Abstract
Recently, a sequence-based approach has been developed for the fast isolation and characterization of class II aryl-hydroxylating dioxygenase activities (S. Kahl and B. Hofer, Microbiology 149:1475-1481, 2003). It comprises the PCR amplification of segments of alpha subunit genes of unknown sequence that encode the catalytic center and their fusion with sequences of the bphA gene cluster of Burkholderia xenovorans LB400. One of the resulting chimeric enzymes, harboring the core segment of a dioxygenase from Pseudomonas sp. strain B4-Magdeburg, has now been characterized with respect to the oxidation of chlorobiphenyls (CBs). Its substrate and product specificities differed favorably from those of the parental dioxygenase of strain LB400. The hybrid possessed a higher regiospecificity and yielded less unproductive dioxygenations at meta and para carbons. It attacked ortho-, meta-, and para-chlorinated rings with comparable efficiencies. It gave significantly higher yields in ortho,meta-dioxygenation of recalcitrant congeners containing a doubly ortho-chlorinated ring. While the parental enzyme yielded mainly unproductive meta, para dioxygenation of 2,5,4′-CB, the hybrid predominantly converted this congener into an ortho,meta-dioxygenated product. The subsequent enzymes of the LB400 catabolic pathway were able to transform most of the metabolites formed by the novel dioxygenase, indicating that the substrate ranges of these biocatalysts are not adapted to that of their initial pathway enzyme. Some of the catabolites, however, were identified as problematic for further degradation. Our results demonstrate that the outlined approach can successfully be applied to obtain novel dioxygenase specificities that favorably complement or supplement known ones.
Industrial mixtures of polychlorobiphenyls (PCBs) constitute an important class of persistent and potentially carcinogenic pollutants. Certain aerobic bacteria are able to oxidize some of the more lightly substituted PCB congeners through pathways that are basically identical in the different organisms (Fig. 1) (1, 8, 11, 13, 26). However, commercial PCB mixtures pose a huge problem to catabolic pathways, as they typically consist of dozens of different congeners. Even if broad in substrate range, no single pathway is able to metabolize all PCBs in such mixtures. Moreover, the characterized pathways convert a fraction of the PCBs into dead-end metabolites (18). Thus, enzymes with novel specificities that are useful to replace or supplement known ones are of particular interest.
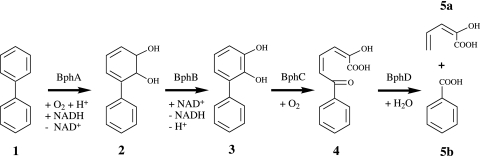
FIG. 1.
Upper pathway for catabolism of biphenyls encoded by the bph locus of B. xenovorans LB400. Compounds: 1, biphenyl; 2, biphenyl-2,3-dihydro-2,3-diol (BDHD); 3, 2,3-dihydroxybiphenyl (DHB); 4, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (HOPDA); 5a, 2-hydroxypenta-2,4-dienoic acid; 5b, benzoic acid. Enzymes: BphA, biphenyl 2,3-dioxygenase; BphB, 2,3-dihydro-2,3-dihydroxybiphenyl 2,3-dehydrogenase; BphC, 2,3-dihydroxybiphenyl 1,2-dioxygenase; BphD, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase.
The initial pathway enzyme, biphenyl dioxygenase (BphA), is of crucial importance for the successful breakdown of PCBs. First, as shown in previous publications (15, 42) as well as in the present work, its substrate range frequently is narrower than that of subsequent pathway enzymes. Second, its regiospecificity of dioxygenation is a critical parameter, as it determines the (potential) site(s) of attack by the subsequent enzymes of the metabolic route (Fig. 1). In this way, it controls whether and how further enzymatic degradation of a given congener may take place.
Two approaches to obtain “missing” enzymatic activities appear particularly promising. One is the generation of altered enzymes through protein engineering and strategies of artificial evolution (14, 42). The other is the detection and isolation of larger numbers of naturally occurring enzymatic activities. The development of high-throughput formats enables the screening of large clone libraries generated by these methods (16). Using the first approach, we recently were able to generate BphAs with improved dioxygenation of PCBs by segmental random mutagenesis (42). For the second approach, we developed a sequence-based strategy for the fast isolation and characterization of BphA and other class II aryl-hydroxylating dioxygenase activities (20). According to the latter method, the part of the alpha subunit gene encoding the catalytic center is amplified by PCR and is fused with sequences of a bphA gene cluster that is efficiently expressed in an appropriate host (Fig. 2). This method was initially tested using the DNA of cultivated microorganisms as a template for PCR amplification. However, it can likewise be applied as a metagenomic approach (C. Standfuβ-Gabisch, D. Al-Halbouni, and B. Hofer, unpublished data), thereby circumventing the cultivation of organisms (23, 31). Depletion assays with different aromatic compounds had indicated that the substrate spectra of the generated hybrid dioxygenases were largely determined by their BphA1 core segment. However, a characterization of their catabolic potential with respect to chlorobiphenyls (CBs) and its comparison to that of the parental enzyme had not been carried out. Now the properties of one of the resulting chimeric enzymes have been investigated with a selection of CBs as potential substrates. This hybrid (BphA-B4h) harbors the core segment of a dioxygenase from Pseudomonas sp. strain B4-Magdeburg, a bacterium isolated from a polluted sediment of the Elbe River near Magdeburg, Germany (7, 10). The other sequences were provided by the BphA of Burkholderia xenovorans (17) (formerly Burkholderia sp.) LB400 (BphA-LB400), a metabolically very well characterized dioxygenase (2, 18, 24, 33, 35, 36, 40, 41). The present investigations showed in detail how substrate and product ranges of the hybrid enzyme differed from those of its parental BphA. They revealed that, with several CBs, the newly generated dioxygenase showed complementing or improved degradative properties.
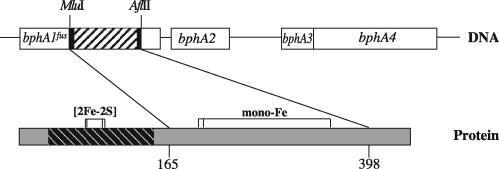
FIG. 2.
