pH 7.5)-100 mM NaCl-10% glycerol in order to remove the imidazole. The purified protein was kept at −20°C for several weeks without loss of activity. The native molecular mass of the NBOR was determined by gel permeation chromatography on a HiLoad 16/60 Superdex 200 prepgrade column equilibrated with 40 Mm HEPES (pH 7.5)-100 mM NaCl. The column was calibrated using aldolase (molecular mass, 158 kDa), bovine albumin (molecular mass, 67 kDa), ovalbumin (molecular mass, 43 kDa), and chymotrypsinogen (molecular mass, 25 kDa).
Western blotting of A. nicotinovorans extracts.
Purified His-tagged NBOR protein was used to raise an antiserum in rabbits by using standard protocols. Lysates of bacterial pellets from 400-ml cultures of A. nicotinovorans grown in the presence or absence of nicotine were obtained by passage through a French pressure cell at 132 MPa and centrifugation for 30 min at 12,000 × g as described previously (12). The extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% polyacrylamide gels and blotted onto Optitran BA-S 85 nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany). The incubation of the membranes with the NBOR antiserum and with alkaline phosphatase- conjugated goat anti-rabbit immunoglobulin G (Sigma) and the detection of the immunocomplexes with bromochloroindolyl phosphate-nitroblue tetrazolium were performed by using standard protocols (16).
Preparation of nicotine blue.
NB was produced in vitro in the dihydroxypyridine hydroxylase (DHPH) reaction (4) performed in 50 mM K-phosphate buffer (pH 7.1) with various amounts of DHP and NADH, as required, in the presence of purified His-tagged DHPH. The formation of NB was monitored by the increase in A578 as determined by an Ultrospec 3100 Pro UV/visible spectrophotometer (Amersham). At the end of the reaction, DHPH was removed by passing the assay mixture containing the NB through a Vivaspin MWCO 3000 membrane (Vivascience, Hanover, Germany). The amount of NB formed was expressed as a function of the amount of DHP in the assay mixture. Increasing amounts of DHP were added to the DHPH assay mixture, and the A578 was recorded after 20 min and was plotted against the concentration of DHP. The dependence was found to be linear in the range of 1 to 200 mM DHP. The complete turnover of DHP into NB was checked by the addition of fresh DHPH and NADH to the assay mixture and monitoring of the change in absorption at 578 nm, which was zero when DHP was consumed. The depletion of NADH was determined similarly by adding fresh DHP and DHPH to the assay mixture and monitoring the change in absorption at 578 nm. The molar absorbance of NB was calculated to be 7.154 × 103 M−1 cm−1.
Determination of NBOR enzyme activity.
The standard reaction assay mixture consisted of 100 μM NB in 50 mM K-phosphate buffer, pH 7.1, and 200 μM NADH or 200 μM NADPH in a final volume of 1 ml. Since the product of the reaction rapidly reacts with oxygen, reforming the blue substrate, the reaction cuvette was degassed using N2 and kept under anaerobic conditions. The enzymatic reaction was initiated by the addition of 0.6 μg of NBOR into the sealed cuvette. After a 10-s delay, the reduction of NB was monitored by the decrease in A578 due to the formation of the reduced, leuco form of NB. Enzyme activity was expressed as nanomoles of NB reduced per minute per microgram of protein.
The determination of the specific activity of NBOR was performed by monitoring at 340 nm the oxidation of NADH in the presence of various quinones which did not require anaerobic conditions. The specific activity of the NBOR (1.5 μg) was determined by using 1-ml assay mixtures with K-phosphate buffer (pH 7.1), 200 μM NADH, and a 100 mM concentration of one of the following compounds: 1,4-naphthoquinone (10 mM stock solution in dimethyl sulfoxide [DMSO]), 1,4-benzoquinone (10 mM stock solution in water), 2,6-dichlorophenol-indophenol (10 mM stock solution in water), 2,5-dihydroxy-1,4-benzoquinone (10 mM stock solution in DMSO), menadione (methylnaphthoquinone; 10 mM stock solution in ethanol), duraquinone (10 mM stock solution in ethanol), and chromate (10 mM stock solution in water). Control assays were performed in the absence of the enzyme or with the enzyme but in the presence of DMSO or ethanol without a substrate.
