Abstract
A new primary model based on a thermodynamically consistent first-order kinetic approach was constructed to describe non-log-linear inactivation kinetics of pressure-treated bacteria. The model assumes a first-order process in which the specific inactivation rate changes inversely with the square root of time. The model gave reasonable fits to experimental data over six to seven orders of magnitude. It was also tested on 138 published data sets and provided good fits in about 70% of cases in which the shape of the curve followed the typical convex upward form. In the remainder of published examples, curves contained additional shoulder regions or extended tail regions. Curves with shoulders could be accommodated by including an additional time delay parameter and curves with tails shoulders could be accommodated by omitting points in the tail beyond the point at which survival levels remained more or less constant. The model parameters varied regularly with pressure, which may reflect a genuine mechanistic basis for the model. This property also allowed the calculation of (a) parameters analogous to the decimal reduction time D and z, the temperature increase needed to change the D value by a factor of 10, in thermal processing, and hence the processing conditions needed to attain a desired level of inactivation; and (b) the apparent thermodynamic volumes of activation associated with the lethal events. The hypothesis that inactivation rates changed as a function of the square root of time would be consistent with a diffusion-limited process.
When microbes are exposed to lethal agents such as heat, gamma radiation, or chemical disinfectants, the concentration of surviving organisms decreases more or less exponentially with time. This behavior has been interpreted in two different ways. The first views the inactivation process as being similar to a first-order reaction such that lethal events occur at random over time within a population of cells that are similar in their susceptibility to the agent (6). The second interpretation, sometimes referred to as the “vitalist” approach, supposes that the observed differences in survival time are the result of differences in resistance among individual cells (41).
The simple first-order model has been applied very successfully for many years in the food industry to define safe thermal processes for canned foods (38). For cells heated under isothermal conditions, a plot of the logarithm of the surviving fraction against time yields a straight line, and inactivation rates are expressed in terms of the decimal reduction time, or D value, which is the reciprocal of the specific inactivation rate at a particular temperature. The relationship between D value and temperature is given by the z value, which is the temperature increase needed to change the D value by a factor of 10. These two parameters are also used to describe microbial inactivation by UV or ionizing radiation. However, there are many exceptions to the simple first-order type kinetics, especially when cells are exposed to relatively mild inactivation treatments, and many investigators regard the classic log-linear inactivation curve as an exception rather than the rule (4, 26, 27).
High hydrostatic pressure is regarded as one of the more promising of the emerging technologies that are being explored as means of providing foods that are safe but retain fresh attributes (31). Commercialization of high-pressure technology would be helped if mathematical models were available that could predict microbial inactivation at different pressures and allow process criteria and resistance parameters to be defined. Although simple first-order-type inactivation curves do occur with pressure-treated cells (10, 22, 23, 28, 29), the inactivation curves often deviate from the classical log-linear pattern, even more so than for heat (11, 37). Even where a reasonable fit is obtained using a log linear plot, better results have been obtained if curvature is accommodated (7). Various shapes of pressure inactivation curve have been reported when the logarithm of the surviving fraction is plotted against time, including curves with shoulders, curves with tails, or sigmoid curves. The type of inactivation kinetics may vary depending on the conditions during pressure treatment. For example, van Opstal et al. (40) observed simple first-order inactivation curves of Escherichia coli MG1655 in carrot juice but biphasic curves in buffer. Despite this variation, a very common and characteristic feature of many reported curves is that the specific inactivation rate appears to decrease markedly with time leading to pronounced curvature (upward concavity) on a log-linear plot (20, 24, 35). It is therefore difficult in many cases to fit curves to the primary data and even more difficult to derive secondary models that describe how inactivation rates vary with pressure (11).
Ideally the mathematical form of a model should reflect the basic underlying physical mechanism of the inactivation process, at least in general terms. In the case of UV irradiation, the observed first-order kinetics are consistent with the physical nature of the process. When a uniform suspension of cells is irradiated, quanta of radiant energy interact with cells in a random stochastic manner which, from first principles, means that lethal “hits” are distributed among cells in a Poissonian fashion. If all cells in a population were equally susceptible and death resulted from a single hit, simple first-order inactivation kinetics would result. The process of thermal inactivation may be interpreted as having a similar statistical basis. When microbial cells are exposed to moist heat, they do not all receive the same dose of energy per unit time because, at the microscopic level, the kinetic energy (speed of molecules) especially of water molecules is distributed according to the Maxwell-Boltzmann distribution. For an inactivation event to occur, the interacting molecules need a minimum amount of energy, the activation energy (Eact). The proportion of molecules that statistically have kinetic energy above a certain critical level increases with temperature. According to this model, cells would receive a lethal hit at random according to whether they encountered a water molecule of sufficient kinetic energy to cause denaturation of a critical target event. McKee and Gould (18) presented a mathematical analysis showing that the conventional thermal resistance parameters D and z were consistent with these underlying assumptions. Thermal inactivation, thus interpreted, has its basis in a random statistical distribution of lethal hits within the population.
Curves with shoulders are usually explained on the basis that is more than one critical target per cell, and/or the critical targets may require more than one “hit” before being inactivated. Miles (21) recently presented a modified critical target theory which relates loss of viability to the number of critical components per cell, the number of such components required for viability, and the probability of a critical component sustaining critical damage. Tails on survival curves are usually attributed to differences in resistance among cells of a population or adaptation of cells to the imposed stress treatment during exposure. Although these model assumptions allow shoulders and tails on survival curves to be accommodated, the underlying mechanism can still be interpreted as a stochastic process.
Pressure inactivation can also be viewed as a stochastic process since, as with temperature and other macroscopic properties, the effect is a result of averaging over a great number of molecules in constant movement at different speeds (8). The number of molecules present in a given volume at any time fluctuates and, consequently, so does the instantaneous pressure. Based on the thermodynamic principles of Eyring's transition state theory, the effect of pressure on the rate of the reaction will depend upon the volume of activation (ΔVact), thus playing a similar role in pressure-driven systems as Eact in thermal processes. The rate of reaction will be controlled by the rate of formation of the activated complex and is a function of the Gibbs free energy change in going from the normal to the activated state. Rodriguez et al. (32) proposed in their work a model based on classic thermodynamic and kinetic principles for the inactivation of bacterial spores by heat and high pressure.
The vitalistic approach assumes that the shape of a survival curve largely reflects the way differences in resistance are distributed among individual cells within a population. Several distribution functions have been applied to inactivation curves including normal, log normal, log logistic, and Weibull (26). While such distributions provide excellent fits to experimental data, the parameters of the primary models do not always vary regularly with intensity of the applied stress, and in some cases it can be difficult to derive simple secondary models (12, 39, 40, 42).
The aim of this work was to develop a model for the inactivation of cells by high hydrostatic pressure that was based on an underlying first-order process but could still accommodate curvature on log linear survival curves. The model that was developed assumes that high-pressure inactivation kinetics are fundamentally first order but that the specific rate varies with time. It emerged that the experimental data were consistent with the rate varying inversely with the square root of time.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
E. coli NCTC 8164 was stored at −70°C in bead vials (Protect Technical Service Consultants Ltd., Lancashire, United Kingdom). This strain was used in previous studies of the mechanisms of inactivation by pressure (3). To activate the strain, one frozen bead was transferred to 9 ml of tryptic soy broth (Oxoid, Basingstoke, United Kingdom) and incubated in shaken culture (140 rpm) at 37°C for approximately 6 h. The culture was then diluted 1:1,000 into 100 ml of fresh tryptone soy broth and incubated in shaken flasks (250 ml) at 37°C for approximately 18 h. The resulting stationary phase culture contained approximately 3 × 109 cells/ml.
