Abstract
Dimethylsulfide (DMS)-degrading enrichment cultures were established from samples of coastal seawater, nonaxenic Emiliania huxleyi cultures, and mixed marine methyl halide-degrading enrichment cultures. Bacterial populations from a broad phylogenetic range were identified in the mixed DMS-degrading enrichment cultures by denaturing gradient gel electrophoresis (DGGE). Sequences of dominant DGGE bands were similar to those of members of the genera Methylophaga and Alcanivorax. Several closely related Methylophaga strains were obtained that were able to grow on DMS as the carbon and energy source. Roseobacter-related populations were detected in some of the enrichment cultures; however, none of the Roseobacter group isolates that were tested were able to grow on DMS. Oxidation of DMS by Methylophaga sp. strain DMS010 was not affected by addition of the inhibitor chloroform or methyl tert-butyl ether, suggesting that DMS metabolism may occur by a route different from those described for Thiobacillus species and other unidentified marine isolates. Addition of DMS and methanethiol to whole-cell suspensions of strain DMS010 induced oxygen uptake when strain DMS010 was grown on DMS but not in cells grown on methanol. The apparent Kms of strain DMS010 for DMS and for methanethiol were 2.1 and 4.6 μM, respectively, when grown on DMS. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the biomass of strain DMS010 and analysis of peptide bands by mass spectrometry techniques and N-terminal sequencing provided the first insight into the identity of polypeptides induced during growth on DMS. These included XoxF, a homolog of the large subunit of methanol dehydrogenase for which a biological role has not been identified previously.
Dimethylsulfide (DMS) is a volatile organosulfur compound that is emitted from the ocean into the atmosphere, where it represents the most abundant organic sulfur gas (31). Atmospheric oxidation of DMS generates sulfur aerosols that backscatter heat radiation, promote cloud formation, and as a result, cause negative radiative forcing (2). It has been suggested that the effects of DMS-derived aerosols provide a global climate feedback loop that could result in climate cooling (7). DMS is produced mainly by enzymatic cleavage of dimethylsulfoniopropionate (DMSP), an algal metabolite which may have a role in osmoregulation (54) or may constitute an antioxidant system in microalgae (46). Sinks of DMS include photochemical degradation to dimethyl sulfoxide (DMSO) (19), bacterial oxidation of DMS to DMSO (55), and its utilization as a sulfur source by microorganisms (15, 27). Microbial degradation of DMS, however, appears to be the main sink for DMS in the marine environment, often leading to the oxidation of 90% or more of DMS in the ocean surface (3, 26). Bacterial degradation of DMS therefore significantly reduces the amount of DMS in the mixed surface layer that is available for sea-to-air transfer.
Growth on DMS as a carbon source has been described for a range of prokaryotes, including anaerobic degradation by methanogens (28) and sulfate-reducing bacteria (48). Aerobic bacterial DMS oxidation was first demonstrated for some members of the genera Hyphomicrobium and Thiobacillus (9, 24, 40a, 47). In these bacteria, DMS monooxygenase was identified as a key enzyme in DMS metabolism, producing methanethiol and formaldehyde. DMS monooxygenase activity was also found in Hyphomicrobium sp. strain S growing on DMSO (9) and in strains of Hyphomicrobium sulfonivorans that were isolated on dimethyl sulfone as the carbon source (4, 37).
The phylogenetic diversity of marine DMS-degrading prokaryotes is still largely unexplored. Alphaproteobacteria, especially members of the Roseobacter clade, have often been implicated in the metabolism of organosulfur compounds in the marine environment (16, 38, 56), but it is not clear whether these bacteria are able to grow on DMS. Marine isolates growing on DMS as the carbon source, obtained from marine sediments, included Rhodovulum sulfidophilum SH1, Thiobacillus sp. strain ASN-1, Thiobacillus thioparus T5, Thiocapsa roseopersicina M11, Methylophaga sulfidovorans, and the unidentified isolate BIS-6 (17, 23, 36, 50, 51). Less is known about the diversity of DMS-degrading bacteria in the pelagic marine environment. Recently, Vila-Costa and colleagues (49) reported the detection of Methylophaga spp. by denaturing gradient gel electrophoresis (DGGE) and clone library analysis of DMS enrichment cultures from seawater samples. Unfortunately, isolates were not obtained and so the assumption that the detected populations of Methylophaga were indeed able to grow on DMS could not be substantiated. Previously reported DMS-degrading bacterial isolates from pelagic marine samples that could grow on DMS were not identified by sequencing of 16S rRNA genes (18, 20), further highlighting the need to cultivate and identify DMS-degrading bacteria from seawater.
Given the phylogenetic diversity of DMS-degrading bacteria thus far identified, and the fact that closely related isolates of DMS-degrading strains may be unable to grow on DMS, the identification of DMS-degrading populations in environmental samples based on 16S rRNA genes is difficult. Functional molecular markers, i.e., PCR primers and probes targeting genes encoding key enzymes of DMS degradation pathways, would therefore be invaluable tools with which to study the abundance and distribution of DMS-degrading bacteria in environmental samples and to characterize the diversity of genes and enzymes involved in this globally relevant process. However, the genes encoding DMS monooxygenases, DMS methyltransferases, or other key enzymes of DMS metabolism from organisms growing on DMS as a carbon source have not yet been identified.
The aims of this study were (i) to identify bacterial populations in marine DMS-degrading enrichment cultures, (ii) to identify isolates capable of growth on DMS, and (iii) to identify polypeptides involved in metabolism of DMS. These were achieved by analyzing enrichment cultures by denaturing gradient gel electrophoresis analysis, sequencing 16S rRNA genes of isolates, testing the ability of isolates to grow on DMS, and characterizing the genetic diversity of DMS-degrading Methylophaga isolates by BOX-PCR (42). Finally sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of cell extracts from biomass of a Methylophaga isolate revealed polypeptides induced during growth on DMS which were identified by mass spectrometry techniques and N-terminal sequencing.
MATERIALS AND METHODS
Sampling and enrichment conditions.
Seawater samples were obtained at low tide from Achmelvich Bay (water depth, 1 m; sampling depth, 0.2 m; Sutherland, Scotland, United Kingdom; on 9 September 2004), from a tidal rock pool at Coral Beach (Isle of Skye, Scotland, United Kingdom, on 11 September 2004), and from sampling station L4 in the English Channel off the coast of Plymouth (50°15′N, 04°13′W; water depth, 55 m; sampling depth, 10 m; Devon, England, United Kingdom, on 1 November 2004, 16 May 2005, and 20 June 2005). Seawater (2.5 liters) from Achmelvich Bay was filtered through 0.2-μm-pore filters (type OS; Millipore), and the biomass retained on the filter was resuspended in 10 ml of seawater sample. Water (250 ml) from the rock pool was processed similarly and resuspended in 10 ml of the sample water. One-milliliter aliquots of the suspensions were used to inoculate 25 ml sterile marine ammonium mineral salt (MAMS) medium in 125-ml crimp-top vials sealed with blue Teflon-coated butyl rubber bungs as previously described (44). The carbon sources used for enrichment were DMS (50 μM), formate (10 mM), methylamine (5 mM), and methanol (5 mM). In addition, the membranes used for filtration of seawater samples from Achmelvich Bay and the rock pool were also used as inocula for 25-ml cultures as described above and amended with 50 μM DMS. For the November 2004 sample, 3 liters of seawater from station L4 was filtered and resuspended in 3 ml of L4 water. Aliquots (0.4 ml) of the suspension were used to inoculate 25 ml MAMS medium, as described above, with DMS (50 μM), methanol, methylamine, formate, and acrylate (all 5 mM) as the carbon sources. The membrane used for filtration was also used as inoculum for an enrichment culture with 50 μM DMS as described above. For the May and June 2005 samples from station L4, enrichment cultures were inoculated with filters through which 200 ml of seawater had been filtered. For the May 2005 samples, DMS enrichments were also set up with the extra addition of thiosulfate (2.5 mM) and bicarbonate (4 mM) and with bicarbonate only (4 mM).