In order to determine the Km of NADH in the NBOR reaction, the standard conditions specified above were employed but increasing amounts of the coenzymes were added to the assay. For the determination of the Kms of NB and other quinones as substrates of the NBOR, the reaction was performed at concentration ranges of 30 to 600 μM NB and 5 to 1,000 μM for the quinones. However, by using NB as the substrate, no saturation levels could be reached in the assays because the pigment precipitated during the enzymatic reaction.
Photoreduction of NBOR.
The photoreduction of NBOR was performed according to methods described in reference 14. The assay mixture consisted of 8.8 μM NBOR in 1 ml of 100 mM Tris-HCl (pH 7. 5)-1.5 mM EDTA and was kept protected from light in a dark cuvette made anaerobic by flushing it with N2. Assay mixtures were exposed to a 50-W halogen lamp source for different periods of time (10 to 100 s), and the UV/visible spectra were registered. Control reactions were performed with the enzyme kept in the dark.
EPR measurements.
Electron paramagnetic resonance (EPR) measurements were conducted with a Bruker EMX 1/6 spectrometer operating at the X band (9.2 GHz). Superoxide radical formation was detected at room temperature by scavenging with the phosphorylated nitrone type spin trap compound DEPMPO. The DEPMPO-OOH adduct radical is easily detected by EPR spectroscopy due to its long half-life (∼8 min) and its distinct superoxide trapping rate (29). The assay mixture consisted of a 1-ml NB preparation, 20 μl of 20 mM NADH, and 1 μl of DEPMPO, and the container was made anaerobic by flushing with N2. Two micrograms of NBOR was added to bleach the NB to the leuco form. Aliquots (10 μl) were withdrawn from the solutions and placed into small quartz tubes preflushed with N2, and the EPR spectra were recorded. O2 was introduced into the sample to reoxidize the NB, and a new aliquot was taken for spectroscopy. Spin trapping with DEPMPO under the experimental conditions described here allows the measurement of less than 0.5 μM superoxide (25). The Fenton reaction was used as a control to generate superoxide radicals (28).
RESULTS
The amine oxidase gene (maO) and the ORF204 gene (nboR) form one operon.
In the gene cluster responsible for the catabolism of γ-N-methylaminobutyrate to citric cycle intermediates, it was intriguing to find a gene (nboR) for an ORF204 protein with a high level of similarity to NAD(P)H-quinone oxidoreductases, and we asked whether the gene forms a functional unit with the rest of the nicotine-induced genes (the nic regulon). RT-PCR with RNA extracted from A. nicotinovorans nicotine-induced and noninduced bacteria showed that this was the case (Fig. 2B, left panel). Transcripts of the nboR gene were present only in RNA of nicotine-induced bacteria (Fig. 2B, left panel, compare lanes 3 and 5). The analysis with primer pairs 1 and 2 also showed that maO, orf114, and nboR form one transcriptional unit (Fig. 2B, left panel, lanes 3). The function, if any, of the small orf114 is unknown.
Expression of the maO-orf114-orf204 operon is regulated by the transcriptional activator PmfR.
Additional support for the functional unity of the NAD(P)H-quinone oxidoreductase with the enzymes of nicotine catabolism came from the finding that the expression of the maO-orf114-orf204 operon is regulated by PmfR. In an A. nicotinovorans strain with pmfR disrupted by the insertion of a chloramphenicol cassette, no transcripts of the operon could be detected (Fig. 2B, right panel, primer pair 2, lane 3, and primer pair 1, lane 3). As a positive control, transcripts of the ketone dehydrogenase gene (which has nicotine-dependent expression but is not under PmfR control) in the cDNA preparation were identified with specific primers (Fig. 2B, right panel, kdh primers).