Pressure treatment.
Samples of stationary phase E. coli NCTC 8164 were centrifuged at 2,800 × g for 15 min at 5°C, resuspended in an equal amount of phosphate buffered saline (PBS; pH 7.2) and dispensed in volumes of 2 ml in plastic sachets, heat sealed, and placed on ice before treatment. Samples were treated in a 300-ml pressure vessel (Foodlab Plunger Press model S-FL-850-9W; Stansted Fluid Power, Stansted, Essex, United Kingdom). The pressure-transmitting fluid was ethanol:castor oil (80:20). The come-up rate was approximately 330 MPa/min, and the deviation at targeted pressure was ±10 MPa. After treatment, the pressure was released quickly in two steps. In the first step the pressure decreased to 30 MPa in about 15 seconds. The total decompression took about 35 s. The transient increase in temperature of the pressurization fluid due to adiabatic heat during the treatment was measured with a thermocouple located near the vessel closures attached to the inside of the vessel lid. Experiments were carried out at an initial temperature of ca. 20°C. The average temperature rise during compression was 4.3 (±0.4)°C/100 MPa, giving a total increase of 21.5°C at 500 MPa.
Viable counts.
Sample bags were opened with sterile scissors, and cell suspensions were diluted 10-fold in maximum recovery diluent (Oxoid, Basingstoke, United Kingdom). Appropriate dilutions were plated on tryptone soy agar plus 0.1% sodium pyruvate as recovery medium, and colonies were counted after incubation at 37°C for 24 and 48 h. Two to four counts at relevant dilutions were performed for each sample. The mean was calculated and expressed as the number of CFU/ml (CFU per ml of sample). The lower limit of accurate measurements was 25 CFU/ml. Because inactivation experiments extended over 5 h at low pressures, it was necessary to construct composite survival curves from several independent time course experiments in order to avoid prolonged exposure of samples to chilling conditions. Each inactivation curve was based on data from between 5 and 15 separate experiments.
Curve fitting.
In modeling terminology a primary model is a mathematical expression that describes changes in microbial numbers with time, while a secondary model describes how the parameters of the primary model change with changes in environmental factors, in this case pressure. The primary model was fitted to survival data, and parameters were estimated by nonlinear least-squares regression using the SPSS statistical package (release 11.5; SSPS Inc., Chicago, IL). The linear secondary models were fitted to parameter data using Microsoft Excel (from Microsoft Office XP).
Goodness of fit and model performance.
Goodness of fit and model performance were assessed by mean square error (MSE), residual analysis, coefficient of determination (R2), analysis of the parameter estimates, and accuracy (Af) and bias (Bf) factors (see references 19 and 33).
Testing the new primary model against external data.
To test the model against independent data, curves were fitted to 138 sets of high-pressure inactivation data taken from the literature. Fitting was done by a nonlinear least squares procedure using the SPSS statistical package. Raw data were obtained by measuring the coordinates of data points from published graphs using a ruler. The error in reading data from graphs was estimated using a set of 40 data points from graphs with known values. A correlation coefficient, r, of 0.9996 and a nonsignificant chi-squared test showed high accuracy of data values read from graphs.
RESULTS
Development of a new primary model.
Under batch conditions, the assumption of a death process following first-order kinetics with a time-varying parameter kd(t) gives
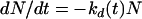 |
(1) |
where N is the viable cell concentration and t is time. In the “classical” first-order process, kd(t) is a constant.
Preliminary observations on our own data showed that the logarithm of the surviving fraction appeared to depend on t0.5. We therefore set kd(t) at any time t to be the following: kd(t) = kt−0.5, where k is a constant. Replacing kd(t) in equation 1 gives the following:
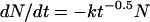 |
(2) |
Integration of equation 2 with N = N0 at t = 0 gives
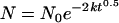 |
(3) |
or
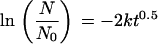 |
(4) |
or
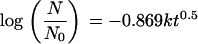 |
(5) |
Testing the new primary model against our own experimental data.
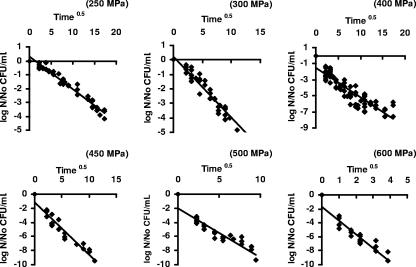
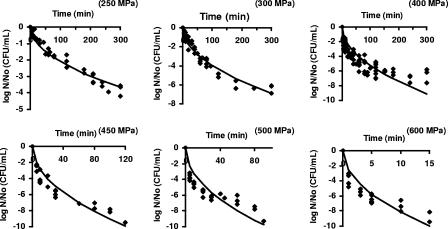
Figure 1 shows a good correlation between the logarithm of the surviving fraction and t, as suggested by equation 5 (data for 700 MPa not shown). The k values for each pressure were calculated using equation 5, via nonlinear regression, with R2 values between 0.81 and 0.96. The fitted curves for the new primary model at different pressures are shown in Fig. 2; k values and the results of the analysis of regression and performance are given in Table 1. The primary model had only slight difficulties at the pressures 400, 500, and 600 MPa, as reflected in the higher MSE and Af and lower R2 values at these three pressures. At 400 MPa the model overestimated inactivation in the tail region. At 500 and 600 MPa, the model systematically underestimated the curvatures slightly and overestimated the tails of the survivor curves. At all other pressures, the model was a good fit.
FIG. 1.
Correlation between log10 of the surviving fraction and the square root of time for pressure-treated E. coli cells.
FIG. 2.
The linear primary model fitted to inactivation curves of pressure-treated cells of E. coli.
TABLE 1.
Goodness of fit and performance of the new primary model
| Parametera | Value at indicated pressure (MPa)
|
Avg | ||||||
|---|---|---|---|---|---|---|---|---|
| 250 | 300 | 400 | 450 | 500 | 600 | 700 | ||
| k(min−0.5) | 0.242 | 0.458 | 0.605 | 1.044 | 1.187 | 2.984 | 5.084 | |
| 95% CI | 0.231-0.252 | 0.438-0.477 | 0.571-0.639 | 0.980-1.109 | 1.089-1.285 | 2.772-3.195 | 4.760-5.409 | |
| MSE | 0.080 | 0.212 | 1.250 | 0.736 | 1.489 | 1.119 | 0.592 | 0.783 |
| R2 | 0.95 | 0.94 | 0.61 | 0.88 | 0.61 | 0.77 | 0.90 | 0.81 |
| Af | 1.24 | 1.22 | 1.33 | 1.17 | 1.30 | 1.18 | 1.12 | 1.22 |
| Bf | 1.17 | 1.12 | 0.81 | 0.90 | 0.83 | 0.90 | 1.00 | 0.96 |
See Materials and Methods. CI, confidence interval.