Enrichment cultures were also set up using unialgal Emiliania huxleyi cultures as the inoculum. Microscopic observation showed that none of the E. huxleyi strains was axenic (M. Cox, personal communication). Two-milliliter culture aliquots of E. huxleyi strains 92A, 371, 373, 373UEA, and 1516 were pooled and gently vacuum filtered through a 0.2-μm-pore SUPOR membrane filter (Pall, Farlington, United Kingdom). The filter was rinsed by filtering 15 ml of MAMS through the membrane before the biomass retained on the filter was resuspended in 4 ml of MAMS, and 400 μl of the suspension was used to inoculate 25 ml of MAMS in sealed, crimp-top vials. A second culture of strain 1516 that had previously been axenic was used separately as an inoculum due to its markedly higher turbidity.
Aliquots of 2.5 ml from each of six different methyl halide-degrading enrichment cultures (44) were pooled and used to inoculate enrichment cultures which were amended with DMS, methanol, formate, and methylamine as described above.
All vials were sealed using sterile blue Teflon-coated butyl rubber septa. Filtered, sterile DMS solution was added aseptically through the septa of crimp-top vials with a syringe and needle to a final concentration of 50 μM from a 5 mM stock solution prepared with MAMS. Enrichment cultures preenriched on substrates other than DMS were later subcultured (10% inoculum) on DMS only (50 μM). Cultures were incubated at room temperature (20 to 25°C). Chemical controls consisting of medium supplemented with DMS were set up alongside enrichment cultures to account for chemical breakdown of DMS. The concentration of DMS in headspace gas was monitored by gas chromatography (GC) analysis. Enrichments were respiked with additional doses of DMS upon depletion of headspace DMS.
GC analysis.
Determination of DMS in headspace gas was carried out by injecting 100 μl of a headspace gas sample into a GCD gas chromatograph (PYE Unicam Ltd., Cambridge, United Kingdom) fitted with a 1 m-by-4 mm glass column containing Poropak Q (Phase Separations Ltd., Deeside, United Kingdom), and nitrogen as the carrier gas (flow rate, 30 ml min−1) at 200°C. A flame ionization detector was used to detect compounds, and peak areas were integrated with a model 3390A integrator (Hewlett Packard, Berkshire, United Kingdom). DMS concentrations were calculated by regression analysis based on a four-point calibration with standard DMS solutions in MAMS.
Isolation of bacterial strains and screening for DMS oxidation activity.
Samples of enrichment cultures were serially diluted in sterile MAMS medium, and 100 μl of sample was spread onto MAMS plates (MAMS solidified with 15 g liter−1 Bacto agar [Difco]). Plates were incubated for at least 2 weeks in gas-tight jars to which DMS was added (concentration of approximately 200 μM). Gas jars were regularly vented and replenished with DMS. Colonies were isolated and incubated as described above. Biomass from isolation plates was taken with a wire loop and resuspended in 1 ml of sterile MAMS and injected with sterile syringes through stoppers into 27-ml crimp-top vials containing 5 ml of sterile MAMS medium. DMS was added to a final concentration of 50 μM, and the degradation of DMS was monitored by GC analysis of headspace gas.
Test for growth on DMS.
Isolates were tested for their ability to grow on DMS on MAMS medium plates in gas-tight jars which contained DMS in the atmosphere (approximately 0.1% volume). To verify that isolates grew at the expense of DMS consumption and not on traces of organic compounds present in the solidified medium, degradation of DMS (50 μM) by isolates was also tested in liquid culture by monitoring headspace concentrations of DMS by gas chromatography. In addition, the growth of Methylophaga isolates was also tested at DMS concentrations of 500 μM and 1 mM. No growth was observed when Methylophaga strains were inoculated into medium lacking a carbon source. Isolates were also inoculated onto marine agar (2216; Difco) or into liquid MAMS medium to which peptone and yeast extract (44) were added, to test for the ability to grow on a complex medium.
PCR amplification of 16S rRNA-encoding genes, identification of isolates, and BOX-PCR of Methylophaga isolates.
Amplification and sequencing of bacterial 16S rRNA genes were done as described previously (44). For isolates, single colonies were taken from an agar plate with a sterile loop, resuspended in 50 μl of PCR-grade water, and boiled for 5 min. Lysates (1 to 5 μl) were used as the template for amplification of 16S rRNA genes by PCR, using primers 27F and 1492R (30). PCR products were obtained for all isolates, including gram-positive isolates. Two milliliters of enrichment cultures was pelleted at 13,000 × g at 4°C for 15 min in a microcentrifuge, and the pellet was resuspended in 10 μl of PCR-grade water and boiled for 5 min in a water bath. PCR products suitable for DGGE analysis were obtained as described previously, using primers 341F-GC and 926RM (45). Sequences were analyzed using BLAST (1) at the NCBI database (http://ncbi.nlm.nih.gov/BLAST) and added to those with the highest-scoring BLAST hits, to an alignment of bacterial 16S rRNA sequences (33) using the aligning tool included in ARB software (32). Phylogenetic trees were calculated using maximum-likelihood, parsimony, and distance methods. Bootstrap values were determined on 1,000 resampled data sets using PHYLIP programs SEQBOOT, DNADIST (with settings Kimura 2-parameter, transition/transversion ration of 2.0), NEIGHBOR, and CONSENSE (14). Genomic fingerprinting of Methylophaga isolates was carried out using BOX-PCR as described previously, using primer BOXA1R (42). The BOX-PCR method exploits conserved and repeated sequence motifs present in bacterial genomes that were first discovered in Streptococcus pneumoniae (35). Using the conserved sequence motif as a primer target site, a specific pattern of amplicons is generated that can be used for genomic fingerprinting of bacterial isolates (41).
DGGE and sequencing of DGGE bands.
DGGE was carried out as described previously (45), using gradients of 30 to 70% denaturants. DGGE staining with SYBR green I (Invitrogen, Paisley, United Kingdom) and image acquisition were carried out as described previously (39), using a FujiFilm FLA-5000 scanner. DGGE bands were sampled using sterile pipette tips and reamplified using primers 341F-GC and 926RM, as described previously (45). Bands were sequenced directly from purified PCR products using primer 926RM. If sequencing data were ambiguous due to mixed templates, PCR products were cloned using a TOPO-TA cloning kit (Invitrogen, Paisley, United Kingdom), and individual clones were reanalyzed by DGGE parallel to the original PCR product to identify comigrating clones, which were sequenced using standard M13 primers.