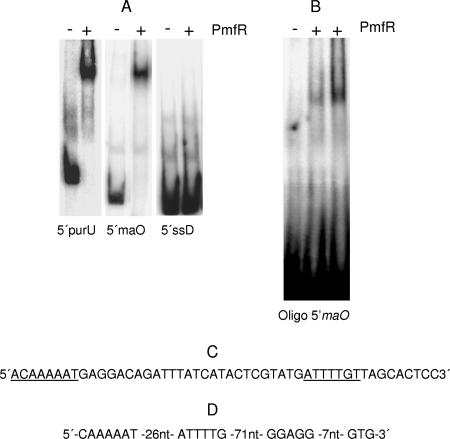
EMSA with a 279-bp 32P-labeled DNA fragment 5′ to maO showed a gel shift with purified PmfR protein (Fig. 3A, middle column) similar to the one obtained with a DNA fragment 5′ to purU (Fig. 3A, left column) (12). However, PmfR did not bind to a DNA fragment derived from the 5′ region of the ssD gene (Fig. 3A, right column). Dissection of the 279-bp fragment with restriction endonucleases revealed that the PmfR binding site was located on a 50-bp RsaI-BfaI fragment (not shown). A double-stranded oligonucleotide containing an imperfect inverted repeat (Fig. 3C) was derived and shown in EMSA to contain the binding site of PmfR (Fig. 3B). The position of the PmfR binding oligonucleotide with respect to the translational start codon GTG and the ribosome binding site of the maO gene is shown in Fig. 3D.
FIG. 3.
EMSA with PmfR and DNA fragments. Assays were performed as described in Materials and Methods. (A) EMSA with 308-bp (12), 279-bp, and 228-bp 5′ DNA fragments derived with primer pairs shown in Table 1 from the 5′ regions of the purU, maO, and ssD genes, respectively. +, present; −, absent. (B) EMSA with the double-stranded oligonucleotide shown in panel C derived from the 5′ maO DNA region. (C) 5′ maO oligonucleotide. Underlined sequences indicate the PmfR binding site. (D) Position of the PmfR binding oligonucleotide with respect to the ribosome binding site and the GTG translational start codon of the maO gene. nt, nucleotides.
The ORF204 protein is an FMN-dependent NAD(P)H-nicotine blue oxidoreductase.
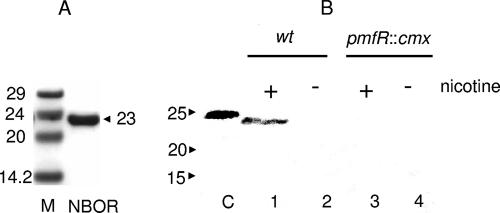
The obvious transcriptional and regulatory link of the ORF204 gene with the gene cluster of γ-N-methylaminobutyrate catabolism determined us to elucidate its biochemical function. To this end, the ORF204 protein was purified as an N-terminally His-tagged recombinant protein from E. coli XL-1 Blue transformed with pH6EX3nboR (see Materials and Methods). The fusion with the His tag of the vector resulted in the N-terminal sequence HHHHHLVPRGSL, with the final L indicating the leucine encoded in this context by the TTG translational start codon of the gene. The purified protein migrated on the SDS-PAGE gels in good accordance with its calculated molecular mass (Fig. 4A). Antiserum corresponding to the ORF204 protein was raised in a rabbit, and Western blotting with proteins of extracts prepared from nicotine-grown wild-type and pmfR::cmx strains was performed. The ORF204 protein was present in the wild-type extracts and migrated with a somewhat smaller molecular mass than the recombinant protein with the N-terminal His extension (Fig. 4B). No NBOR was detected in the extracts of the wild-type strain grown without nicotine (Fig. 4B, lane 2) or in the extracts of the pmfR-disrupted strain irrespective of the presence of nicotine in the growth medium (Fig. 4B, lanes 3 and 4). Despite many efforts, it was not possible to inactivate the nboR gene by homologous recombination with an antibiotic resistance cassette.
FIG. 4.