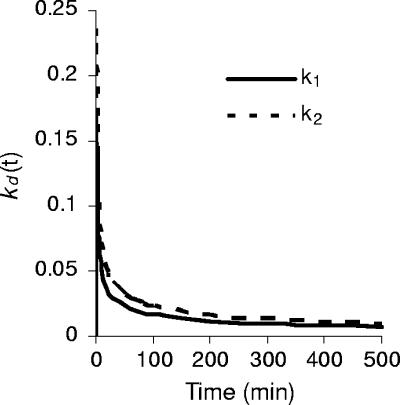
The parameter kd(t) decreases with time. This departure from the simple, constant-parameter linear model implies that the decimal reduction time D is not constant and that the total time to achieve a given number of decimal reductions is no longer proportional to the number of reductions. The characteristic shape of the kd(t) versus time curve is shown in Fig. 3 using two arbitrary k values as examples. Thus, in the graph kd(t) = kt−0.5 is plotted using k = k1 = 0.163 min−0.5 and k = k2 = 0.23 min−0.5, respectively. This example shows that initially the parameter falls rapidly and then more slowly with time. From equation 5, the time, measured from the initial concentration N0, for the first decimal reduction, D1, is 1.324/k2. From equation 5 the total time elapsed for subsequent successive decimal reductions (i.e., to 0.01N0, 0.001N0, etc.) is 4D1, 9D1, 16D1, etc. The total time needed for n decimal reductions from N0 is n2D1, compared with nD for the simple (kd = constant) linear model. The decimal reduction times for each successive 10-fold reduction (i.e., from N0 to 0.1N0, 0.1N0 to 0.01N0, etc.) are D1, 3D1, 5D1, 7D1,… (2n − 1)D1, respectively. D1 thus plays the role of a characteristic decimal reduction time. Note that the corresponding reduction times for two systems with different characteristic reduction times, D11 and D12, say, are always in a fixed ratio, that is, Dn1/Dn2 = D11/D12. The total times to achieve a given reduction are in the same ratio. Table 2 shows the calculated times to achieve specified numbers of decimal reductions at the pressures tested here. The time to achieve one decimal reduction, i.e., D1, has been calculated from D1 = 1.342/k2, using the values of k from Table 1. Subsequent times are calculated using the fact that the time for n decimal reductions is n2D1.
FIG. 3.
Variation of the first-order rate constant kd(t) with time. The values of k used in this illustration were k1 = 0.163 min−0.5 (solid line) and k2 = 0.230 min−0.5 (dashed line).
TABLE 2.
The calculated time to achieve the specified number of decimal reductionsa
| No. of decimal reductions | Time (min) required at indicated pressure (MPa)
|
||||||
|---|---|---|---|---|---|---|---|
| 250 | 300 | 400 | 450 | 500 | 600 | 700 | |
| 1 | 22.9 | 6.4 | 3.7 | 1.2 | 0.95 | 0.15 | 0.052 |
| 2 | 91.7 | 25.6 | 14.7 | 4.9 | 3.8 | 0.60 | 0.21 |
| 3 | 206.3 | 57.6 | 33.0 | 11.1 | 8.6 | 1.4 | 0.47 |
| 4 | 366.6 | 102.4 | 58.7 | 19.7 | 15.2 | 2.4 | 0.83 |
| 5 | 572.9 | 160.0 | 91.7 | 30.8 | 23.8 | 3.8 | 1.3 |
| 5b | 114.6 | 32 | 18.3 | 6.2 | 4.8 | 0.75 | 0.26 |
The time to achieve one decimal reduction has been calculated from D1 = 1.342/k2, using the values of k(min−0.5) from Table 1. Subsequent times are calculated using the fact that the time for n decimal reductions is n2D1.
Values represent the time to reach five decimal reductions with the same values of D1 and assuming that this was then constant (i.e., time = 5D1).
Development of a secondary model.
From a thermodynamic perspective it was anticipated that k = k0e−(ΔG/RT) where ΔG is the free energy of inactivation. The effect of pressure on k enters the exponential term as PΔVact/RT (for R and T, see below) and thus should be characterized by the (in)activation volume, ΔVact. For an isothermal process, the following applies:
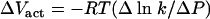 |
(6) |
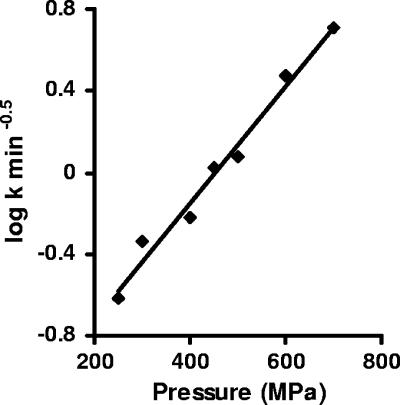
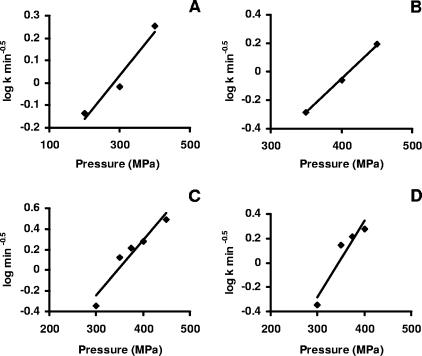
where R = 8.314 × 10−6 m3 mol−1 MPa−1 K−1 and T is absolute temperature (K). If equation 6 holds, a plot of ln k versus pressure should yield a straight line. Figure 4 shows that the relationship between the logarithm of the k values and pressure was indeed linear with the inactivation volume ΔVact = −1.63 × 10−5 m3/mol.
FIG. 4.
Log-linear plot of the death rate at different pressures.
Given ΔVact, it is now possible to calculate the pressure change needed to change the decimal reduction time by a factor of 10. We recall that although the decimal reduction time does vary with time, its value at any point is always proportional to the characteristic decimal reduction time, D1. Hence we define the zp value here as the pressure change needed to change the characteristic decimal reduction time by a factor of 10. Since D1 = 1.324/k2 a 10-fold change in D1 implies a change by a factor of 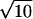 in the rate constant k. Since, for an isothermal system, k = k0e−PΔVact/RT, the pressure change needed to increase or decrease k by a factor of
in the rate constant k. Since, for an isothermal system, k = k0e−PΔVact/RT, the pressure change needed to increase or decrease k by a factor of 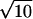 is
is
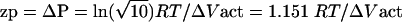 |
(7) |
Substituting ΔVact = −1.63 × 10−5 m3/mol in equation 7 gives a zp value of 172 MPa.
Note that the zp value for the nth decimal reduction has the same value. We can conclude that the concepts of decimal reduction time and z value, when carefully defined, are useful with our model. However, care must be taken in comparing z values with those derived from a constant parameter linear model.
The times required to obtain a fivefold log-reduction in bacterial counts at different pressures are presented in Table 3. The times predicted by the secondary model were close to the observed times.
TABLE 3.