Effect of inhibitors on DMS metabolism by Methylophaga sp. strain DMS010.
An inhibition assay was carried out using biomass of strain DMS010 grown on DMS to an optical density (OD) (at 540 nm) of 0.3. Two hundred fifty milliliters of the culture was harvested by centrifugation at 17,700 × g at 15°C in a JA-10 rotor in a Beckman centrifuge. The cells were washed with sterile MAMS and resuspended in 25 ml of fresh medium. The assay was set up in triplicate in 27-ml crimp-top vials containing 5 ml of MAMS, 400 μl of a 3 mM DMS solution prepared in MAMS, 100 μl of inhibitor (50 mM chloroform or methyl tert-butyl ether [MTBE] in sterile distilled water or sterile water for controls, see below), and 500 μl of cell suspension (final optical density at 540 nm of approximately 0.3). Prior to the addition of cell suspension (or water for controls), the DMS-containing vials were left to equilibrate for 1 h. Uninoculated controls were set up in parallel to assess chemical losses of DMS.
Substrate-induced oxygen uptake of resting cell suspensions.
Methylophaga sp. strain DMS010 was grown in batch culture at 25°C in a shaking incubator at 150 rpm in 1.1-liter sealed crimp-top bottles in 250 ml MAMS medium and either 25 mM methanol or 1 mM DMS as the carbon source. Multiple cultures were grown on DMS and repeatedly respiked with DMS in order to obtain enough biomass for oxygen electrode experiments with DMS-grown cells. Cells were harvested by centrifugation at approximately 10,000 × g (15°C, 20 min) in a Beckman centrifuge using a JA-10 rotor and resuspended in 50 ml of sterile MAMS medium. The harvested cells were incubated for 2 h on a shaking incubator as described above before being used for the measurement of substrate-induced oxygen uptake rates, using a Clark-type oxygen electrode (Rank Brothers, Bottisham, United Kingdom) and a cell volume of 2 ml. The assay temperature was kept constant at 25°C by using a recirculating water bath. Substrates were added by using gas-tight syringes from concentrated stock solutions. Signals were recorded with a Philips PM8521A one-line recorder.
Analysis of polypeptides by SDS-PAGE, mass spectrometry, and N-terminal sequencing.
Methylophaga sp. strain DMS010 was grown on methanol (25 mM) and DMS (1 mM), and the biomass was harvested by centrifugation at 17,700 × g using a JA-10 rotor in a Beckman centrifuge at 4°C for 20 min. SDS-PAGE analysis of biomass from methanol- and DMS-grown Methylophaga sp. strain DMS010 was carried out using various percentages of acrylamide/bis-acrylamide as described previously (44). Polypeptide bands were excised from the gels and analyzed by mass spectrometry using matrix-assisted laser desorption ionization-mass spectrometry and in-line electrospray ionization tandem mass spectrometry at the Biological Mass Spectrometry and Proteomics Facility, Department of Biological Sciences, University of Warwick, as described previously (44). For N-terminal sequencing, SDS-PAGE gels were electroblotted onto polyvinylidene difluoride membrane (Amersham, United Kingdom) using a Novex Xcell blot module (Invitrogen) following the manufacturer's instructions. Blots were stained with Ponceau S (0.1% [wt/vol] in 1% [vol/vol] acetic acid), briefly rinsed in sterile deionized water, and air dried before target bands were cut out for N-terminal sequence analysis at Alta Bioscience (Birmingham, United Kingdom).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been deposited in the EMBL Nucleotide Sequence Database under accession numbers DQ660911 to DQ660973.
RESULTS
Enrichment of DMS-degrading bacteria.
Twenty-four DMS-degrading enrichment cultures were established from enrichments inoculated with E. huxleyi cultures, from pooled methyl halide enrichments, from filters with biomass retained from seawater obtained from Achmelvich Bay (NW Scotland), a rock pool in the seaweed-colonized intertidal zone from the Isle of Skye (NW Scotland), and from sampling station L4, which is situated 10 miles offshore from Plymouth in the English Channel. These cultures had an initial DMS concentration of 50 μM and generally depleted the headspace of DMS completely within 2 weeks of inoculation. Enrichment cultures on carbon sources other than DMS, i.e., formate, methylamine, and methanol (used to preenrich methylotrophic bacteria), showed a slight increase in turbidity (OD was increased but kept below 0.1). Enrichment cultures initially amended with substrates other than DMS were subcultured (10% inoculum) and amended with 50 μM DMS. Subcultured methanol enrichments from E. huxleyi cultures, pooled methyl halide enrichments, and Achmelvich Bay and the subculture of the formate enrichment from the rock pool depleted an initial addition of DMS (50 μM) and were given further additions of DMS to increase biomass. This was done to avoid potential toxicity of higher DMS concentrations. All other subcultures did not oxidize DMS and were not analyzed further. DMS-degrading enrichment cultures were also established with samples from the English Channel and included samples initially amended with methanol, acrylate, and thiosulfate.
PCR-DGGE analysis of DMS enrichment cultures and sequencing of DGGE bands.
DGGE analysis (Fig. 1) showed all enrichment cultures to be mixed cultures with common DGGE bands between enrichments obtained from the same sample. Several predominant bands were identical between DGGE profiles of enrichments obtained from different samples. E. huxleyi isolate-derived DMS-enrichment cultures had almost identical electrophoretic patterns, with a common dominant band observed for genetic fingerprints of all cultures. The DGGE profiles of samples from Achmelvich Bay and the rock pool that had been exposed to 500 μM DMS also had similar predominant bands.
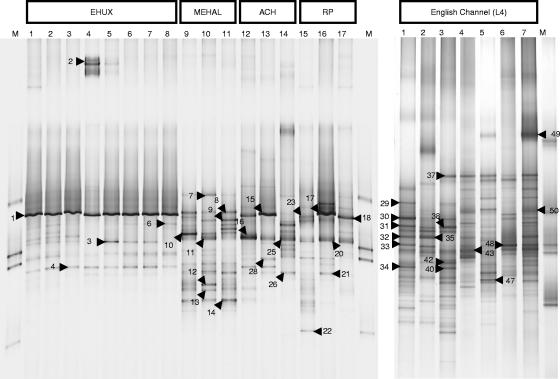
FIG. 1.