Purification of NBOR and identification of the protein on Western blots with extracts of nicotine-grown bacteria. (A) Recombinant NB oxidoreductase was purified from E. coli XL-1 Blue transformed with pH6EX3nboR as detailed in Materials and Methods and analyzed by SDS-PAGE on 12% gels. M, molecular mass markers with sizes expressed in kilodaltons; NBOR, 5 μg of purified protein with the indicated molecular mass in kilodaltons. (B) Western blots prepared as described in Materials and Methods with protein extracts of nicotine-induced (+; lanes 1 and 3) and non-nicotine-induced (−; lanes 2 and 4) A. nicotinovorans wild-type (wt) and A. nicotinovorans pmfR::cmx bacteria, respectively, were decorated with anti-NBOR antiserum and developed with alkaline phosphatase-coupled secondary antibody.
The yellow protein contained noncovalently bound FMN as identified by thin-layer chromatography (not shown). The nicotine-dependent expression of this protein and its similarity to NAD(P)H-quinone oxidoreductases raised the question as to what compound derived from nicotine catabolism could be the substrate of this enzyme. The only logical candidate was nicotine blue [4,4′,5,5′-tetrahydroxy-3,3′-diazadiphenoquinone-(2,2′)], which is produced by the dimerization of trihydroxypyridine formed in the dihydroxypyridine hydroxylase reaction from dihydroxypyridine. In its quinone form, this compound shows the characteristic color of nicotine blue. When it is reduced, it changes into the colorless leuco form (Fig. 5A). NB was prepared in vitro from DHP with DHPH in the presence of NADH as described in Materials and Methods. The molar concentration of NB was expressed as a function of the DHP initially present in the assay mixture. The NB preparation, made hydroxylase free by filtration through a Vivaspin MWCO 3000 membrane, was employed in a reaction assay mixture with NADH or NADPH and NBOR under anaerobic conditions. When the NB concentration dependence of the NBOR reaction was monitored, it was not possible to reach saturation levels with NB because the pigment precipitated at higher concentrations. Therefore, the Km of NB as the substrate in the reaction could not be accurately determined. The apparent Km of NADH in the reaction mixture was found to be 35 ± 2.88 μM. With NADPH, the speed of the reaction was still 70% of that observed with NADH. Thus, the enzyme may be defined as a NAD(P)H-NB oxidoreductase.
FIG. 5.
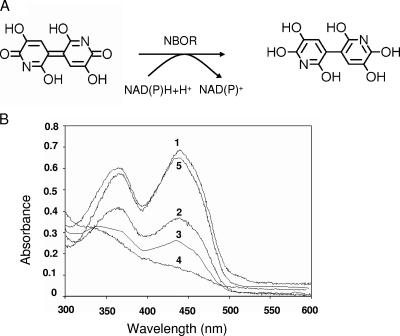
Reaction performed by the NBOR and the photoreduction of its FMN cofactor. (A) Reaction scheme of the NAD(P)H-dependent reduction of NB to its leuco form. (B) Time-dependent photoreduction of NBOR (time of exposure to light: 1, 0 s; 2, 10 s; 3, 20 s; 4, 100 s; 5, 100 s, followed by the addition of O2) performed as described in Materials and Methods.
The flavin nucleotide of the enzyme was photochemically reduced without the formation of a blue or red flavin radical (Fig. 5B). The reduction performed with NADH followed the same pattern (data not shown), an indication that the enzyme performed two-electron transfers. The oxidation of the flavin by NB could not be monitored as the color of the pigment interfered with the determination.
Substrate specificity of the oxidoreductase.
Besides NB, the enzyme accepted several quinones as substrates. A comparison of the kinetic parameters of the NBOR obtained with NB and that obtained with various quinone substrates is shown in Table 2. The highest specific activity was reached with NB, which correlated with the highest Vmax/Km ratio. The specific activity of NBOR decreased and the Km of the quinone substrate increased in the order 1,4-naphthoquinone, 1,4-benzoquinone, 2,6-dichlorophenol-indophenol, menadione, and duraquinone. The Vmax for the last two compounds could not be reached with reasonable concentrations. The reduced Vmax/Km values with quinones as substrates indicated that NBOR preferentially turned over NB. Apparently, NBOR is enzymatically less efficient with methylated quinone substrates as indicated by the kinetic data for 1,4-naphthoqionone versus menadione and for 1,4-benzoquinone versus duraquinone.
TABLE 2.