Generation of the fusion dioxygenase. The top line shows a representation of the bphA gene cluster of B. xenovorans LB400, encoding alpha and beta subunits (bphA1/bphA2), ferredoxin (bphA3), and ferredoxin reductase (bphA4). The hatched bphA1 segment between the restriction sites was exchanged with a PCR amplicon. The bottom line shows a representation of the alpha subunit. Catalytic and Rieske domains are shown in gray or are hatched, respectively. Horizontally connected vertical bars indicate sites encoding amino acid ligands of the Rieske iron-sulfur cluster ([2Fe-2S]) and of the active site mononuclear iron (mono-Fe). The part encoded by the amplicon and the flanking amino acid positions of the recipient subunit are indicated.
MATERIALS AND METHODS
Chemicals.
CB congeners (99% purity) were obtained from Lancaster Synthesis (White Lund, Morecombe, England), Promochem (Wesel, Germany), or Restek (Sulzbach, Germany). Chlorobenzoates (CBAs) (98% purity) were purchased from Fluka AG (Buchs, Switzerland) or Lancaster Synthesis.
Bacterial strains, plasmids, and culture conditions.
The Escherichia coli strain used in this study was BL21(DE3)(pLysS) (38) harboring either pAIA111, pAIA6100, pAIA1104, pAIA6104, or pAIA51. These plasmids are based on the phage T7 expression vector pT7-6. pAIA111 (27) carries bphA1A2A3A4 (collectively referred to as bphA), and pAIA6100 (20) harbors bphABC of B. xenovorans LB400. pAIA6104 contains a bphA1 gene that is a fusion of the bphA1 genes of B. xenovorans LB400 and Pseudomonas sp. strain B4-Magdeburg (20) and genes bphA2A3A4BC of B. xenovorans LB400. pAIA1104 was obtained from pAIA6104 by cleavage with PpuMI and recircularization. This deleted most of genes bphBC. pAIA51 (42) carries the bphD gene of strain LB400. Bacteria were grown in Luria-Bertani medium (30) at 37°C. Chloramphenicol and ampicillin at concentrations of 20 and 50 mg/ml, respectively, were used for selection.
Preparation of resting cells.
Preparation of resting cells was carried out as previously described (34) with some modifications. Cells of E. coli BL21(DE3)(pLysS) harboring the respective bph-containing plasmid were grown in Luria-Bertani medium at 30°C. At an optical density at 600 nm (OD600) of 0.6 to 1.0, 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added and the incubation was continued for another 30 to 60 min. Cells were harvested, washed with 1 volume of 50 mM sodium phosphate buffer (pH 7.5), and resuspended in the same buffer to give the concentrations specified below.
Biotransformations.
Substrates were dissolved in hexane or cyclohexane. They were dispensed into Teflon-sealed Erlenmeyer flasks. After evaporation of the solvent, reactions were started by addition of typically 10 ml of resting cells. Individual details of the different transformation experiments are given below.
Transformations of CBs by BphA-B4h or BphA-LB400 and analysis of products.
Resting cell suspensions (OD600 of 18) of E. coli BL21(DE3)(pLysS) harboring either pAIA111 or pAIA1104 were incubated with a CB congener at a nominal concentration of 1 mM on a gyratory shaker for 6 h at 30°C. Reaction mixtures were extracted with ethyl acetate, and the dried extracts were derivatized with n-butylboronic acid as described before (36). After derivatization, mixtures were evaporated to dryness and dissolved in 20 μl of hexane. Samples (1 μl) were injected in the splitless mode into a gas chromatography-mass spectrometry (GC-MS) system consisting of a Autosystem XL gas chromatograph (Perkin-Elmer, Boston, MA) with an MDN-1 column (Supelco, Bellefonte, PA) coupled to a Perkin-Elmer Turbo Mass mass spectrometer. Helium served as the carrier gas. The mass spectrometer was operated in the electron impact ionization mode at 70 eV.
Transformations of CBs by BphA-B4h and BphBC-LB400 or by BphABC-LB400 and analysis of products.
Resting cell suspensions (OD600 of 2.0) of E. coli BL21(DE3)(pLysS) harboring either pAIA6104 or pAIA6100 were supplemented with glucose to 0.5% and were incubated with a CB at a nominal concentration of 125 μM on a rotary shaker at 30°C. The formation of chlorinated 2-hydroxy-6-oxo-6-phenyl-hexa-2,4-dienoates (HOPDAs) was monitored at intervals up to 24 h by spectral scanning of the supernatants with a UV-2401 PC spectrophotometer (Shimadzu, Kyoto, Japan).
Transformations of CBs by BphA-B4h and BphBCD-LB400 or by BphABCD-LB400 and analysis of products.
Equal volumes of resting cell suspensions (OD600 of 2.0) of E. coli strains BL21(DE3)(pLysS)(pAIA6104) and BL21(DE3)(pLysS)(pAIA51) or BL21(DE3)(pLysS)(pAIA6100) and BL21(DE3)(pLysS)(pAIA51), respectively, were supplemented with glucose to 0.5% and were incubated as described above with a CB at a nominal concentration of 125 μM for 16 h. Cell-free supernatants were analyzed by high-performance liquid chromatography as described previously (34). CBAs were identified and quantitated by comparison with authentic standards.
RESULTS
It had been shown that Pseudomonas sp. strain B4-Magdeburg is able to grow with biphenyl as the sole source of carbon and energy (7, 10). However, its abilities to metabolize CBs had so far not been investigated. The construction of a hybrid bphA1 gene carrying a sequence from strain B4-Magdeburg in its core region (Fig. 2) has been described previously (20). The respective plasmid, pAIA6104, also harbored genes bphBC from strain LB400. In order to avoid further catabolism of the products formed through catalysis by the initial pathway dioxygenase, major parts of the latter genes were deleted, as described in Materials and Methods. This yielded pAIA1104. E. coli strains harboring either pAIA1104 (encoding BphA-B4h) or pAIA111 (encoding BphA-LB400) were incubated with a selection of CBs. As a first group, the three symmetrical di-CBs were chosen to compare the attack at monosubstituted rings chlorinated at all possible positions. The dioxygenation products were analyzed by GC-MS (Table 1).