Negative images of SYBR green-stained DGGE gels showing the profiles of PCR products obtained from DMS-degrading enrichment cultures. Numbered arrows (top) indicate bands that were sequenced (see Table 1). (Left panel) M, marker; 1, Emiliania huxleyi strain 1516 culture on 50 μM DMS subculture of methanol enrichment; 2, E. huxleyi strain culture on 50 μM DMS; 3, E. huxleyi strain 500 μM DMS subculture of 50 μM DMS enrichment; 4, pooled E. huxleyi strains under conditions of increasing DMS concentration (50 μM to 250 μM); 5, 250 μM subculture of pooled E. huxleyi with increasing DMS concentration; 6, pooled E. huxleyi 50 μM DMS; 7, pooled E. huxleyi 500 μM DMS subculture of 50 μM DMS; 8, pooled E. huxleyi 50 μM DMS subculture of methanol enrichment; 9, pooled methyl halide enrichment 50 μM DMS; 10, pooled methyl halide enrichment 500 μM DMS subculture of 50 μM culture; 11, pooled methyl halide enrichment 50 μM DMS subculture of methanol enrichment; 12, Achmelvich Bay (filter) 50 μM DMS; 13, Achmelvich Bay 500 μM DMS subculture of 50 μM DMS culture; 14, Achmelvich Bay 50 μM DMS subculture of methanol enrichment; 15, rock pool (filter) 50 μM DMS; 16, rock pool 500 μM DMS subculture of 50 μM DMS culture; 17, rock pool 50 μM DMS subculture of formate enrichment; M, molecular marker. (Right panel) DGGE analysis of DMS-degrading enrichment cultures derived from samples from the English Channel (L4). Lane 1, sample taken Nov 2004 preenriched on formate; 2, sample taken Nov 2004 preenriched on acrylate; 3, sample taken Nov 2004 50 μM DMS; 4, sample taken May 2004 50 μM DMS; 5, sample taken July 2004 50 μM DMS; 6, sample taken May 2004 50 μM DMS and 4 mM bicarbonate; 7, sample taken May 2004 (50 μM DMS, 4 mM bicarbonate, 2.5 mM thiosulfate); M, marker.
Sequencing of DGGE bands indicated that the populations present in the enrichment cultures were from a wide phylogenetic range, including members of the classes Alpha-, Beta-, Gamma- and Deltaproteobacteria, the phyla Bacteroidetes, Firmicutes and Actinobacteria (Table 1), and further sequences of uncertain affiliation. E. huxleyi isolate-derived enrichment cultures appeared to be dominated by Methylophaga, e.g., as shown in Table 1, band 1 and comigrating bands, and a less dominant band affiliated with the Roseobacter clade (Table 1, band 4) was also present. Some of the E. huxleyi isolate-derived enrichment cultures also contained members of the family Flavobacteriaceae (Table 1, band 2), Alcanivorax sp.-related populations (Table 1, band 3), and relatives of other unclassified Gammaproteobacteria (Table 1, band 6, clone 2).
TABLE 1.
Summary of DGGE band sequencing analysis
| Sample and band no. | Enrichment conditions | Genus affiliation (RDP II classifier)a | Best database match (GenBank accession no.)b | Identity (%)c | Characteristic/origin of closest database hitd | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Emiliania huxleyi cultures | ||||||||||
| 1 | Strain 1516, methanol preenrichment, 50 μM DMS | Methylophaga (Gammaproteobacteria) | Methylophaga thalassica (X95460) | 537/548 (97) | Marine methylotrophic bacterium | |||||
| 4 | Strain 1516, 50 μM DMS, then subcultured with 500 μM DMS | Unclassified Rhodobacteraceae | Uncultured bacterium clone E4aB11 (DQ103616) | 553/559 (98) | Hypersaline endoevaporitic microbial mat | |||||
| 2 | Pooled strains, increasing DMS concn | Unclassified Flavobacteriaceae | Uncultured Flavobacteria bacterium clone V1_026 (AY907285) | 495/499 (99) | Culture of Thalassiosira rotula 04 | |||||
| 3 | Pooled strains, increasing DMS concn | Alcanivorax (Gammaproteobacteria) | Fundibacter jadensis (AJ001150) | 581/586 (99) | Intertidal North Sea sediment; hydrocarbon degrader | |||||
| 6, clone 1 | Pooled strains, methanol preenrichment, 50 μM DMS | Methylophaga (Gammaproteobacteria) | Methylophaga thalassica (X95460) | 573/586 (97) | Marine methylotrophic bacterium | |||||
| 6, clone 2 | Pooled strains, methanol preenrichment, 50 μM DMS | Unclassified Gammaproteobacteria | Uncultivated gammaproteobacterium clone YC499B15_AB (AY701432) | 448/482 (92) | Culture of Gymnodinium catenatum YC499B15 | |||||
| Pooled methyl halide enrichments | ||||||||||
| 10 | 50 μM DMS | Unclassified Betaproteobacteria | Uncultured betaproteobacterium clone 139_16sGbFL1 (AY786220) | 521/534 (97) | Guaymas Basin background deep water | |||||
| 7 | 50 μM DMS, then subcultured with 500 μM DMS | Unclassified bacteria | Uncultured proteobacterium clone IAFDn69 (AY090126) | 289/290 (99) | Marine methanol-fed denitrification bioreactor | |||||
| 11 | 50 μM DMS, then subcultured with 500 μM DMS | Alcanivorax (Gammaproteobacteria) | Alcanivorax sp. strain DG813 (AY258105) | 518/535 (96) | Culture of Gymnodinium catenatum | |||||
| 12 | 50 μM DMS, then subcultured with 500 μM DMS | Unclassified Myxococcales | Uncultured bacterium clone BG.c4 (DQ228369) | 533/570 (93) | Bench Glacier | |||||
| 13 | 50 μM DMS, then subcultured with 500 μM DMS | Unclassified Sphingomonadaceae | Uncultured forest soil bacterium clone DUNssu153 (AY913360) | 490/506 (96) | 0-20-cm bulk soil, Duennwald forest | |||||
| 8 | Methanol preenrichment, then 50 μM DMS | Unclassified bacteria | Unidentified bacterium clone 1959 (AF097803) | 539/549 (98) | Activated sludge | |||||
| 9 | Methanol preenrichment, then 50 μM DMS | Sphingopyxis | Sphingomonas sp. strain SA-3 (AF327069) | 524/526 (99) | ||||||
| 14 | Methanol preenrichment, then 50 μM DMS | Unclassified Alphaproteobacteria | Alphaproteobacterium strain AP-25 (AY145562) | 451/474 (95) | Dilution culture (10−6) from marine section of Weser estuary | |||||
| Achmelvich Bay | ||||||||||
| 16 | 50 μM DMS | Alcanivorax (Gammaproteobacteria) | Alcanivorax sp. strain DG813 (AY258105) | 533/533 (100) | Culture of Gymnodinium catenatum | |||||
| 15 | 50 μM DMS, then subcultured with 500 μM DMS | Methylophaga (Gammaproteobacteria) | Methylophaga thalassica (X95460) | 518/529 (97) | Marine methylotrophic bacterium | |||||
| 25 | Methanol preenrichment, then 50 μM DMS | Alcanivorax (Gammaproteobacteria) | Alcanivorax sp. strain DG813 (AY258105) | 584/586 (99) | Culture of Gymnodinium catenatum | |||||
| 26 | Methanol preenrichment, then 50 μM DMS | Unclassified Alphaproteobacteria | Marine bacterium SCRIPPS_94 (AF359545) | 557/557 (100) | Culture of Scrippsiella trochoidea NEPCC 15 | |||||
| Coral Beach, rock pool | ||||||||||