Predicted time to achieve a 105-fold inactivation calculated by the secondary model relating the k parameter of the primary model to pressure
| Value type | Time (min) required for a 105-fold inactivation at indicated pressure (MPa)
|
||||||
|---|---|---|---|---|---|---|---|
| 250 | 300 | 400 | 450 | 500 | 600 | 700 | |
| Predicted | 460 | 235 | 62 | 32 | 16 | 4.3 | 1.1 |
| Observed | >300.0 | 140 (±35) | 90 (±30) | 27 (±6) | 13 (±5) | 2 (±1) | 1.2 (±0.4) |
Testing the new primary model against external data.
The model was tested against data from published high-pressure inactivation curves. Most of the 138 curves examined (70%) showed a simple concave upward curvature (Table 4), but shouldered curves (15%) and curves with pronounced tails (15%) were also encountered. In the following sections the performance criteria for the model are summarized in Tables 5, 6, and 7. The estimated parameters for the individual experimental runs are posted as Tables S5.1, S6.1, and S7.1 in the supplemental material.
TABLE 4.
Sets of independent data used for model testing
| Data set no. | Source | No. of survivor curves | Microorganism(s) | Substrate(s) |
|---|---|---|---|---|
| 1 | Erkmen and Dogan (9) | 16 | L. monocytogenes, aerobic bacteria | Peach and orange juice, raw milk |
| 2 | Hauben et al. (13) | 10 | E. coli strains | Bis-Tris-HCl buffer |
| 3 | Patterson et al. (24) | 13 | E. coli strains, L. monocytogenes strains | PBS |
| 4 | Simpson and Gilmour (35) | 24 | L. monocytogenes strains | PBS and PBS with added glucose, bovine serum albumin, and olive oil |
| 5 | Patterson et al. (25) | 8 | Yersinia enterolitica, E. coli O157:H7 | PBS and food |
| 6 | Benito et al. (2) | 3 | E. coli strains | PBS |
| 7 | Kalchanand et al. (16) | 3 | Staphylococcus aureus, Salmonella enterica serovar Typhimurium, E. coli O157:H7 | Peptone water |
| 8 | Masschalk et al. (17) | 1 | E. coli MG 1655 | PBS |
| 9 | Metrick et al. (20) | 9 | Salmonella enterica serovar Typhimurium strains | PBS and baby food |
| 10 | Schreck et al. (34) | 3 | E. coli strains | Nutrient broth |
| 11 | Casadei et al. (3) | 2 | E. coli NCTC 8164 | PBS |
| 12 | Smelt et al. (36) | 3 | L. monocytogenes, Lactobacillus plantarum | Milk, phosphate buffer |
TABLE 5.
Goodness of fit of the new primary model to survivor data showing simple concave upward curvature
| Data set | Pressure (MPa) | Treatment time (min) | MSE
|
R2
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Avg | Median | Minimum | Maximum | Avg | Median | Minimum | Maximum | |||
| 1 | 200-400 | 5-110 | 0.45 | 0.49 | 0.83 | 0.04 | 0.95 | 0.94 | 0.99 | 0.91 |
| 2 | 270-600 | 60-120 | 0.21 | 0.17 | 0.49 | 0.01 | 0.94 | 0.95 | 0.98 | 0.89 |
| 3 | 350-600 | 30 | 0.30 | 0.23 | 0.97 | 0.02 | 0.91 | 0.92 | 0.99 | 0.77 |
| 4 | 350-450 | 30 | 0.22 | 0.16 | 0.88 | 0.02 | 0.94 | 0.94 | 1.00 | 0.84 |
| 5 | 225-700 | 30 | 0.24 | 0.17 | 0.68 | 0.01 | 0.94 | 0.95 | 0.99 | 0.89 |
| 6 | 500 | 30 | 0.17 | 0.04 | 0.45 | 0.01 | 0.89 | 0.88 | 0.92 | 0.87 |
| 7 | 207 | 30 | 0.06 | 0.05 | 0.12 | 0.01 | 0.94 | 0.94 | 0.98 | 0.91 |
| 8 | 300 | 60 | 0.06 | 0.96 | ||||||
| 9 | 238-340 | 20-40 | 0.05 | 0.02 | 0.26 | 0.01 | 0.95 | 0.95 | 0.99 | 0.88 |
| 10 | 250-300 | 15-30 | 0.27 | 0.30 | 0.37 | 0.13 | 0.95 | 0.94 | 0.97 | 0.93 |
| 11 | 400 | 20 | 0.04 | 0.03 | 0.04 | 0.93 | 0.95 | 0.90 | ||
| 12 | 250-500 | 15-120 | 0.37 | 0.37 | 0.53 | 0.19 | 0.94 | 0.94 | 0.94 | 0.92 |
TABLE 6.
Goodness of fit of the new primary model to curves with shoulders: effect of delay time adjustment (τ)
| Reference | Microorganism(s) | Substrate | Pressure (MPa) | Time course (min) | Original model | Delay time model | ||
|---|---|---|---|---|---|---|---|---|
| Patterson et al. (24) | L. monocytogenes strains | PBS, UHT-treated milk, sterile raw poultrya | 300 | 30 | 0.48 | 0.67 | 0.06 | 0.94 |
| 375 | 30 | 0.14 | 0.52 | 0.04 | 0.90 | |||
| 375 | 30 | 1.24 | 0.73 | 0.10 | 0.97 | |||
| 600 | 30 | 0.86 | 0.79 | 0.08 | 0.97 | |||
| Simpson and Gilmour (35) | L. monocytogenes strains | PBS with glucose, bovine serum albumin, olive oil | 300 | 30 | 0.37 | 0.71 | 0.02 | 0.98 |
| 350 | 30 | 0.65 | 0.80 | 0.09 | 0.95 | |||
| 375 | 30 | 0.24 | 0.74 | 0.04 | 0.96 | |||
| 375 | 30 | 0.12 | 0.73 | 0.02 | 0.90 | |||
| 375 | 30 | 0.30 | 0.83 | 0.30 | 0.97 | |||
| 375 | 30 | 0.14 | 0.88 | 0.01 | 0.99 | |||
| 375 | 30 | 0.47 | 0.83 | 0.02 | 0.86 | |||
| 375 | 30 | 0.61 | 0.79 | 0.14 | 0.90 | |||
| 375 | 30 | 0.31 | 0.81 | 0.03 | 0.86 | |||
| 375 | 30 | 0.25 | 0.77 | 0.00 | 1.00 | |||
| 375 | 30 | 0.61 | 0.79 | 0.14 | 0.90 | |||
| 375 | 30 | 0.52 | 0.73 | 0.08 | 0.99 | |||
| Masschalk et al. (17) | E. coli M91655 strains | PBS | 400 | 60 | 0.23 | 0.89 | 0.12 | 0.94 |
| 400 | 60 | 0.33 | 0.53 | 0.07 | 0.94 | |||
| Schreck et al. (34) | E. coli ATCC 10303 | Nutrient broth | 250 | 40 | 0.33 | 0.86 | 0.05 | 0.97 |
| Casadei et al. (3) | E. coli NCTC 8164 | PBS | 200 | 20 | 0.40 | 0.79 | 0.19 | 0.90 |
UHT, ultra-high temperature.
TABLE 7.