Substrate specificity of NBOR
| Substrate | Sp acta (nmol of NADH min−1 μg−1 of protein) | Km (μM) | Vmax/Km s−1 |
|---|---|---|---|
| NB | 893 ± 61.56 | NDb | 310.8 |
| 1,4-Naphthoquinone | 733 ± 17.62 | 6.25 ± 0.144 | 3.712 |
| 1,4-Benzoquinone | 536.75 ± 15.19 | 8.83 ± 0.73 | 2.219 |
| 2,6-Dichlorophenol-indophenol | 368.43 ± 6.95 | 23 ± 0.57 | 0.947 |
| Menadione | 102.57 ± 5.94 | ND | 0.576 |
| Duraquinone | 4.26 ± 0.34 | ND | 0.048 |
Determined at a 100 μM concentration of each substrate.
ND, not determined.
EPR measurements of O2− radical formation.
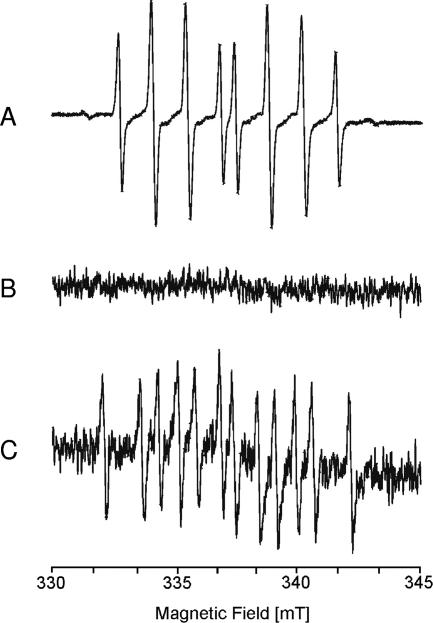
EPR measurements showed no formation of superoxide during the reduction of NB by NBOR (Fig. 6), an indication that no longer-lived NB semiquinone radical was formed during the reaction and therefore no redox cycling with the formation of ROS occurred. However, the reoxidation of the reduced NB in the presence of O2 was accompanied by the release of O2− (Fig. 6).
FIG. 6.
Analysis by EPR of superoxide formation during the reduction and reoxidation of NB. The spectra were recorded with aliquots of the assay mixture in the presence of 100 mM DEPMPO. The spectra show the Fenton reaction as a positive control (A), the reduction of NB by NBOR (B), and the nonenzymatic reoxidation of reduced NB in the presence of O2 (C). EPR conditions were as follows: microwave frequency, 9.65 GHz; modulation amplitude, 0.1 mT; time constant, 0.164 s; scan rate, 5.4 mT/min.
DISCUSSION
The ability of A. nicotinovorans to use nicotine as a carbon, nitrogen, and energy source is linked to the presence of the 165-kb catabolic plasmid pAO1 (19). The genetic organization of the pathway exhibits a modular structure (19). One of these modules is dedicated to the catabolism of γ-N-methylaminobutyrate (11, 13). As shown here, it contains, besides the purU-mabO-folD-nepAB operon, the operon maO-nboR. The operons are integrated into one functional unit by the transcriptional activator PmfR (12). This arrangement was clearly shown by the results of the EMSA. The 5′-AGTTTA-3′ and 5′-AAAACCA-3′ sequences protected by DNase I footprinting in the purU 5′ sequence (12) are also found in the PmfR binding oligonucleotide derived from the 5′ maO sequence in the form of 5′-ACAAAAA-3′ and 5′-AGATTTA-3, but in inverted orientation. The absence in the nicotine-grown A. nicotinovorans pmfR::cmx strain of maO-nboR transcripts and of NB oxidoreductase protein on Western blots indicated that PmfR functions as an activator of the expression of the operon. Apparently not under the control of PmfR was the gene for the succinate semialdehyde dehydrogenase, which uses NAD+ to oxidize the semialdehyde to succinate and leaves NAD in its reduced NADH form. The same situation applies to the permease gene. How the expression of these genes is regulated remains to be established. The entire gene cluster apparently represents one genetic block inserted in pAO1 by transposition. How these genes were assembled and where they originated are interesting aspects of the evolution of pAO1 as a catabolic plasmid.