TABLE 1.
Characterization of CB metabolites formed by dioxygenation catalyzed by BphA-B4h or BphA-LB400
| Substrate | No. theoretically possiblea
|
Meta- bolite no. | Retention time (min) by GC of derivative | Molecular mass (Da) by MS of derivative | No. of Cl | Type of compound | Apparent absolute yield (area units)b
|
Apparent relative yieldb (%)
|
Assignment of oxidized carbonsc | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BDHDs | DHBs | BphA-B4h | BphA-LB400 | BphA-B4h | BphA-LB400 | |||||||
| 2,2′-CB | 3 | 1 | 1 | 21.3 | 286 | 1 | DHB | NOd | 80.8 ± 8.5 | NO | 93 | 2,3 |
| 2 | 22.1 | 322 | 2 | BDHD | 25.6 ± 5.7 | NO | 100 | NO | 5,6 | |||
| 3 | 22.6 | 322 | 2 | BDHD | NO | 6.1 ± 1.0 | NO | 7 | 3,4 | |||
| 3,3′-CB | 2 | 2 | 1 | 22.5 | 286 | 1 | DHB | 0.34 ± 0.12 | NO | 4 | NO | 2,3 |
| 2 | 23.7 | 322 | 2 | BDHD | 9.5 ± 3.6 | 6.7 ± 0.3 | 96 | 68 | 5,6 | |||
| 3 | 24.1 | 322 | 2 | BDHD | NO | 3.1 ± 0.1 | NO | 32 | 4,5 | |||
| 4,4′-CB | 1 | 1 | 1 | 24.1 | 322 | 2 | BDHD | 20.0 ± 4.6 | 4.4 ± 1.6 | 100 | 100 | 2,3 |
| 2,6-CB | 3 | 1 | 1 | 22.4 | 322 | 2 | BDHD | 4.46 ± 0.47 | NO | 25 | NO | 2′,3′ |
| 2 | 22.6 | 322 | 2 | BDHD | NO | 0.51 ± 0.31 | NO | 23 | 3,4 or 3′,4′ | |||
| 3 | 23.0 | 322 | 2 | BDHD | 13.6 ± 4.3 | 1.71 ± 1.00 | 75 | 77 | 3,4 or 3′,4′ | |||
| 2,6,4′-CB | 2 | 2 | 1 | 23.4 | 320 | 2 | DHB | NO | 0.05 ± 0.03 | NO | 16 | 2,3 |
| 2 | 24.1 | 356 | 3 | BDHD | 11.6 ± 4.2 | NO | 100 | NO | 2′,3′ | |||
| 3 | 24.4 | 356 | 3 | BDHD | NO | 0.26 ± 0.14 | NO | 84 | 3,4 | |||
| 2,5,4′-CB | 2 | 4 | 1 | 24.4 | 356 | 3 | BDHD | 72.5 ± 38.5 | 14.4 ± 0.46 | 99 | 16 | 2′,3′ |
| 2 | 25.0 | 356 | 3 | BDHD | 0.97 ± 0.68 | 72.9 ± 16.3 | 1 | 84 | 3,4 | |||
Not taking into account dioxygenations involving C-1 or C-1′, respectively. These would not yield cyclic boronates and thus would not be detected in the analysis. To our knowledge, dioxygenations at these carbons have so far not been observed.
Deduced from total ion chromatogram peak areas.
For details, see text.
NO, not observed.
The ortho-monochlorinated ring.
The dioxygenation of 2,2′-CB by the two enzymes was targeted to completely different positions. The parental BphA-LB400 yielded two metabolites, a monochlorinated dihydroxybiphenyl (DHB) as a major product and a dichlorinated biphenyldihydrodiol (BDHD) as a minor product, in accordance with previous findings (18). It has been shown before that the DHB is hydroxylated at carbons 2 and 3 (18, 35). For the BDHD, hydroxylation at carbons 5 and 6 has been proposed (18), which, however, is in conflict with our results (see below). BphA-B4h formed a different BDHD as the sole product.
In order to monitor the further degradation of the dioxygenation products and to obtain additional evidence for the assignment of dioxygenation sites, congeners were incubated with cells also harboring the subsequent pathway enzymes, BphB, BphC, and BphD, of strain LB400 (Fig. 1).
The results on the formation of HOPDAs are shown in Table 2. It should be noted that BphC of strain LB400 does not cleave 3,4-DHB (12). Thus, the formation of ring cleavage products suggests dioxygenation at ortho- and meta-carbons.
TABLE 2.
Chlorinated HOPDAs formed from CBs by BphA-B4h or BphA-LB400 and BphBC-LB400
| Substrate | BphA | λmax (nm) of HOPDA |
|---|---|---|
| 2,2′-CB | B4h | 395 ± 2 |
| LB400 | 393 ± 2 | |
| 3,3′-CB | B4h | 420 ± 3a |
| LB400 | 425 ± 3a | |
| 4,4′-CB | B4h | 434 ± 2 |
| LB400 | 432 ± 2 | |
| 2,6-CB | B4h | 392 ± 2 |
| LB400 | NOb | |
| 2,6,4′-CB | B4h | NO |
| LB400 | 403 ± 5 | |
| 2,5,4′-CB | B4h | 397 ± 3 |
| LB400 | 413 ± 3c and | |
| 397 ± 3c |
Shifted to 395 nm during the incubation.
NO, not observed.
For details, see the text.