Goodness of fit of the new primary model to curves without shoulders but having pronounced tails: effect of truncating tail region
| Reference | Microorganism(s) | Substrate | Pressure (MPa) | Time course | Original model
|
Constrained tail model
|
||
|---|---|---|---|---|---|---|---|---|
| MSE | R2 | MSE | R2 | |||||
| Hauben et al. (13) | E. coli strains | Bis-Tris-HCl buffer | 300 | 120 | 3.41 | 0.47 | 0.92 | 0.82 |
| 300 | 120 | 1.50 | 0.71 | 0.16 | 0.96 | |||
| 270 | 120 | 0.94 | 0.81 | 0.33 | 0.92 | |||
| 600 | 60 | 0.44 | 0.87 | 0.12 | 0.96 | |||
| 600 | 60 | 1.88 | 0.69 | 0.80 | 0.85 | |||
| 600 | 60 | 3.26 | 0.64 | 0.13 | 0.98 | |||
| 400 | 60 | 2.74 | 0.63 | 0.20 | 0.96 | |||
| 400 | 60 | 1.55 | 0.79 | 0.45 | 0.93 | |||
| Simpson and Gilmour (35) | L. monocytogenes | PBS added with glucose | 375 | 30 | 0.37 | 0.85 | 0.14 | 0.90 |
| Patterson et al. (25) | E. coli 0157:H7 | UHT-treated milka | 700 | 30 | 0.04 | 0.87 | 0.02 | 0.96 |
| Benito et al. (2) | E. coli 0157:H7 | PBS | 500 | 30 | 0.80 | 0.81 | 0.24 | 0.96 |
| Masschalk et al. (17) | E. coli M91655 | PBS | 250 | 60 | 0.10 | 0.89 | 0.01 | 0.92 |
| Kalchayanand et al. (16) | L. monocytogenes Scott A | Peptone water | 207 | 0.23 | 0.86 | 0.01 | 0.89 | |
| Schreck et al. (34) | E. coli | Culture medium | 250 | 40 | 1.26 | 0.82 | 0.34 | 0.95 |
| Casadei et al. (3) | E. coli NCTC 8164 | PBS | 200 | 0-12 | 0.40 | 0.79 | 0.19 | 0.90 |
| 200 | 0-20 | 0.41 | 0.86 | 0.11 | 0.97 | |||
| 400 | 0-16 | 1.01 | 0.83 | 0.83 | 0.87 | |||
| 400 | 0-12 | 1.87 | 0.82 | 0.91 | 0.92 | |||
| Smelt et al. (36) | L. monocytogenes | Phosphate buffer | 400 | 0-40 | 0.99 | 0.87 | 0.27 | 0.96 |
| 450 | 0-20 | 1.99 | 0.77 | 0.44 | 0.96 | |||
UHT, ultra-high temperature.
Survivor curves with upward concavity.
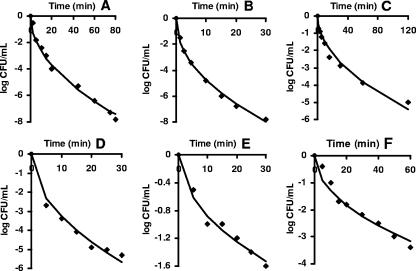
The model performance parameters with the data sets shown in Table 4 are given in Table 5, which shows that satisfactory fits were obtained in all cases. The examples of fitted curves shown in Fig. 5 represent sets of published data with the highest number of recorded experimental points.
FIG. 5.
New primary model fitted to data from the literature showing typical upwardly concave survival curves. (A) L. monocytogenes at 300 MPa (9). (B) Aerobic bacteria at 400 MPa (9). (C) E. coli MG 1655 at 300 MPa (13). (D) L. monocytogenes poultry isolate at 375 MPa (35). (E) E. coli O157:H7 at 207 MPa (16). (F) E. coli MG 1655 at 300 MPa (17).
Survivor curves with shoulders.
During the validation process, fitting difficulties became evident with 15% of survivor curves that exhibited shoulders. In this study a simple approach to deal with shouldering survivor curves was explored. The new model was transformed to a pure time delay model with an additional parameter, the time delay constant (τ) that was also susceptible of a mechanistic interpretation. In this case it was assumed that the kinetic constant at time t had the value that it had in the original model at time t − T.
The delay time for the shoulder region was easily estimated, since it corresponded to the intersection of the curve with a horizontal line through log N/N0 = 0. To calculate the value τ the original model with the additional parameter intercept (b), log N/N0 = −0.869kt0.5 + b, was fitted to the experimental data excluding the data of the shoulder. The estimated values of the intercept (b) and the slope (k) were used to calculate τ according to the following equation: τ = (b/k)2.
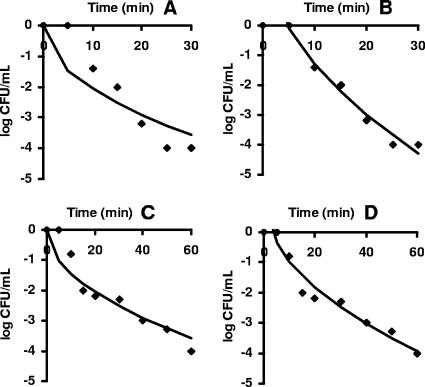
Once τ was determined, the predicted curves could be fitted to the whole experimental data of the shouldered curves. Table 6 presents the regression analysis of the original model and the improvement of the fitting with the time delay model. In all the cases analyzed, the introduction of the time delay constant τ enhanced the fitting of the new model substantially, providing a proper description of the experimental data. The sets of data shown in Fig. 6 were chosen to represent curves with a good number of observed data points and an obvious shoulder.
FIG. 6.
New primary model (A and C) and delay time model (B and D) fitted to survivor data with upward concavity and shoulders. Data are for L. monocytogenes NCTC 11994 at 375 MPa (35) in panels A and B and for E. coli M91655 at 400 MPa (17) in panels C and D.
Survivor curves with pronounced tails.
The new model also experienced problems fitting curves to experimental data showing pronounced asymptotic tails (15%). However, survivor curves with tails could be properly described by the model by omitting data points of the tail regions where the survivor fractions (log10 N/N0) diminished very slowly. The tails were restricted in the regression analysis by including only two points after the establishment of the tail. The points to be omitted were chosen subjectively by visual inspection. Nevertheless, this admittedly empirical method resulted in a much improved fit to the data.
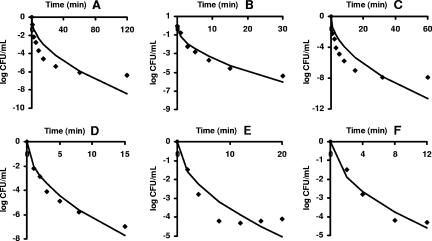
Table 7 shows the regression analysis for the model fitting unrestricted and restricted tails in survivor curves with upward concavity. In all the sets analyzed, the fitness of the model improved markedly. The examples presented in Fig. 7 were selected based on the number of experimental data points and the presence of an obvious extended tail.
FIG. 7.
New primary model (A, C, and E) and truncated version (B, D, and F) fitted to survivor data with upward concavity and pronounced tails. Data are for E. coli MG 1655 at 300 MPa (13) in panels A and B, E. coli LMM 1010 at 400 MPa (13) in panels C and D, and E. coli NCTC 8164 at 200 MPa (3) in panels E and F.
Construction of secondary models based on data from the literature.