It came as a surprise that a gene encoding a NAD(P)H-quinone oxidoreductase is part of the gene cluster of γ-N-methylaminobutyrate catabolism and part of the pAO1 nic regulon. Apparently, there was evolutionary pressure to have a gene for an oxidoreductase with specificity for NB within the gene cluster. A BLAST search with the amino acid sequence of NBOR revealed a high degree of similarity to soluble NAD(P)H-quinone and chromate reductases. Soluble NAD(P)H-quinone acceptor oxidoreductases are flavoenzymes that can catalyze the two-electron reduction of quinones to hydroquinones. In this way, they prevent the reduction of quinones by one-electron reductases that would result in the formation of reactive oxygen species generated by the redox cycling of semiquinones in the presence of molecular oxygen (6). The predicted NAD(P)H-quinone reductase of Rhizobium etli (accession number Q2JZ68) with 56% amino acid identity was the closest relative to NBOR. The quinone and chromate reductase YieF of E. coli (accession number POAGE6) characterized recently in more detail (2) showed 35% amino acid identity to NBOR over the entire length of the protein. Typical for this family of soluble bacterial NAD(P)H-quinone oxidoreductases is the presence, like in NBOR, of FMN as a prosthetic group. The signature sequence LFVTPEYNXXXXXXLKNAIDXXS, where X's represent variable residues, of this NADH-dh2 family of flavin binding quinone reductases (15) is found between amino acids 74 and 96. It is believed to represent the FMN binding sites of these enzymes. The NAD(P)H binding amino acid motif AXGXXG present in several quinone oxidoreductases (22) is not present in this form in NBOR. However, in a similar position in the C-terminal part of NBOR, the amino acid sequence SXGXXG, which is highly conserved among proteins related to NBOR, was found (not shown). These enzymes may form homodimers (1) or homotetramers (23). Similarly, NBOR forms homotetramers with a minor proportion of homodimers, as determined by gel permeation chromatography. Together with the YieF of E. coli, which in its native state is a homodimer, it belongs to the class I family of chromate reductases (2). YieF appears to be a two-electron reducing enzyme (1) similar to mammalian DT-diaphorase (24) which obligatorily performs two-electron transfers to electrophiles (9, 21). Diaphorase-like activity in bacterial extracts of Arthrobacter oxydans was observed previously (17) and is probably identical with that of the NBOR characterized in this work. NBOR apparently also performs two-electron transfers to its substrate, since no formation of a flavin semiquinone was observed. The kinetic data show the enzyme to have relatively broad quinone substrate specificity, possibly an indication of the recent recruitment of the NBOR gene to the nic regulon. However, different from other soluble bacterial quinone reductases, NBOR showed insignificant chromate reductase activity. The enzyme preferred NADH over NADPH, in accord with a role in nicotine catabolism and not in a biosynthetic pathway.
Why should this NAD(P)H-dependent NB oxidoreductase be part of the nicotine catabolic pathway? Our proposal is because it prevents the formation of NB semiquinone radicals by cellular enzymes which may perform one-electron transfers to NB (3). NBOR apparently reduces NB by two-electron transfer, without the formation of a flavin or NB semiquinone radical, as indicated by the photoreduction of NBOR and the EPR measurements. The formation of these radicals would lead to redox cycling in the presence of oxygen, with the formation of highly toxic ROS (15, 22). If this consequence was not prevented, the large amounts of NB produced intracellularly during nicotine degradation would result in considerable oxidative stress to the bacteria. We assume that NB in its reduced form is secreted out of the bacteria into the growth medium, where it auto-oxidizes in the presence of O2, turning it to the characteristic blue. This reaction, however, was accompanied by O2− formation. This finding may represent a selective advantage for A. nicotinovorans bacteria in competition with other soil community bacteria sensitive to oxygen radicals. A. nicotinovorans itself appeared not to be affected by the generation of these radicals during growth on nicotine, and growth in the presence of nicotine was identical to that in the absence of nicotine (M. Mihasan and R. Brandsch, unpublished data).