| 17 | 50 μM DMS, then subcultured with 500 μM DMS | Methylophaga (Gammaproteobacteria) | Methylophaga thalassica (X95460) | 571/584 (97) | Marine methylotrophic bacterium | |||||
| 18 | Formate preenrichment, then 50 μM DMS | Methylophaga (Gammaproteobacteria) | Methylophaga thalassica (X95460) | 506/534 (94) | Marine methylotrophic bacterium | |||||
| 20 | 50 μM DMS, then subcultured with 500 μM DMS | Alcanivorax (Gammaproteobacteria) | Alcanivorax sp. strain DG813 (AY258105) | 532/532 (100) | Culture of Gymnodinium catenatum | |||||
| 21 | 50 μM DMS | Unclassified Alphaproteobacteria | Marine bacterium SCRIPPS_94 (AF359545) | 510/510 (100) | Culture of Scrippsiella trochoidea NEPCC 15 | |||||
| 22 | 50 μM DMS | Unclassified bacteria | Uncultured Actinomycetales bacterium (DQ228712) | 414/445 (93) | Cave rock | |||||
| English Channel | ||||||||||
| 29 | Nov 2004, formate preenrichment, 50 μM DMS | Sphingobacteria (Bacteroidetes) | Uncultured bacterium clone 72-ORF19 (DQ376575) | 407/423 (96) | Aerobic sequencing batch reactor | |||||
| 30 | Nov 2004, formate preenrichment, 50 μM DMS | Methylophaga (Gammaproteobacteria) | Uncultured Methylophaga sp. clone JL-ECS-X17 (AY663963) | 525/536 (97) | East China Sea | |||||
| 31 | Nov 2004, formate preenrichment, 50 μM DMS | Gammaproteobacteria | Uncultured bacterium clone WIM-Mm-3 (AY309182) | 352/385 (91) | Peat | |||||
| 32 | Nov 2004, formate preenrichment, 50 μM DMS | Alcanivorax (Gammaproteobacteria) | Alcanivorax sp. strain DG813 (AY258105) | 513/535 (95)e | Culture of Gymnodinium catenatum | |||||
| 33 | Nov 2004, formate preenrichment, 50 μM DMS | Rhodobacteraceae (Alphaproteobacteria) | Uncultured bacterium clone ELB16-059 (DQ015815) | 500/504 (99) | Lake Bonney (Antarctica) | |||||
| 34 | Nov 2004, formate preenrichment, 50 μM DMS | Clostridia (Firmicutes) | Uncultured bacterium clone s101 (AY171344) | 482/495 (97) | Marine sediment | |||||
| 35 | Nov 2004, acrylate preenrichment, 50 μM DMS | Alcanivorax (Gammaproteobacteria) | Alcanivorax sp. strain DG813 (AY258105) | 484/484 (100) | Culture of Gymnodinium catenatum | |||||
| 37 | Nov 2004, 50 μM DMS | Bacteroidetes | Uncultured alphaproteobacterium clone 131637 (AY922203) | 527/528 (99) | Gray whale bone, Pacific Ocean | |||||
| 38 | Nov 2004, 50 μM DMS | Gammaproteobacteria | Uncultured bacterium clone WIM-Mm-3 (AY309182) | 352/385 (91) | Peat | |||||
| 40 | Nov 2004, 50 μM DMS | Clostridiales (Firmicutes) | Uncultured bacterium clone s101 (AY171344) | 503/513 (98) | Marine sediment | |||||
| 42 | Nov 2004, 50 μM DMS | Alphaproteobacteria | Uncultured alphaproteobacterium clone P_wp0211 (AY186195) | 501/502 (99) | Deep-sea sediment | |||||
| 43 | May 2005, 50 μM DMS | Alphaproteobacteria | Kordiimonas gwangyangensis strain GW14-5 (AY682384) | 417/450 (92) | Degrader of polycyclic aromatic hydrocarbons | |||||
| 47 | May 2005, 50 μM DMS, 4 mM bicarbonate, 2.5 mM thiosulfate | Erythrobacter (Alphaproteobacteria) | Uncultured Erythrobacter sp. clone JL-ETNP-Y43 (AY726907) | 457/458 (99) | 1,000-m depth of tropical eastern North Pacific | |||||
| 48 | May 2005, 50 μM DMS, 4 mM bicarbonate | Rhodobacteraceae (Alphaproteobacteria) | Uncultured bacterium clone ELB16-059 (DQ015815) | 505/510 (99) | Lake Bonney (Antarctica) | |||||
| 49 | May 2005, 50 μM DMS, 4 mM bicarbonate, 2.5 mM thiosulfate | Gelidibacter (Bacteroidetes) | Uncultured CFB group bacterium clone DBS2 (AF466705) | 513/515 (99) | Sludge sample of a digestion basin of a mariculture system | |||||
| 50 | May 2005, 50 μM DMS, 4 mM bicarbonate, 2.5 mM thiosulfate | Piscirickettsiaceae (Gammaproteobacteria) | Uncultured gammaproteobacterium clone HMMVCen-15 (AJ704664) | 474/483 (98) | Marine sediment of Haakon Mosby mud volcano | |||||
Using the program “classifier” at the Ribosomal Database Project II (RDP II) website (http://rdp.cme.msu.edu/classifier/classifier.jsp) and the default confidence threshold of 80%.
Using BLASTn against NR database at NCBI.
Identities according to BLASTn output; gaps were ignored.
Source data based on GenBank entry.
Poor sequence quality.
DMS-degrading enrichment cultures established from pooled methyl halide-degrading enrichments harbored a variety of phylotypes; however, none of the dominant DGGE bands was related to Methylophaga. Sequencing showed that in DGGE profiles of these enrichments, the bands migrating to positions close to those identified as Methylophaga in other DGGE profiles were related to Sphingopyxis spp. (Table 1, band 9) or other unclassified bacteria (Table 1, band 8, GenBank accession number AF097803, clone 1959 from activated sludge). Other dominant DGGE bands in these enrichments were identified as Alcanivorax spp. (Table 1, band 11), members of the family Sphingomonadaceae (band 13), members of the order Myxococcales (Table 1, band 12), an unclassified alphaproteobacterium (Table 1, band 14), an unclassified betaproteobacterium (Table 1, band 10), and other unclassified bacteria (Table 1, bands 7 and 8).
DMS-degrading enrichment cultures from Scottish coastal seawater samples, Achmelvich Bay, and the rock pool shared a number of phylotypes related to Methylophaga (Achmelvich, Table 1, band 15; rock pool, Table 1, bands 17 and 18), Alcanivorax (Achmelvich, Table 1, bands 15 and 25; rock pool, Table 1, band 20), and bacterial 16S rRNA genes identical to those of SCRIPPS_94, a sequence type identified in cultures of Scrippsiella sp. algae (Achmelvich, Table 1, band 26; rock pool, Table 1, band 21). In addition, one of the rock pool enrichments (50 μM DMS) contained a population related to an uncultured Actinomycetales bacterium (Table 1, band 22).
DGGE profiles of enrichments with samples from the English Channel (L4) had a higher number of bands than those from other samples. Affiliation of the sequences from dominant bands included Methylophaga (Table 1, band 30), a Gammaproteobacteria clade related to Methylophaga found in methane-rich environments (Table 1, band 50, enrichment with DMS, bicarbonate and thiosulfate; the best BLAST hit was clone HMMVCen-15, accession number AJ704664; T. Loesekann, T. Nadalig, H. Niemann, K. Knittel, A. Boetius, and R. Amann, unpublished data), Alcanivorax, members of the phyla Bacteroidetes and Firmicutes, members of the Roseobacter group, and Erythrobacter-like bacteria.
Isolation of bacterial strains from DMS enrichment cultures.