Fig. 8 shows secondary models that were obtained using data from the literature. The death rates showed a linear relationship to pressure in four sets of data for Listeria monocytogenes. From the secondary models the pressure resistance descriptors ΔVact and zp for each L. monocytogenes data set were calculated and are presented in Table 8.
FIG. 8.
Secondary models relating pressure to death rate. (A) L. monocytogenes in BHIB (9). (B) L. monocytogenes NCTC 11994 in PBS (35). (C) L. monocytogenes poultry isolate in PBS (35). (D) L. monocytogenes 2433 in PBS (24).
TABLE 8.
Pressure resistance descriptors ΔVact and zp calculated for published sets of data
| Data seta | ΔVact (m3/mol) | zp (MPa) |
|---|---|---|
| A | −1.07 × 10−5 | 262 |
| B | −2.69 × 10−5 | 104 |
| C | −2.97 × 10−5 | 94 |
| D | −3.54 × 10−5 | 79 |
A, data of Erkmen and Dogan (9) for L. monocytogenes in brain heart infusion broth; B, data of Simpson and Gilmour (35) for L. monocytogenes NCTC 11994 in PBS; C, data of Simpson and Gilmour (35) for L. monocytogenes poultry isolate in PBS; D, data of Patterson et al. (25) for L. monocytogenes 2433 in PBS.
DISCUSSION
It is clear from perusal of the literature that the precise shape of pressure inactivation curves depends on numerous factors including the type of organism, the composition of the suspending medium, the pressure intensity, and the treatment temperature. Classical first-order-type pressure inactivation curves do occur, but they are not commonly encountered. Curves with upward concavity and tails are more common and appear to represent typical behavior under a range of conditions.
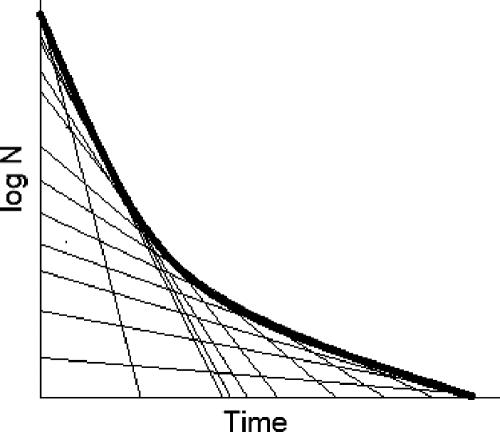
The new primary model described here assumes that inactivation by pressure is a first-order inactivation process in which the specific rate changes with the reciprocal of the square root of time. In this model, the observed survival curve can be thought of as being composed of a series of first-order inactivation curves with progressively decreasing inactivation rates, as shown in Fig. 9. Fair agreement was found between this model and data obtained in this work with E. coli and also with data reported in the literature for other organisms.
FIG. 9.
Interpretation of nonlinear survivor curves as time-dependent changes in the instantaneous inactivation rate constant (tangential slopes).
The assessment of the goodness of fit and performance showed that the new primary model gave a reasonable overall fit to experimental data (Table 1). However, the goodness of fit and performance were not uniform at all pressures tested. The values of the accuracy parameter (Af) for curves at 400 and 500 MPa (1.33 and 1.30, respectively) were slightly greater than the value of 1.10 recommended as an acceptable value when one independent variable (time) is involved (33). The MSE values at these pressures were consequently above 1 log10 CFU/ml. At pressures of 500 and 600 MPa, the new model experienced slight difficulties describing the initial rapid decrease in viable numbers, and the analysis of residuals showed some clustering of residuals of the same sign. This initial rapid decrease was almost certainly due to the temperature increase during compression, and this effect would be expected to have a stronger influence on the inactivation kinetics at the higher pressures. Some workers have been able to minimize this complicating factor by precooling the sample to below the treatment temperature so that adiabatic heat brings the sample up to the final temperature (5). Unfortunately, this stratagem did not work with our particular pressure rig. Nevertheless, despite these limitations, the primary model predicted survivor counts that were quite close to the experimental ones, as shown in the graphical analysis.
The new primary model delivered a simple linear secondary model from which the pressure-resistance descriptors ΔVact and zp could be calculated. The value of ΔVact for E. coli NCTC 8164 pressurized in PBS was −1.63 × 10−5 m3/mol, somewhat higher than the values reported for E. coli KUEN (10) which were as follows: −1.15 × 10−5 m3/mol in broth, −1.22 × 10−5 m3/mol in milk, −1.215 × 10−5 m3/mol in peach juice, and −1.06 × 10−5 m3/mol in orange juice (10). The calculated ΔVact values for L. monocytogenes (Table 8) were between −1.07 and −3.54, similar to published values for L. monocytogenes Scott A, which were −2.11 × 10−5 m3/mol in milk and −3.43 × 10−5 m3/mol in fresh pork chops (22, 23). The values for L. monocytogenes tended to be higher than those for E. coli, implying greater pressure sensitivity. The corresponding zp value for E. coli 8164 (172 MPa) was similar to the zp value reported for E. coli MG1655 in carrot juice (213 MPa) (40), whereas the value for E. coli KUEN in broth of 460.7 MPa was substantially higher (10). It should be pointed out that the published values for ΔVact and zp were obtained from log-linear curve fits and are not strictly comparable with those obtained using the present approach, and there is clearly also some variability depending on organism and conditions of pressurization. Nevertheless, it is interesting that all the above ΔVact values for inactivation of vegetative bacteria fall within the range for protein denaturation of −0.6 to −8.5 × 10−5 m3/mol (30).
The new primary model was tested against 138 sets of independent data covering a considerable range of situations. All sets of inactivation data having typical concave upward survivor curves were satisfactorily described by the new model. A proportion of data sets that showed survivor curves with additional shoulders (15%) or tails (15%) could not be described by the primary model in its simplest form. Curves with shoulders, in which inactivation was initially absent or very slow, could be described adequately by the delay time model. This implies that the remaining inactivation phase followed the simple concave upwards pattern and reinforces the view that this is typical behavior for pressure inactivation.
Curves with “tails,” in which the inactivation rate slows to almost zero, represent another class of curve that is not uncommon. Table 7 showed that most of the published sets of data with tailing conditions arose from low-pressure and/or short time course experiments or the use of a baroresistant strain. In our work with E. coli NCTC 8164, an apparent tail with little inactivation was also observed at low pressures if experiments were terminated soon after the loss of viability became very slow. However, if sampling was continued for much longer, it became apparent that the stable resistant fraction did decrease, and the curve as a whole followed the upward concave form. Most of the published data did not include survivor curves generated at different pressures under otherwise similar conditions, but, where such date were available, the secondary models showed a linear relationship between log10 k and pressure, allowing the calculation of pressure resistance descriptors. Based on testing the model against external data we may conclude that it can be considered to provide “good enough” estimations of bacterial survivors as discussed by Baranyi and Roberts (1).
The dependence of inactivation rates on a simple function of time, namely, its square root, was unexpected though fortuitous for modeling purposes. The fact that consistent secondary models could also be derived from the rate parameter of the primary model also provides support for the model. While great caution should be exercised in inferring mechanism from inactivation kinetics, the observed relationship would be consistent with inactivation's being dependent on a diffusion process. The rate at which solute particles migrate through a solvent depends on the size and shape of the particles. The parameter that describes the movement of a particle through the solvent is the diffusion coefficient (D). It is inversely proportional to the square of the radius of the particle (D ∝ 1/r2). As molecular radius increases, D decreases because of the increase in frictional interactions between solute and solvent.