A. nicotinovorans is an aerobic gram-positive soil bacterium. Under these conditions, the NADH produced in the succinate semialdehyde dehydrogenase reaction should be mainly reoxidized by the respiratory chain. However, it is tempting to speculate that under conditions of low O2 concentrations in the soil, NB quinone may act as a terminal electron acceptor for the reoxidation of NADH (10) and the regeneration of NAD+ needed in the succinate semialdehyde dehydrogenase reaction. Thus, the gene cluster of pAO1 responsible for the catabolism of γ-N-methylaminobutyrate may also represent an incidence biochemical unity.
Acknowledgments
We thank Veronika Erichsen for excellent technical assistance.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to R.B. and to T.F.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Ackerley, D. F., C. F. Gonzalez, C. H. Park, R. Blake II, M. Keyhan, and A. Martin. 2004. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl. Environ. Microbiol. 70:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerley, D. F., C. F. Gonzalez, M. Keyhan, R. Blake, and A. Matin. 2004. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ. Microbiol. 6:851-860. [DOI] [PubMed] [Google Scholar]
- 3.Argyrou, A., S. Guangxing, B. A. Palfey, and J. S. Blanchard. 2003. Catalysis of diaphorase reactions by Mycobacterium tuberculosis lipoamide dehydrogenase occurs at the EH4 level. Biochemistry 42:2218-2228. [DOI] [PubMed] [Google Scholar]
- 4.Baitsch, D., C. Sandu, and R. Brandsch. 2001. A gene cluster on pAO1 of Arthrobacter nicotinovorans involved in the degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine-3-hydroxylase. J. Bacteriol. 183:5262-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthold, H., M. Scanarini, C. C. Abney, B. Frorath, and W. Northemann. 1992. Purification of recombinant antigenic epitopes of the human 68-kDa (U1) ribonucleoprotein antigen using the expression system pH6EX3 followed by metal chelating affinity chromatography. Protein Expr. Purif. 3: 50-56. [DOI] [PubMed] [Google Scholar]
- 6.Bianchet, M. A., M. Faig, and L. M. Amzel. 2004. Structure and mechanism of NAD(P)H:quinone acceptor oxidoreductases (NQO). Methods Enzymol. 382:144-174. [DOI] [PubMed] [Google Scholar]
- 7.Brandsch, R. 2006. Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69:493-498. [DOI] [PubMed] [Google Scholar]
- 8.Brühmüller, M., A. Schimz, L. Messmer, and K. Decker. 1975. Covalently bound FAD in d-6-hydroxynicotine oxidase. J. Biol. Chem. 250:7747-7751. [PubMed] [Google Scholar]
- 9.Cenas, N., Z. Anusevicius, H. Nivinskas, L. Miseviciene, and J. Sarlauskas. 2004. Structure-activity relationships in two-electron reduction of quinones. Methods Enzymol. 382:258-277. [DOI] [PubMed] [Google Scholar]
- 10.Cervantes, F., S. van der Velde, G. Lettinga, and J. Field. 2000. Quinones as terminal electron acceptors for anaerobic microbial oxidation of phenolic compounds. Biodegradation 11:313-321. [DOI] [PubMed] [Google Scholar]
- 11.Chiribau, C.-B., C. Sandu, M. Fraaije, E. Schiltz, and R. Brandsch. 2004. A novel γ-N-methylaminobutyrate demethylating oxidase involved in the catabolism of the tobacco alkaloid nicotine by Arthrobacter nicotinovorans pAO1. Eur. J. Biochem. 271:4677-4684. [DOI] [PubMed] [Google Scholar]
- 12.Chiribau, C. B., C. Sandu, G. L. Igloi, and R. Brandsch. 2005. Characterization of PmfR, the transcriptional activator of the pAO1-borne purU-mabO-folD operon of Arthrobacter nicotinovorans. J. Bacteriol. 187:3062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiribau, C.-B., M. Mihasan, P. Ganas, G. L. Igloi, V. Artenie, and R. Brandsch. 2006. Final steps in nicotine dgradation. Deamination versus demethylation of gamma-N-methylaminobutyrate. FEBS J. 273:1528-1536. [DOI] [PubMed] [Google Scholar]