Both the wild-type and hybrid pathways converted 2,2′-CB into HOPDAs. This indicated that BphA-B4h also generated an ortho,meta-dioxygenated metabolite. Thus, the BDHD produced by BphA-B4h must be 5,6 dioxygenated. It follows that the BDHD formed by BphA-LB400 was meta,para dioxygenated. This is in agreement with the results of Barriault et al. (4), who assigned, on the basis of its nuclear magnetic resonance spectrum, hydroxylation at carbons 3 and 4 to a metabolite, produced by a BphA variant, that showed the same retention time as the BDHD formed by BphA-LB400.
The results of the conversion of congeners into CBAs in the presence of recombinant cells synthesizing the hydrolase BphD of B. xenovorans LB400 are shown in Table 3.
TABLE 3.
CBAs formed from CBs by BphA-B4h or BphA-LB400 and BphBCD-LB400
| Substrate | BphA | Result for CBA
|
|||
|---|---|---|---|---|---|
| Retention time (min) | λmax (nm) | Identification | Concn (μM) | ||
| 2,2′-CB | B4h | 3.40 ± 0.01 | 204 ± 2 | 2-CBA | 44.8 ± 15.4 |
| LB400 | 3.38 ± 0.01 | 204 ± 2 | 2-CBA | 121.0 ± 13.0 | |
| 3,3′-CB | B4h | 5.80 ± 0.05 | 231 ± 2 | 3-CBA | 25.4 ± 4.6 |
| LB400 | 5.75 ± 0.04 | 231 ± 2 | 3-CBA | 9.9 ± 2.7 | |
| 4,4′-CB | B4h | NOa | NO | NO | NO |
| LB400 | NO | NO | NO | NO | |
| 2,6-CB | B4h | 2.98 ± 0.05 | 206 ± 5 | 2,6-CBAb | 2.9 ± 1.4 |
| LB400 | NO | NO | NO | NO | |
| 2,6,4′-CB | Bh4 | NO | NO | NO | NO |
| LB400 | 5.58 ± 0.04 | 240 ± 2 | 4-CBA | 0.5 ± 0.3 | |
| 2,5,4′-CB | B4h | 5.84 ± 0.05 | 200 ± 5 | 2,5-CBAb | 0.5 ± 0.3 |
| LB400 | NO | NO | NO | NO | |
NO, not observed.
Identification based mainly on the retention time due to the uncharacteristic spectrum of the respective CBA.
Extradiol fission between carbons other than 1 and 2 or 1 and 6 (equivalent to 1′ and 2′ or 1′ and 6′, respectively) would not lead to benzoates. Thus, the formation of 2-chlorobenzoate (CBA) from the different dioxygenation products of 2,2′-CB generated by the two BphAs provided additional evidence for their attacks at ortho- and meta-carbons. It furthermore demonstrated that the subsequent pathway enzymes were able to convert metabolites that were not formed via their cognate dioxygenase.
The meta-monochlorinated ring.
The major site of attack of 3,3′-CB was identical for both enzymes and yielded a BDHD. Studies with BphA-LB400 had shown that this metabolite is dioxygenated at carbons 5 and 6 (36). This agrees with a decreasing effect of ortho substituents on the retention time (29), as shown in Table 1, and with the finding that both the BphA-LB400- and -B4h-initiated pathways converted 3,3′-CB into significant amounts of HOPDA and 3-CBA (Table 3). The observed instability of the extradiol fission product agrees with the slow spontaneous hydrolytic dehalogenation, followed by a tautomeric shift to the 2-oxo form, that has been described for HOPDA chlorinated at carbon 4 (32). BphA-B4h additionally only formed trace amounts of a second metabolite, identified as a monochlorinated DHB. Based on the high preference of the hybrid for ortho,meta dioxygenation, we tentatively assign oxidation at carbons 2 and 3. The LB400 enzyme catalyzed formation of a second BDHD to which we assign dioxygenation at positions 4 and 5, in agreement with its larger retention time and with previous results (18, 36).
The para-monochlorinated ring.
Both enzymes showed identical regiospecificity in the dioxygenation of 4,4′-CB. They formed the same BDHD as the only metabolite, which has previously been identified to be dioxygenated at carbons 2 and 3 (36). However, yields were approximately fivefold higher for the hybrid enzyme. A low level of HOPDA formation from 4,4′-CB, but no conversion of this HOPDA into 4-CBA, was detected, in agreement with previous results (34).
Three CBs that were not efficiently or not productively dioxygenated by BphA-LB400 were selected for further comparison of the two enzymes. “Not productively” is used in the sense that congeners were mainly converted into dead-end metabolites (18).
CBs possessing a doubly ortho-chlorinated ring.
With 2,6-CB, no DHB was detected as a metabolite, ruling out a dechlorinating attack at an ortho-carbon. Both enzymes yielded two BDHDs, of which the major one was common to both dioxygenases. However, yields with the hybrid enzyme were about eightfold higher.
Conversion of 2,6-CB into a HOPDA was not detected after dioxygenation by BphA-LB400. However, it was observed after hydroxylation by the hybrid enzyme. This suggests that the BDHD exclusively formed by BphA-B4h was dioxygenated at positions 2′ and 3′. Moreover, it is in keeping with the observation that this BDHD showed the smallest retention time of the three dioxygenation products. The formation of a small quantity of a metabolite with a UV spectrum and retention time consistent with those of 2,6-CBA agrees with 2′,3′ dioxygenation by BphA-B4h. The major product formed by BphA-B4h could be either 3,4 or 3′,4′ dioxygenated. We favor the latter possibility, because in this case the two attacks observed with this enzyme would not require two completely different orientations of the substrate at the active site. One of the best ways to position a substrate for catalysis is hydrogen bonding. Chloroaromatics, however, can at best act as very weak hydrogen bond acceptors. Thus, a certain oscillation between similar orientations of such a less tightly bound substrate is not unexpected. In the case of BphA-B4h, a position of the activated dioxygen close to C-3′ of 2,6-CB in conjunction with some motion around the axis perpendicular to the plane of the oxidized ring could lead to the involvement of either C-4′ or C-2′ as the second site of attack. The probably best characterized BphA in terms of regiospecificity with CBs is the enzyme from strain LB400. A survey of available data indicates that about 50% of the reported cases of relaxed regiospecificity could be explained by the described rationale.