In such processes the progression of an event with time follows a law like 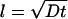 , where l is the distance diffused in time t and D is the diffusion coefficient. This relation shows that the time required for a diffusion process increases with the square of the distance over which diffusion occurs. A 10-fold increase in distance increases diffusion time by a factor of 100.
, where l is the distance diffused in time t and D is the diffusion coefficient. This relation shows that the time required for a diffusion process increases with the square of the distance over which diffusion occurs. A 10-fold increase in distance increases diffusion time by a factor of 100.
Diffusion coefficients of molecules in liquids are typically around 10−9 to 10−11 m2/s; hence, if a diffusion distance of the size of a bacterial cell is considered (ca. 1 μm), then a time of about t ∼ l2/D or t ∼ 10−12/10−11 ∼ 10−1 s is obtained.
If distance is of the order of 100 μm, then the calculated time is around 103 s. Many studies report the release of cellular material (cations, anions, amino acids, and nucleotides) from cells with damaged membranes. However, the calculated times for diffusion of material out of a bacterial cell are very much smaller than treatment times needed for appreciable bacterial inactivation at most pressures, suggesting that this is not the rate-limiting event in microbial inactivation and may be an effect rather than a cause of cell death.
When diffusion in solids or quasi-solids is examined, diffusion coefficients are lower, typically of the order 10−13 m2/s and smaller. These values would be further reduced somewhat under pressure (15). The time required for a diffusion distance of bacterial dimensions is then about 100 s, which would fit with the idea of critical molecules (e.g., signal molecules) moving inside the cell.
The question arises of how a diffusion phenomenon inferred from observations of a population of millions of cells can be reconciled with a stochastic interpretation of inactivation at the single-cell level. One possibility is that cellular resistance might increase with time of exposure, due to some physical diffusion-limited process within the cell. For example water loss from the cell or from a critical target within cell (e.g., the membrane or a critical protein) or a slow change of state in the cell membrane, such as a phase transition between liquid crystalline and gel states or a phase separation between phospholipid domains or between protein and lipid domains. These putative changes might result in increased pressure resistance such that stochastic events of the same frequency have a decreasing probability of causing cell death as time of exposure increases because the susceptibility of the target changes. The physical stochastic energy fluctuations remain the same but the probability of their causing a lethal change in a critical target decreases with time.
Different views as to the most appropriate model to describe microbial inactivation curves were voiced nearly a century ago (6, 41), and the topic continues to excite vigorous discussion today (14). We have described a model that takes stochastic lethal events as its starting point and have suggested that diffusion-limited phenomena could be involved in determining the shape of pressure survival curves. However, we are fully aware that the square root dependence could simply be coincidence and that explanations other than diffusion may account for the observed deviation from simple first-order kinetics. The notion that stochastic events underlie inactivation kinetics is not at variance with the idea of a distribution of resistances within the population. In fact, we regard it as physiologically inevitable that differences in resistance will affect the outcome of stochastic pressure events at the cellular level and may thus influence the shape of population inactivation curves. However, we have not yet found a statistical distribution that, of itself, would account entirely for the observed square root time dependence.
Apart from these theoretical considerations, the model in its simplest form provided a reasonable description of 70% of a sample of published inactivation data. With relatively simple (empirical) modifications the remainder of the data could be accounted for. Given the wide diversity of form among inactivation curves, it seems unlikely that a single model will account for all circumstances, irrespective of the theoretical starting point. The primary model described here provides a reasonable fit for much of the data and has the advantage of providing simple secondary models that should be of practical use. This study has also shown that the concepts of decimal reduction time and z value are still applicable to situations following our specific time-varying model. The time for the initial decimal reduction, D1, can be taken to be the characteristic decimal reduction time for the system: the time for the nth decimal reduction is given by Dn = (2n − 1)D1. The overall time for n decimal reductions is n2D1 as opposed to nD1 for a first-order system with constant specific rate, illustrating the significance of deviations from the simple first-order model for microbial food safety assurance.
Supplementary Material
Acknowledgments
B.K. thanks the European Union ALFA Network II and the Universidad de La Sabana, Colombia, for the financial support for her doctoral studies. D.L.P thanks the Leverhulme Trust for fellowship support during this work.
Footnotes
Published ahead of print on 9 February 2007.
Supplemental material for this article is available at http://aem.asm.org/.
REFERENCES
- 1.Baranyi, J., and T. A. Roberts. 2004. Predictive microbiology-quantitative microbial ecology. Culture. http://www.ifr.ac.uk/safety/comicro/Culture_25pdf.
- 2.Benito, A., G. Ventoura, M. Casadei, T. Robinson, and B. Mackey. 1999. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl. Environ. Microbiol. 65:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadei, M. A., P. Mañas, G. Niven, E. Needs, and B. Mackey. 2002. Role of membrane fluidity in pressure resistance of Escherichia coli NCTC 8164. Appl. Environ. Microbiol. 68:5965-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casolari, A. 1998. Microbial death, p. 1-44. In M. J. Bazin and J. I. Prosser (ed.), Physiological models in microbiology, vol. 2. CRC Press, Boca Raton, FL. [Google Scholar]
- 5.Chen, H., and D. G. Hoover. 2003. Modeling the combined effect of high hydrostatic pressure and mild heat on the inactivation kinetics of Listeria monocytogenes Scott A in whole milk. Innov. Food Sci. Emerg. Technol. 4:25-34. [Google Scholar]
- 6.Chick, H. 1910. The process of disinfection by chemical agencies and hot water. J. Hyg. Camb. 10:237-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey, K. R., and S. T. G. Phua. 2005. Re: Erkmen and Dogan 2004 Food Microbiology 21, 181-185. Food Microbiol. 22:483-487. [Google Scholar]
- 8.Denbigh, K. 1966. Principles of chemical equilibrium: with applications in chemistry and chemical engineering. Cambridge, United Kingdom.
- 9.Erkmen, O., and C. Dogan. 2004. Effects of ultra high hydrostatic pressure on Listeria monocytogenes and natural flora in broth, milk and fruit juices. Int. J. Food Sci. Technol. 39:91-97. [Google Scholar]
- 10.Erkmen, O., and C. Dogan. 2004. Kinetic analysis of Escherichia coli inactivation by high hydrostatic pressure in broth and foods. Food Microbiol. 21:181-185. [Google Scholar]
- 11.Farkas, D. F., and D. H. Hoover. 2000. High pressure processing. J. Food Sci. 65(Suppl.):47-64. [Google Scholar]
- 12.Fernández, A., J. Collado, L. M. Cunha, M. J. Ocio, and A. Martinez. 2002. Empirical model building based on Weibull distribution to describe the joint effect of pH and temperature on the thermal resistance of Bacillus cereus in vegetable substrate. Int. J. Food Microbiol. 77:147-153. [DOI] [PubMed] [Google Scholar]
- 13.Hauben, K. J. A., K. Bernaerts, and C. W. Michiels. 1998. Protective effect of calcium on inactivation of Escherichia coli by high hydrostatic pressure. J. Appl. Microbiol. 85:678-684. [DOI] [PubMed] [Google Scholar]
- 14.Heldman, D. R., and R. L. Newsome. 2003. Kinetic models for microbial survival during processing. Food Technol. 57:40-46. [Google Scholar]
- 15.Isaacs, N. S. 1981. Liquid phase high pressure chemistry. John Wiley, Chichester, United Kingdom.