- 14.Deller, S., S. Sollner, R. Trenker-El-Toukhy, I. Jelesarov, G. M. Gübitz, and P. Macheroux. 2006. Characterization of a thermostable NADPH:FMN oxidoreductase from the mezophilic bacterium Bacillus subtilis. Biochemistry 45:7083-7091. [DOI] [PubMed] [Google Scholar]
- 15.Gonzales, C. F., D. F. Ackerley, S. V. Lynch, and A. Martin. 2005. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J. Biol. Chem. 280:2590-22595. [DOI] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies, p. 471-510. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Holmes, P. E., and S. C. Rittenberg. 1972. The bacterial oxidation of nicotine. VII. Partial purification and properties of 2,6-dihydroxypyridine oxidase. J. Biol. Chem. 247:7622-7627. [PubMed] [Google Scholar]
- 18.Holmes, P. E., and S. C. Rittenberg. 1972. The bacterial oxidation of nicotine. VIII. Synthesis of 2,3,6-trihydroxypyridine and accumulation and partial characterization of the product of 2,6-dihydroxypyridine oxidation. J. Biol. Chem. 247:7628-7633. [PubMed] [Google Scholar]
- 19.Igloi, G. L., and R. Brandsch. 2003. Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J. Bacteriol. 185:1976-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knackmuss, H.-J., and W. Beckmann. 1973. The structure of nicotine blue from Arthrobacter oxydans. Arch. Mikrobiol. 90:167-169. [DOI] [PubMed] [Google Scholar]
- 21.Lind, C., P. Hochstein, and L. Ernster. 1982. DT-diaphorase as a quinone reductase: a cellular control device against semiquinone and superoxide radical formation. Arch. Biochem. Biophys. 216:178-185. [DOI] [PubMed] [Google Scholar]
- 22.Maruyama, A., Y. Kumagai, K. Morikawa, K. Taguchi, H. Hayashi, and T. Ohta. 2003. Oxidative-stress-inducible qorA encodes an NADPH-dependent quinine oxidoreductase catalysing a one-electron reduction in Staphylococcus aureus. Microbiology 149:389-398. [DOI] [PubMed] [Google Scholar]
- 23.Patridge, E. V., and J. G. Ferry. 2006. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 188:3498-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross, D., and D. Siegel. 2004. NAD(P)H:quinone oxidoreductase 1 (NQO1, DT-diaphorase), functions and pharmacogenetics. Methods Enzymol. 382:115-144. [DOI] [PubMed] [Google Scholar]
- 25.Roubaud, V., S. Sankarapandi, P. Kuppusamy, P. Tordo, and J. Zweier. 1997. Quantitative measurement of superoxide generation using the spin trap 5-(diethoxyphosphoryl)-4-methyl-1-pyrroline-N-oxide. Anal. Biochem. 247:404-411. [DOI] [PubMed] [Google Scholar]
- 26.Sachelaru, P., E. Schiltz, G. L. Igloi, and R. Brandsch. 2005. An alpha/beta-fold C-C bond hydrolase is involved in a central step of nicotine catabolism by Arthrobacter nicotinovorans. J. Bacteriol. 187:8516-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandu, C., C. Chiribau, and R. Brandsch. 2003. Characterization of HdnoR, the transcriptional repressor of the 6-hydroxy-d-nicotine oxidase gene of Arthrobacter nicotinovorans pAO1, and its DNA-binding activity in response to l- and d-nicotine derivatives. J. Biol. Chem. 278:51307-51315. [DOI] [PubMed] [Google Scholar]
- 28.Sgherri, C. L. M., C. Pinzino, E. Samaritani, and F. Navari-Izzo. 1999. Activated oxygen generation from thylakoids: a novel spin trap. Free Radic. Res. 31:199-204. [DOI] [PubMed] [Google Scholar]
- 29.Vasquez-Vivar, J., B. Kalyanaraman, and M. C. Kennedy. 2000. Mitochondrial aconitase is a source of hydroxyl radical. An electron spin resonance investigation. J. Biol. Chem. 275:14064-14069. [DOI] [PubMed] [Google Scholar]