When incubated with BphA-LB400, the structurally related 2,6,4′-CB yielded a trichlorinated BDHD and very small amounts of a dichlorinated DHB, resulting from a partly dechlorinating attack at carbons 2 and 3 (42). The former metabolite had not been detected after overnight incubations (42), presumably due to the known tendency of some DHDs to readily rearomatize by elimination of water. In contrast to the parental enzyme, the hybrid formed only a single metabolite, a BDHD that was different from the LB400 product. The yield of dioxygenation was approximately 40-fold higher than with BphA-LB400. As only two BDHDs are theoretically possible (Table 1), the retention times suggest that BphA-B4h formed the 2′,3′-dioxygenated isomer, whereas BphA-LB400 attacked carbons 3 and 4. However, the former metabolite was not converted into a HOPDA. In contrast, the 2,3-dioxygenated DHB formed by BphA-LB400 was further transformed into 4-CBA by the LB400 pathway (Table 3).
The para-monochlorinated ring in combination with the 2,5-dichlorinated ring.
There are several indications that both chlorination patterns are problematic for the catabolism through the LB400 pathway. As shown above, dioxygenation at the para-chlorinated ring is productive but gives only minor yields. Hydroxylation of the 2,5-disubstituted ring is usually efficient (2, 9, 18) but typically leads to dead-end metabolites.
The two BphAs behaved complementarily towards 2,5,4′-CB. They yielded the same two BDHDs as the only products, however, in roughly inverse amounts. As only two BDHDs are theoretically possible (Table 1), this indicates dioxygenation at positions 3 and 4 or 2′ and 3′, respectively. Their retention times suggest that the major metabolite of BphA-LB400 is dioxygenated at carbons 3 and 4. This is in accordance with the reported preference of this enzyme for the 2,5-dichlorinated ring.
Significantly higher yields of a HOPDA with a wavelength at the absorption maximum (λmax) of 397 nm were observed after dioxygenation of 2,5,4′-CB by BphA-B4h. Moreover, in the presence of BphD, a low concentration of a product with a UV spectrum and retention time consistent with those of 2,5-CBA was found. These results confirm the above assignment of 2′,3′ dioxygenation.
When HOPDA formation via dioxygenation by BphA-LB400 was monitored early in the reaction, an absorption maximum at 413 nm was initially observed. During the first 3 to 4 h, it was shifted to and then stably remained at 397 nm, the value of the HOPDA also generated via dioxygenation by BphA-B4h (Table 2). This suggests, in the case of BphA-LB400, the transient appearance of an unstable second extradiol fission product, derived from the 3,4-hydroxylated BDHD, which was the only other product formed by this enzyme. Low activities of the BphB of Comamonas testosteroni B-356 and of strain LB400 against the 3,4-DHD derived from 2,5,2′,5′-CB have been described previously (6). Moreover, extradiol cleavage of the resulting catechol by DoxG from Pseudomonas sp. strain C18 has been reported (5). Oxygenolytic ring fission on either side of a 2,5-dichlorinated 3,4-diol would yield a reactive acylchloride. Nucleophilic attack either by its own hydroxy group at carbon 2 (21) or by water (25) would lead to conversion into colorless metabolites, a lactone or a dimuconate, respectively, consistent with the observed disappearance of the long-wavelength absorption maximum.
Time course of HOPDA formation.
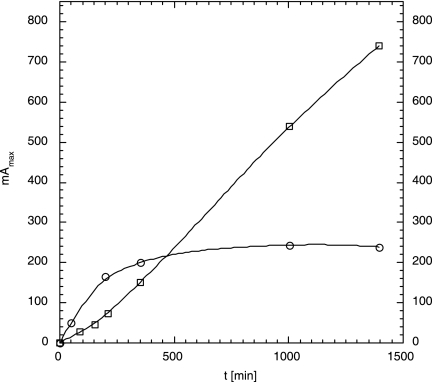
When the time course of HOPDA formation was monitored, generally two types of curves were obtained, depending on both the CB and the BphA involved. If 4,4′- or 2,5,4′-CB was initially dioxygenated by BphA-B4h, the HOPDA concentration showed an essentially linear increase up to the last sample taken (Fig. 3). In all other cases, the increases in the absorptions leveled off after 1 or a few hours. As an example, the degradation of 3,3′-CB via dioxygenation by BphA-B4h is shown in Fig. 3. These apparent decreases in the rate of HOPDA formation cannot be explained by substrate depletion. They are likely due to chemical instability of some ring cleavage products (32) and to enzyme inhibition along the pathway.
FIG. 3.
Time course of HOPDA formation. Absorbance (milliunits) at absorption maxima (mAmax) are shown. The substrates were 3,3′-CB (circles) and 2,5,4′-CB (squares). Both substrates were initially oxidized by BphA-B4h.
DISCUSSION
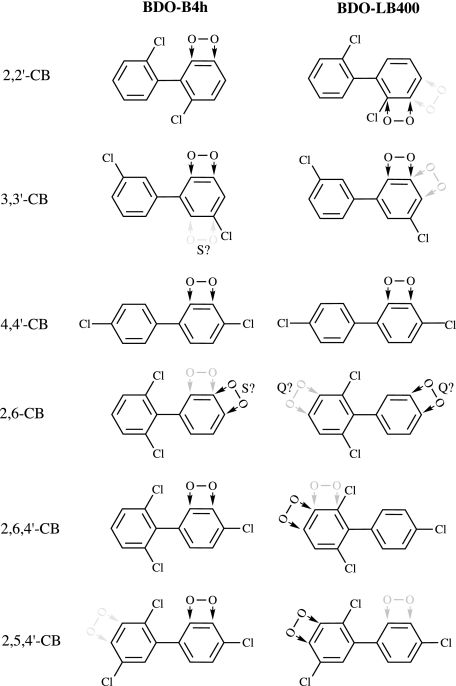
The dioxygenation of various CBs by the two enzymes showed that the substrate acceptance and regiospecificity of dioxygenation by the hybrid BphA investigated in this study differed fundamentally from those of the parental enzyme. This is illustrated in Fig. 4.