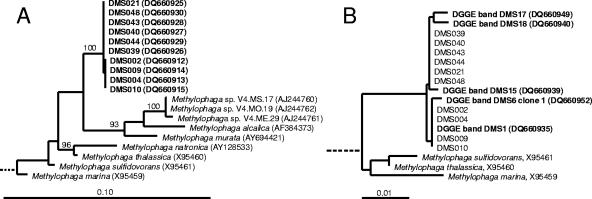
Twenty-four isolates were obtained from the enrichment cultures. These belonged to classes Alpha- and Gammaproteobacteria and to the Actinobacteria phylum. The identity of the isolates obtained by sequencing of 16S rRNA genes and the results of growth experiments with DMS are summarized in Table 2. The relationship of Methylophaga isolates to DGGE band sequences and other Methylophaga species is shown in Fig. 2. The sequences obtained from DGGE bands were all identical to those of Methylophaga strains isolated in this study, except for a few positions of sequence ambiguity. PCR products suitable for DGGE analysis obtained from Methylophaga isolates DMS002, DMS004, DMS009, and DMS010 comigrated with DGGE bands from enrichments cultures identified as Methylophaga populations (results not shown). Of the isolates that were obtained from DMS and methanol enrichment cultures, those related to the genera Methylophaga, Marinobacter, and Glaciecola were capable of oxidizing two consecutive additions of DMS (50 μM). Other cultures did not deplete the headspace of DMS. Growth of Methylophaga isolates DMS002, DMS004, DMS009, and DMS010 on DMS at concentrations of 500 μM and 1 mM was concomitant with an increase in optical density of liquid cultures (data not shown). Unlike the Methylophaga isolates, the Marinobacter and Glaciecola strains did not grow on DMS (50 μM or 500 μM). A number of cultures related to the Roseobacter clade were also tested for DMS oxidation. Leisingera methylohalidivorans strain MB2 has been reported previously to grow on DMS to a limited extent (34, 43); however, GC measurements of headspace concentrations of DMS in this study did not show any evidence of DMS degradation (50 μM). Other Roseobacter group isolates that were tested for DMS oxidation did not degrade DMS (50 μM) either, including the methyl halide-degrading strains 179, 198, and Roseovarius sp. strain 217 (44) and Ruegeria algicola (strain FF3), Roseovarius tolerans (DSM 11457), R. nubinhibens (DSM 15170), R. crassostreae (DSM16950), and R. mucosus (DSM 17069).
TABLE 2.
Properties of isolates obtained in this study
| Strain | Inoculum for enrichment and substrate | DMS oxidation | Phylogenetic group | Closest cultured relative | % Identity |
|---|---|---|---|---|---|
| DMS001 | Emiliania huxleyi (pooled) 50-250 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 98 |
| DMS002 | Emiliania huxleyi (pooled) 50-250 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS003 | Emiliania huxleyi (pooled) 50-250 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 98 |
| DMS004 | Emiliania huxleyi (pooled) 50-250 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS006 | Emiliania huxleyi (pooled) 50-250 μM DMS | − | Alphaproteobacteria | Sphingopyxis flavimaris | 99 |
| DMS007 | Emiliania huxleyi (pooled) 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 98 |
| DMS009 | Emiliania huxleyi (pooled) 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS010 | Emiliania huxleyi (pooled) 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS011 | Emiliania huxleyi (pooled) 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 98 |
| DMS012 | Emiliania huxleyi (pooled) 50 μM DMS | − | Alphaproteobacteria | Stappia stellulata | 99 |
| DMS021 | Coral Beach rock pool 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 98 |
| DMS025 | English Channel, L4 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 98 |
| DMS026 | English Channel, L4 50 μM DMS | − | Alphaproteobacteria | Ruegeria algicola | 97 |
| DMS028 | English Channel, L4 50 μM DMS | − | Actinobacteria | Microbacterium schleiferi | 99 |
| MeOH030 | Emiliania. huxleyi (pool) 5 mM methanol | − | Alphaproteobacteria | Sphingopyxis flavimaris | 99 |
| DMS039 | Achmelvich Bay 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS040 | Achmelvich Bay 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS043 | Achmelvich Bay 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS044 | Achmelvich Bay 50 μM DMS | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS048 | Rock pool 50 μM DMS (formate preenriched) | + | Gammaproteobacteria | Methylophaga thalassica | 97 |
| DMS049 | English Channel (May 2005), L4 50 μM DMS | + | Gammaproteobacteria | Glaciecola mesophila | 99 |
| DMS050 | English Channel (May 2005), L4 50 μM DMS | + | Gammaproteobacteria | Marinobacter sp. strain Splume3.1825c | 99 |
| DMS052 | English Channel (May 2005), L4 50 μM DMS | − | Actinobacteria | Streptomyces sodiiphilus | 97 |
| DMS054 | English Channel (May 2005), L4 50 μM DMS | + | Gammaproteobacteria | Marinobacter sp. strain Splume3.1825c | 99 |
FIG. 2.
(A) Maximum-likelihood tree based on 16S rRNA gene sequences showing the relationship of Methylophaga isolates obtained in this study (prefixed DMS) to other Methylophaga species. The outgroup consisted of nine 16S rRNA gene sequences representing other genera of Gammaproteobacteria and is not shown. Partial sequences of isolates DMS001, DMS003, DMS007, DMS011, and DMS025 were identical to those of strains DMS002, DMS004, DMS009, and DMS010 and were not added to the tree. Bootstrap values were determined using PHYLIP programs (see Materials and Methods); only values above 90% are shown. The scale bar corresponds to 10% sequence divergence. (B) Neighbor-joining tree showing the relationship of partial 16S rRNA gene sequences obtained by sequencing DGGE bands (only those related to Methylophaga) with Methylophaga strains isolated from enrichment cultures. The scale bar indicates 1% sequence divergence.
BOX-PCR fingerprinting of Methylophaga isolates.
Four different electrophoretic patterns were obtained for BOX-PCR products from the nine closely related strains of Methylophaga which had maximum differences of one nucleotide in 16S rRNA gene sequences (result not shown). This demonstrated that these isolates that belonged to the same phylogenetic cluster based on 16S rRNA gene sequence data corresponded to at least four genetically different populations. One group of strains with identical BOX-PCR fingerprints consisted of strains DMS002, DMS004, DMS009, and DMS010; DMS021 had a unique pattern. Furthermore, patterns obtained for strains DMS039 and DMS040 were identical, as were BOX-PCR patterns of strains DMS043 and DMS044.
Effect of inhibitors on DMS oxidation by Methylophaga sp. strain DMS010.
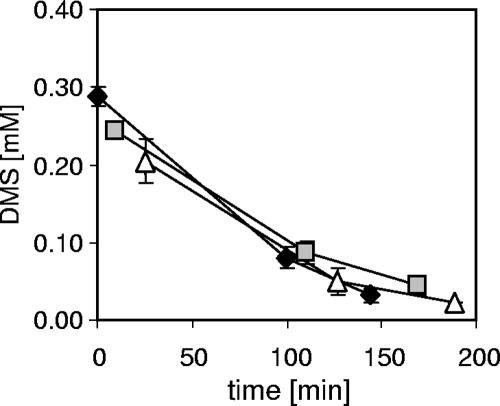
The effect of chloroform and of MTBE on DMS oxidation by Methylophaga sp. strain DMS010 was tested with DMS-grown cells; however, neither chloroform nor MTBE addition affected DMS oxidation rates compared to those of inhibitor-free controls (Fig. 3).