- 16.Kalchayanand, N., T. Sikes, C. P. Dunne, and B. Ray. 1998. Factors influencing death and injury of foodborne pathogens by hydrostatic pressure-pasteurization. Food Microbiol. 15:207-214. [Google Scholar]
- 17.Masschalk, B., C. García-Graells, E. van Haver, and C. W. Michiels. 2000. Inactivation of high pressure resistant Escherichia coli by lysozyme and nisin under high pressure. Innov. Food Sci. Emerg. Technol. 1:39-47. [Google Scholar]
- 18.McKee, S., and G. W. Gould. 1988. A simple mathematical model of the thermal death of microorganisms. Bull. Math. Biol. 50:493-501. [DOI] [PubMed] [Google Scholar]
- 19.Mellefont, L. A., T. A. McMeekin, and T. Ross. 2003. Performance evaluation of a model describing the effects of temperature, water activity, pH and lactic acid concentration on the growth of Escherichia coli. Int. J. Food Microbiol. 82:45-58. [DOI] [PubMed] [Google Scholar]
- 20.Metrick, C., D. Hoover, and D. Farkas. 1989. Effects of high hydrostatic pressure on heat-resistant and heat-sensitive strains of Salmonella. J. Food Sci. 54:1547-1549. [Google Scholar]
- 21.Miles, C. A. 2006. Relating cell killing to the inactivation of critical components. Appl. Environ. Microbiol. 72:914-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mussa, D. M., H. S. Ramaswamy, and J. P. Smith. 1999a. High pressure destruction kinetics of Listeria monocytogenes on pork. J. Food Prot. 62:40-45. [DOI] [PubMed] [Google Scholar]
- 23.Mussa, D. M., H. S. Ramaswamy, and J. P. Smith. 1999b. High pressure destruction kinetics of Listeria monocytogenes Scott A in raw milk. Food Res. Int. 31:343-350. [DOI] [PubMed] [Google Scholar]
- 24.Patterson, M. F., M. Quinn, R. Simpson, and A. Gilmour. 1995a. Sensitivity of vegetative pathogens to high hydrostatic pressure treatment in phosphate-buffered saline and foods. J. Food Prot. 58:524-529. [DOI] [PubMed] [Google Scholar]
- 25.Patterson, M. F., M. Quinn, R. Simpson, and A. Gilmour. 1995b. Effects of high pressure on vegetative pathogens, p. 47-63. In D. A. Ledward, D. E. Johnston, R. G. Earnshaw, A. P. M. Hastings (ed.), High pressure processing of foods. Nottingham University Press, Leicestershire, United Kingdom.
- 26.Peleg, M. 2003. Microbial Inactivation curves: interpretation, mathematical modeling, and utilization. Comments Theor. Biol. 8:357-387. [Google Scholar]
- 27.Peleg, M., and M. B. Cole. 1998. Reinterpretation of microbial survival curves. Crit. Rev. Food Sci. 38:353-380. [DOI] [PubMed] [Google Scholar]
- 28.Ponce, E., R. Pla, M. Capellas, B. Guamis, and M. Mor-Mur. 1998. Inactivation of Escherichia coli inoculated into liquid whole egg by high hydrostatic pressure. Food Microbiol. 15:265-272. [Google Scholar]
- 29.Ponce, E., R. Pla, M. Mor-Mur, M. Gervilla, and B. Guamis. 1998. Inactivation of Listeria innocua inoculated into liquid whole egg by high hydrostatic pressure. Food Microbiol. 16:119-122. [DOI] [PubMed] [Google Scholar]
- 30.Prehoda, K. E., E. S. Mooberry, and J. L. Markley. 1998. High pressure effects on protein structure, p. 55-86. In J.-F. Lefevre and R. E. Holbrook (ed.), Protein dynamics, function and design. Plenum Press, New York, NY.
- 31.Raso, J., and G. Barbosa-Cánovas. 2003. Nonthermal preservation of foods using combined processing techniques. Crit. Rev. Food Sci. Nut. 43:265-285. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez, A. C., J. W. Larkin, J. Dunn, E. Patazca, N. R. Reddy, M. Alvarez-Medina, R. Tetzloff, and G. J. Fleischman. 2004. Model of the inactivation of bacterial spores by moist heat and high pressure. J. Food Sci. 69:367-373. [Google Scholar]
- 33.Ross, T. 1996. Indices for performance evaluation of predictive models in food microbiology. J. Appl. Bacteriol. 81:501-508. [DOI] [PubMed] [Google Scholar]
- 34.Schreck, C., G. Almsick, and H. Ludwig. 1999. Influence of culturing conditions on the pressure sensitivity of Escherichia coli, p. 313-324. In F. A. R. Oliveira and J. C. Oliveira (ed.), Processing foods: quality optimization and process assessment. CRC Press, Boca Raton, FL.
- 35.Simpson, R. K., and A. Gilmour. 1997. The effect of high hydrostatic pressure on Listeria monocytogenes in phosphate-buffered saline and model food systems. J. Appl. Microbiol. 83:181-188. [DOI] [PubMed] [Google Scholar]
- 36.Smelt, J. P. P. M., P. C. Wouters, and A. G. F. Rijke. 1998. Inactivation of microorganisms by high pressure, p. 398-417. In D. S. Reid, The properties of water in foods, ISOPOW 6. Blackie Academic and Professional Press, London, United Kingdom.
- 37.Smelt, J. P. P. M., J. C. Hellemons, P. C. Wouters, and S. J. C. van Gerwen. 2002. Physiological and mathematical aspects in setting criteria for decontamination of foods by physical means. Int. J. Food Microbiol. 787:57-77. [DOI] [PubMed] [Google Scholar]
- 38.Stumbo, C. R. 1973. Thermobacteriology and food processing, 2nd ed. Academic Press, New York, NY.
- 39.van Boekel, M. A. 2002. On the use of the Weibull model to describe thermal inactivation of microbial vegetative cells. Int. J. Food Microbiol. 74:139-159. [DOI] [PubMed] [Google Scholar]
- 40.van Opstal, I. V., S. C. M. Vanmuysen, E. Y. Wuytack, B. Masschalk, and C. W. Michiels. 2005. Inactivation of Escherichia coli by high hydrostatic pressure at different temperatures in buffer and carrot juice. Int. J. Food Microbiol. 98:179-191. [DOI] [PubMed] [Google Scholar]
- 41.Withel, E. R. 1942. The significance of the variation in shape on time-survivor curves. J. Hyg. Camb. 42:124-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto, K., M. Matsubara, S. Kawasaki, M. L. Bari, and S. Kawamoto. 2005. Modelling the pressure inactivation dynamics of Escherichia coli. Braz. J. Med. Biol. Res. 38:1253-1257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.