FIG. 4.
Overview of the regiospecificity of CB dioxygenation by BphA-B4h and -LB400. Regiospecificities of attack are symbolized by O2 molecules with arrows. Relative quantities are indicated as follows: black arrows, >33% of total dioxygenation; gray arrows, 10 to 33% of total dioxygenation; light gray arrows, <10% of total dioxygenation. For experimental evidence of site assignments and further details, see the text and Table 1. A lack of experimental evidence for the assignment of an oxidation site is indicated by “S?”; a lack of experimental evidence for the assignment of the relative quantity of oxidation is indicated by “Q?”
BphA-LB400 showed major differences in the amounts of productive ortho,meta dioxygenation of the ortho-, meta-, and para-chlorinated rings. Such behavior is typical for BphAs (3, 9, 19, 27). In contrast, BphA-B4h attacked all three rings with comparable efficiencies.
Congeners with a doubly ortho-chlorinated ring are known to generally be recalcitrant to attack by BphAs (9, 24, 39). Thus, the LB400 enzyme showed no ortho,meta dioxygenation of 2,6-CB, although this congener possesses an unchlorinated ring, which normally is easily attacked. Similarly, the enzyme yielded only small amounts of ortho,meta dioxygenation products of 2,6,4′-CB. In contrast, BphA-B4h was able to catalyze significant 2′,3′-dioxygenation of both congeners.
Available data suggest that typically the 2,5-dichlorinated ring is less recalcitrant to CB dioxygenation than the 2,6-dichlorinated ring and that for some BphAs, including the enzyme of strain LB400, the 2,5-dichlorinated ring even is a preferred target (2, 9, 18). Remarkably, the only sites of attack are carbons 3 and 4. Typically, 3,4-BDHDs are dead-end metabolites (12, 18), although a low turnover of the 3,4-DHD derived from 2,5,2′,5′-CB by BphB of Comamonas testosteroni B-356 and by strain LB400 has been reported (6), and our results suggest a low level of dehydrogenation and subsequent extradiol cleavage of the 3,4-DHD formed from 2,5,4′-CB. Moreover, transformation of chlorinated meta,para-BDHDs by BphB and BphC may form acylchlorides, which may inactivate enzymes by attack of side chains in the active site. In contrast, ring cleavage of a chlorinated ortho,meta-DHB between carbons 1 and 2 or 1 and 6, respectively, can never yield an acylchloride. Contrary to the LB400 enzyme, BphA-B4h converted 2,5,4′-CB into only marginal amounts of the 3,4-dioxygenated metabolite but into almost 100-fold-larger amounts of the BDHD resulting from the more favorable ortho,meta dioxygenation of the other ring.
In summary, BphA-B4h showed a higher regiospecificity of dioxygenation. In only one case did the main product represent less than 95% of the dioxygenated CB (Table 1). BphA-B4h also possessed a greater preference for ortho,meta dioxygenation. Thus, BphA-LB400 formed meta,para-dioxygenated products from five of the six substrates (Fig. 4), with an average contribution of 61% (Table 1), while BphA-B4h yielded such metabolites only from three CBs, with a major contribution only in the case of 2,6-CB (Table 1).
Interestingly, the subsequent enzymes of the bph-encoded metabolic pathway of strain LB400 were able to transform most of those metabolites produced by the hybrid BphA, with which they normally are not confronted. Thus, 2′,3′-dioxygenated 2,6-CB was converted at least into the respective HOPDA, and 5,6-dioxygenated 2,2′-CB was transformed to 2-CBA. These results show that subsequent metabolic enzymes can possess a broader substrate spectrum than the initial pathway enzyme. This is particularly remarkable for the hydrolase BphD, which has been reported to constitute a major bottleneck (14, 15, 32).
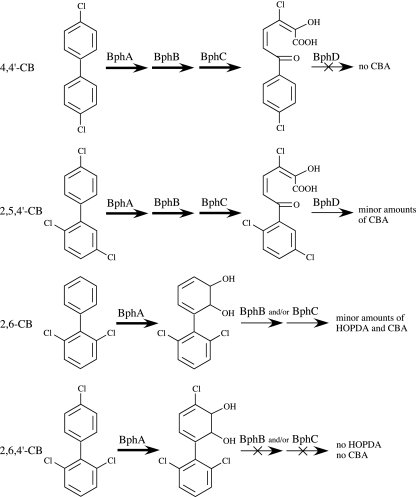
On the other hand, it is directly apparent from a correlation of Tables 1 and 3 that the products of 2,3 dioxygenation of 4,4′-CB and of 2′,3′-dioxygenation of 2,5,4′-, 2,6-, and 2,6,4′-CB were not or only marginally converted into the respective CBAs by the subsequent pathway enzymes. When 4,4′- and 2,5,4′-CB were dioxygenated by BphA-B4h, significant amounts of the meta-fission products were found. This indicates that these HOPDAs were not or were very slowly converted by the hydrolase BphD (Fig. 5). Only small or no detectable quantities of HOPDAs were formed from the BDHDs generated by 2′,3′ dioxygenation of 2,6- and 2,6,4′-CB. This indicates a problem with the dehydrogenase BphB and/or the extradiol dioxygenase BphC (Fig. 5). A comparison of the structures of the respective ortho,meta-dioxygenated BDHDs suggests that it is the double ortho substitution of the nonoxidized ring which prevents efficient turnover by one or both of these enzymes.
FIG. 5.
Bottlenecks in the metabolism of specific ortho,meta-dioxygenated chlorinated BDHDs by enzymes BphB, BphC, and BphD of the pathway from B. xenovorans LB400. Thin arrows symbolize low transformation; crossed-out thin arrows symbolize no detectable transformation.