FIG. 3.
Effect of chloroform and methyl tert-butyl ether on the degradation of DMS by Methylophaga sp. strain DMS010. The DMS concentrations in the headspace were determined by GC; values are means of triplicate samples. DMS headspace concentrations in chemical control incubations in the presence or absence of inhibitors did not change. Symbols: diamonds, DMS only; squares, DMS and chloroform; triangles, DMS and MTBE.
Substrate-induced oxygen uptake of Methylophaga sp. strain DMS010.
DMS strongly induced oxygen uptake in Methylophaga strain DMS010 grown on DMS. However, DMS addition to methanol-grown cell suspensions failed to enhance oxygen uptake. Similarly, methanethiol also induced oxygen uptake of biomass grown on DMS even at low concentrations (5 μM), while the methanethiol-induced oxygen uptake rates of cells grown on methanol were too low to be integrated even at relatively high methanethiol concentrations (500 μM). The apparent Km values for DMS and methanethiol were derived by plotting oxygen uptake data in Eadie-Hofstee plots. The Km values for DMS and methanethiol were 2.1 and 4.6 μM, respectively; the Vmax rates were determined as 62 nmol O2 min−1 mg dry weight−1 for DMS and 114 nmol O2 min−1 mg dry weight−1 for methanethiol.
SDS-PAGE analysis and identification of DMS-induced polypeptides in Methylophaga sp. strain DMS010.
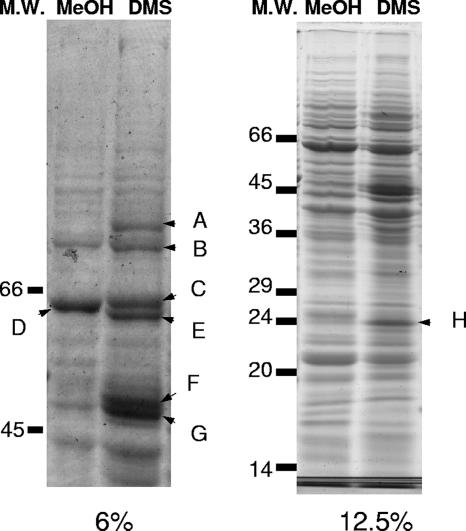
Biomass of Methylophaga sp. strain DMS010 was obtained on methanol and DMS and polypeptides induced under the two growth conditions were analyzed by SDS-PAGE analysis of crude cell extracts (Fig. 4). A number of polypeptides appeared to be more highly expressed during growth on DMS than methanol-grown cells. The results of mass spectrometry analysis of excised polypeptide bands and N-terminal sequencing are reported in Table 3. The large subunit of methanol dehydrogenase (MxaF) was identified in biomass of cells grown on methanol and DMS. However, additional polypeptide bands were observed during growth on DMS (Fig. 4). These polypeptides included transketolase, a thiol-specific alkyl hydroxyperoxide reductase, a protein tentatively identified as a homolog of proteins predicted from the genome sequences of Silicibacter pomeroyi and Methylococcus capsulatus (Bath) as selenium binding proteins, and XoxF, a homolog of MxaF. The function of XoxF in methylotrophs is unknown (8).
FIG. 4.
SDS-PAGE analysis of polypeptides induced during growth of Methylophaga sp. strain DMS010 on methanol and DMS (as indicated at the top of the lanes as MeOH and DMS, respectively). (Left panel) 6% SDS-PAGE; (right panel) 12.5% SDS-PAGE; M.W., molecular weight. Bands were excised as indicated (A to H) and analyzed by in-line electrospray ionization tandem mass spectrometry in order to generate de novo amino acid sequences. In addition, N-terminal sequencing by Edman degradation was performed for bands F, G, and H. Results of de novo and N-terminal sequencing are reported in Table 3.
TABLE 3.
Identification of polypeptides
| Banda | Approximate molecular mass (kDa) | Identificationb | De novo peptides supporting identificationc | N-terminal sequenced |
|---|---|---|---|---|
| A | >66 | No identification possible | ND | ND |
| B | >66 | Transketolase (Vibrio sp. strain MED222) | FDGPSSLVVFSR | ND |
| Transketolase (Vibrio fischeri ES114) | FPEIAAEFTR | |||
| C | 64 | Methanol dehydrogenase large subunit (MxaF) (Methylophaga sp. strain DMS010) | RFKVLEGAHASFVEK | ND |
| D | 64 | Methanol dehydrogenase large subunit | AVACCDVVNR | ND |
| (MxaF) (Methylophaga sp. strain DMS010) | LLTHPDR | |||
| NGIVYTLDR | ||||
| E | 62 | XoxF (methanol dehydrogenase large subunit- | PAVNWSNGVN(I/L)K | ND |
| like protein) | QPAAYSPR | |||
| GELLVAEK | ||||
| F | 50 | Putative selenium binding protein [Silicibacter pomeroyi DSS-3; Methylococcus capsulatus (Bath)] | YLWAGGLDTSK | DET(C?)MSPYMAKISGQe |
| G | 48 | No identification possible | ND | No data; peptide may be blocked N terminally |
| H | 24 | Alkyl hydroperoxide reductase C thiol specific | EINDLGIGR | STLINTEIKPFKTTAf |
Band as labeled in Fig. 5.
Identification based on hits with the in-house database (containing partial methanol dehydrogenase large subunit gene sequences of Methylophaga isolates) and BLASTp searches (using the “search for short nearly exact matches” option).
Amino acid sequences obtained by in-line electrospray ionization tandem mass spectrometry that supported the identification, single-letter amino acid code. ND, not determined.
N-terminal amino acid sequence (single-letter amino acid code). ND, not determined.
No amino acid was detected at position 4, which may be due to a cysteine residue at this position.
N-terminal sequence was obtained from a Western blot of a rerun of the sample on another 12.5% SDS-PAGE gel (result not shown).
DISCUSSION
Despite the global importance of microbially mediated DMS degradation, surprisingly few studies have addressed by enrichment and isolation the identity of marine bacteria capable of using DMS as a carbon source in the marine water column. In previous studies, marine bacterial isolates growing on DMS were of undetermined phylogeny (18, 20, 52), or isolates studied for organosulfur compound transformation were isolated from DMSP enrichments and chosen on the basis of their affiliation with marine Alphaproteobacteria (16). Therefore, there was a clear need to isolate and identify marine bacteria that are able to degrade DMS.
Diversity of DMS-degrading enrichment cultures.
DGGE analysis suggested that the dominant populations in many of the DMS-degrading enrichments were related to Methylophaga and Alcanivorax species. The identity of 16S rRNA gene sequences of Methylophaga DGGE bands and Methylophaga isolates from the same enrichments indicated that the strains represented the populations growing in the enrichment cultures. Some phylotypes (Fig. 1, bands 7 and 50) were present in enrichments that had as their closest relatives sequences obtained from environments characterized by the turnover of one-carbon substrates, e.g., a marine methanol-fed bioreactor (29) and the methane-rich sediments of the Haakon Mosby mud volcano (GenBank accession number AJ704664; Loesekann et al., unpublished data), suggesting their potential involvement in the turnover of DMS or intermediates of C1 metabolism. Other DGGE band sequences (e.g., Fig. 1, bands 2 and 6, clone 2, bands 16, 20, 21, 25, 26, 32, and 35) had the highest similarities to those of phylotypes detected in cultures of a variety of marine phytoplankton, especially those from dinoflagellates, which are key producers of DMSP, the precursor of DMS.