We note that Seah et al. (32) found that HOPDA chlorinated at position 3 was hydrolyzed by BphD-LB400 at a 500-fold lower rate than the unchlorinated metabolite. This is consistent with no or only marginal conversions of the 3,10-dichlorinated or 3,8,11-trichlorinated HOPDA, resulting from 4,4′- or 2,5,4′-CB, respectively, into 4-CBA (Fig. 5). The same authors also reported that HOPDA chlorinated only at position 4 was hydrolytically cleaved at a 10,000-fold-lower rate than the unsubstituted compound (32). Thus, the observed conversion of 3,3′-CB into a 4,9-dichlorinated HOPDA that was further metabolized to 3-CBA suggests that, unexpectedly, the additional chlorine substituent at the nonoxidized ring significantly enhances this rate. A positive electronic effect appears unlikely, as recently, Speare et al. (37) have shown that electron-withdrawing substituents at the nonoxidized ring decrease the rate of HOPDA hydrolysis by BphD-LB400.
The exchange of the BphA1 core segment resulted in 24 amino acid differences between the LB400 and the hybrid sequence (see the supplemental material). Therefore, alterations in catalytic behavior cannot directly be ascribed to single or a few amino acid substitutions. However, several of the replaced residues have previously been exchanged, either singly or in groups of two and three (22, 28, 41, 42). In most cases, this did not result in significant changes of the examined properties. An exception was the region comprising amino acids 335 to 341 (LB400 numbering), where four of the present substitutions are located. However, more than these exchanges are likely to play a role, because seemingly “unimportant” residues may exert significant effects when replaced in concert with additional amino acids. Such a context dependence of substitutions has repeatedly been reported (22, 28).
The fundamental differences in substrate and product spectra between the parent and hybrid enzymes demonstrate that BphAs with novel catabolic potential can be obtained through a rapid approach involving PCR amplification of partial genes encoding the large subunit of aryl-hydroxylating dioxygenases and their fusion with cloned “helper” genes and gene segments to reconstitute a complete aryl-hydroxylating dioxygenase system. The speed of this approach and the possibility of carrying out PCR amplifications directly with metagenomic DNA samples may permit rapid access to a much larger part of natural BphA diversity than was previously possible. Such an approach could thus favorably complement or supplement methods of artificial evolution (14) for the acquisition of novel dioxygenase activities.
Acknowledgments
Support of this work by grants from the Bundesministerium für Bildung und Forschung (WTZ CHL 99/029), CONICYT, FONDECYT (1020221-7020221), USM (130122), and MILENIO P04/007-F (MIDEPLAN) is gratefully acknowledged.
We also thank the reviewers for helpful comments.
Footnotes
Published ahead of print on 23 February 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahmed, M., and D. D. Focht. 1973. Degradation of polychlorinated biphenyls by two species of Achromobacter. Can. J. Microbiol. 19:47-52. [DOI] [PubMed] [Google Scholar]
- 2.Arnett, C. M., J. V. Parales, and J. D. Haddock. 2000. Influence of chlorine substituents on rates of oxidation of chlorinated biphenyls by the biphenyl dioxygenase of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 66:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barriault, D., C. Simard, H. Chatel, and M. Sylvestre. 2001. Characterization of hybrid biphenyl dioxygenases obtained by recombining Burkholderia sp. strain LB400 bphA with the homologous gene of Comamonas testosteroni B-356. Can. J. Microbiol. 47:1025-1032. [PubMed] [Google Scholar]
- 4.Barriault, D., F. Lepine, M. Mohammadi, S. Milot, N. Leberre, and M. Sylvestre. 2004. Revisiting the regiospecificity of Burkholderia xenovorans LB400 biphenyl dioxygenase toward 2,2′-dichlorobiphenyl and 2,3,2′,3′-tetrachlorobiphenyl. J. Biol. Chem. 279:47489-47496. [DOI] [PubMed] [Google Scholar]
- 5.Barriault, D., J. Durand, H. Maaroufi, L. D. Eltis, and M. Sylvestre. 1998. Degradation of polychlorinated biphenyl metabolites by naphthalene-catabolizing enzymes. Appl. Environ. Microbiol. 64:4637-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barriault, D., M. Vedadi, J. Powlowski, and M. Sylvestre. 1999. cis-2,3-Dihydro-2,3-dihydroxybiphenyl dehydrogenase and cis-1, 2-dihydro-1,2-dihydroxynaphathalene dehydrogenase catalyze dehydrogenation of the same range of substrates. Biochem. Biophys. Res. Commun. 260:181-187. [DOI] [PubMed] [Google Scholar]
- 7.Bartels, F., S. Backhaus, E. R. B. Moore, K. N. Timmis, and B. Hofer. 1999. Occurrence and expression of glutathione S-transferase-encoding bphK genes in Burkholderia sp. strain LB400 and other biphenyl-utilizing bacteria. Microbiology 145:2821-2834. [DOI] [PubMed] [Google Scholar]
- 8.Bedard, D. L., and M. L. Haberl. 1990. Influence of chlorine substitution pattern on the degradation of polychlorinated biphenyls by eight bacterial strains. Microb. Ecol. 20:87-102. [DOI] [PubMed] [Google Scholar]
- 9.Bedard, D. L., R. Unterman, L. H. Bopp, M. J. Brennan, M. L. Haberl, and C. Johnson. 1986. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol. 51:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blumenroth, P. 1997. Konstruktion gentechnisch modifizierter Mikroorganismen für den in situ-Abbau polychlorierter Biphenyle (PCBs) in Sediment. Ph.D. thesis. Technical University of Braunschweig, Braunschweig, Germany.
- 11.Catelani, D., A. Colombi, C. Sorlini, and V. Treccani. 1973. 2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoate: the meta-cleavage product from 2,3-dihydroxybiphenyl by Pseudomonas putida. Biochem. J. 134:1063-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eltis, L. D., B. Hofmann, H.-J. Hecht, H. Lünsdorf, and K. N. Timmis. 1993. Purification and crystallization of 2,3-dihydroxybiphenyl 1,2-dioxygenase. J. Biol. Chem. 268:2727-2732. [PubMed] [Google Scholar]
- 13.Furukawa, K., F. Matsumura, and K. Tonomura. 1978. Alcaligenes and Acinetobacter capable of degrading polychlorinated biphenyls. Agric. Biol. Chem. 42:543-548. [Google Scholar]
- 14.Furukawa, K., H. Suenaga, and M. Goto. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J. Bacteriol. 186:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa, K., J. Hirose, A. Suyama, T. Zaiki, and S. J. Hayashida. 1993. Gene components responsible for discrete substrate specificity in the metabolism of biphenyl (bph operon) and toluene (tod operon). J. Bacteriol. 175:5224-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goddard, J. P., and J. L. Reymond. 2004. Enzyme assays for high-throughput screening. Curr. Opin. Biotechnol. 15:314-322. [DOI] [PubMed] [Google Scholar]
- 17.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400(T) and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 18.Haddock, J. D., J. R. Horton, and D. T. Gibson. 1995. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imbeault, N. Y., J. B. Powlowski, C. L. Colbert, J. T. Bolin, and L. D. Eltis. 2000. Steady-state kinetic characterization and crystallization of a polychlorinated biphenyl-transforming dioxygenase. J. Biol. Chem. 275:12430-12437. [DOI] [PubMed] [Google Scholar]
- 20.Kahl, S., and B. Hofer. 2003. A genetic system for the rapid isolation of aromatic-ring-hydroxylating dioxygenase activities. Microbiology 149:1475-1481. [DOI] [PubMed] [Google Scholar]
- 21.Kersten, P. J., P. J. Chapman, and S. Dagley. 1985. Enzymatic release of halogens or methanol from some substituted protocatechuic acids. J. Bacteriol. 162:693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura, N., A. Nishi, M. Goto, and K. Furukawa. 1997. Functional analyses of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J. Bacteriol. 179:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz, P., K. Liebeton, F. Niehaus, and J. Eck. 2002. Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr. Opin. Biotechnol. 13:572-577. [DOI] [PubMed] [Google Scholar]
- 24.Maltseva, O. V., T. V. Tsoi, J. F. Quensen III, M. Fukuda, and J. M. Tiedje. 1999. Degradation of anaerobic reductive dechlorination products of Aroclor 1242 by four aerobic bacteria. Biodegradation 10:363-371. [DOI] [PubMed] [Google Scholar]
- 25.Mars, A. E., T. Kasberg, S. R. Kaschabek, M. H. van Agteren, D. B. Janssen, and W. Reineke. 1997. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J. Bacteriol. 179:4530-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massé, R., F. Messier, L. Péloquin, C. Ayotte, and M. Sylvestre. 1984. Microbial biodegradation of 4-chlorobiphenyl, a model compound of chlorinated biphenyls. Appl. Environ. Microbiol. 47:947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKay, D. B., M. Seeger, M. Zielinski, B. Hofer, and K. N. Timmis. 1997. Heterologous expression of biphenyl dioxygenase-encoding genes from a gram-positive broad-spectrum polychlorinated biphenyl degrader and characterization of chlorobiphenyl oxidation by the gene products. J. Bacteriol. 179:1924-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plomley, J. B., M. Lausevic, and R. E. March. 2000. Determination of dioxins/furans and PCBs by quadrupole ion-trap gas chromatography-mass spectrometry. Mass Spectrom. Rev. 19:305-365. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Schloss, P. D., and J. Handelsman. 2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303-310. [DOI] [PubMed] [Google Scholar]
- 32.Seah, S. Y., G. Labbe, S. Nerdinger, M. R. Johnson, V. Snieckus, and L. D. Eltis. 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 275:15701-15708. [DOI] [PubMed] [Google Scholar]
- 33.Seeger, M., B. Cámara, and B. Hofer. 2001. Dehalogenation, denitration, dehydroxylation, and angular attack of substituted biphenyls and related compounds by a biphenyl dioxygenase. J. Bacteriol. 183:3548-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeger, M., K. N. Timmis, and B. Hofer. 1995. Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorobiphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl. Environ. Microbiol. 61:2654-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeger, M., K. N. Timmis, and B. Hofer. 1995. Degradation of chlorobiphenyls catalyzed by the bph-encoded biphenyl-2,3-dioxygenase and biphenyl-2,3-dihydrodiol-2,3-dehydrogenase of Pseudomonas sp. LB400. FEMS Microbiol. Lett. 133:259-264. [DOI] [PubMed] [Google Scholar]
- 36.Seeger, M., M. Zielinski, K. N. Timmis, and B. Hofer. 1999. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 65:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speare, D. M., S. M. Fleming, M. N. Beckett, J. J. Li, and T. D. Bugg. 2004. Synthetic 6-aryl-2-hydroxy-6-ketohexa-2,4-dienoic acid substrates for C-C hydrolase BphD: investigation of a general base catalytic mechanism. Org. Biomol. Chem. 2:2942-2950. [DOI] [PubMed] [Google Scholar]
- 38.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 39.Williams, W. A., J. H. Lobos, and W. E. Cheetham. 1997. A phylogenetic analysis of aerobic polychlorinated biphenyl-degrading bacteria. Int. J. Syst. Bacteriol. 47:207-210. [DOI] [PubMed] [Google Scholar]
- 40.Zielinski, M., S. Backhaus, and B. Hofer. 2002. The principal determinants for the structure of the substrate-binding pocket are located within a central core of a biphenyl dioxygenase alpha subunit. Microbiology 148:2439-2448. [DOI] [PubMed] [Google Scholar]
- 41.Zielinski, M., S. Kahl, H.-J. Hecht, and B. Hofer. 2003. Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction. J. Bacteriol. 185:6976-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zielinski, M., S. Kahl, C. Standfuβ-Gabisch, B. Cámara, M. Seeger, and B. Hofer. 2006. Generation of novel-substrate-accepting biphenyl dioxygenases through segmental random mutagenesis and identification of residues involved in enzyme specificity. Appl. Environ. Microbiol. 72:2191-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]