DMS degradation by isolated strains.
Of the 24 isolates that were obtained, only those identified as Methylophaga were able to grow on DMS, providing a clear link between the populations detected in enrichments and DMS degradation. Methylophaga is a genus of restricted and obligate methylotrophs (10-13, 21), i.e., obligate methylotrophic isolates that exclusively utilize C1 compounds and restricted methylotrophs that, in addition to C1 substrates, can utilize one or a few multicarbon compounds as growth substrates. Previously, Methylophaga sulfidovorans, isolated from a marine microbial mat, had been shown to degrade and grow on DMS (10). Recently, DGGE bands related to Methylophaga were also detected in marine DMS enrichments by Vila-Costa and colleagues (49); however, DMS-degrading Methylophaga isolates were not obtained in that study. Previously reported Methylophaga isolates were obtained from marine sediments or microbial mats (10, 22), and so the isolation of DMS-degrading Methylophaga strains from samples obtained from coastal water and seawater further offshore in this study demonstrates for the first time that certain Methylophaga species may also play a role in DMS oxidation in pelagic marine environments. In the current study, the presence of a Methylophaga sp. in nonaxenic cultures of E. huxleyi could indicate that Methylophaga may cooccur with E. huxleyi or other DMSP-producing phytoplankton in the environment. This is also suggested by the detection of Methylophaga sp.-related bacteria in marine mesocosms used to study bacterium-alga interactions (40) and in a culture containing the dinoflagellate Gymnodinium catenatum (GenBank accession number AY701420) (D. H. Green and C. J. S. Bolch, unpublished data). BOX-PCR demonstrated that the Methylophaga isolates obtained in this study represented at least four genetically different populations and suggested that the 16S rRNA gene sequences did not reflect the diversity at the strain level.
Other isolates that were obtained did not grow on DMS. While cell suspensions of Marinobacter and Glaciecola isolates degraded DMS (50 μM), DMS did not support the growth of these isolates. This may have been due to the utilization of DMS as a sulfur source or due to its conversion to DMSO. It is likely that additional DMS-degrading bacteria were present in these enrichments that could not be isolated with the culturing conditions used. This is concluded from the observation that DGGE analysis and sequencing of bands suggested that in some enrichments Methylophaga-related bacteria were not present.
Despite the potential of some members of the Roseobacter clade to transform organosulfur compounds such as DMSP, methanethiol, and DMS (6, 16, 38), growth on DMS is clearly not a common phenotype of Roseobacter clade bacteria. This is concluded from the observation that Leisingera methylohalidivorans, several Roseovarius isolates, Ruegeria algicola, Silicibacter pomeroyi, and the methyl halide-degrading strains 179, 198, and 217 (44), all members of the Roseobacter clade, were not able to grow on DMS. The observation that L. methylohalidivorans did not grow on DMS was similar to the findings by Schaefer and coworkers (43), who reported that the strain did not grow on 1.4 or 5 mM DMS but that it was able to increase in cell numbers on 50 μM DMS and was maintained over three subcultures. In the present study, L. methylohalidivorans did not degrade DMS, as determined by GC analysis of headspace gas, suggesting that the limited growth observed on DMS may previously have been due to trace organic constituents present in the medium or that the isolate had lost the ability to degrade DMS during serial transfer in the laboratory.
DMS metabolism of Methylophaga sp. strain DMS010.
Inhibition of DMS oxidation by MTBE and by chloroform has been used as a means to differentiate between the operation of the monooxygenase pathway and the methyltransferase pathway of DMS oxidation (20, 53). Neither MTBE nor chloroform had an inhibitory effect on DMS oxidation by strain DMS010. This was different from observations for Thiobacillus strains, in which a marked inhibition of DMS oxidation by MTBE was observed in Thiobacillus sp. strain T5, while chloroform strongly inhibited DMS oxidation by Thiobacillus sp. strain ASN-1 (53). However, strain DMS010 had a lower apparent Km (2.1 μM) for DMS than Thiobacillus sp. strain T5 (Ks of 90 μM), which opens up the possibility that the higher affinity for DMS in Methylophaga might preclude inhibition by either of the two inhibitors at relatively high DMS concentrations. Based on results with these inhibitors, a metabolic route of DMS oxidation in Methylophaga sp. strain DMS010 cannot be assigned, and so further studies of the biochemistry are essential. With Thiocapsa roseopersicina M11, aerobic DMS degradation was not inhibited by these compounds either (23). The Km for DMS of Methylophaga sp. strain DMS010 was comparable to those determined for Methylophaga sulfidovorans (Ks, 1.5 μM [10]), Thiocapsa roseopersicina M11 (Km, 2 μM [23]), and Hyphomicrobium strain EG (Ks, 3 μM [47]).
The induction of polypeptides during the growth of marine DMS-degrading isolates has not been studied previously. The role of some of the polypeptides detected in biomass of DMS-grown Methylophaga remains unknown in the absence of further genetic and biochemical data, but the peptides identified here are promising candidates for further study. The homolog of the large subunit of methanol dehydrogenase, XoxF, might have a role in the metabolism of DMS or in the degradation of the intermediate methanethiol; this role, however, will need to be investigated in future studies. Previously, mxaF′ knockout mutants of Methylobacterium extorquens (similar to xoxF) were not affected in their ability to grow on methanol or methylamine, and a phenotype associated with this gene has not yet been identified (8). Induction of a thiol-specific alkyl hydroperoxide reductase during growth on DMS may be a consequence of thiol stress due to the production of methanethiol as an intermediate of DMS metabolism.
Conclusions and outlook.
The information presented here strongly suggests that Methylophaga spp. are involved in DMS degradation in seawater and therefore may be part of the population of marine methylotrophs that has been suggested to be responsible for this biogeochemical process (26). The strains of Methylophaga obtained in this study are the first DMS-degrading isolates of this genus obtained from seawater samples. Strain DMS010 differed in its DMS metabolism from that of Thiobacillus species and unidentified isolates based on inhibition assays (20, 53). Strain DMS010 had a low apparent Ks, indicating that it may be able to degrade DMS at typical environmental concentrations (1 to 5 nM) (25) or when DMS concentrations may reach high nM concentrations during the decay of phytoplankton blooms and even μM concentrations as observed in coral mucus (5). Degradation of DMS by bacteria in the upper mixed layer of the oceans is potentially carried out by diverse bacterial populations and metabolic pathways. Clearly, 16S rRNA gene sequences obtained by culture-independent means are of limited use to predict the potential of a given population to degrade DMS, since species closely related to DMS-degrading isolates may lack the potential to degrade DMS. Strains of some species (e.g., Rhodovulum sulfidophilum and Thiocapsa roseopersicina) may also be able to carry out DMS transformations by more than one pathway (23, 36). Studying the phylogenetic and functional diversity of DMS-degrading bacteria in the marine environment will require functional genetic markers that target key enzymes of DMS degradation pathways, such as DMS monooxygenase, methyltransferases, or other enzymes. The Methylophaga strains obtained in this study provide essential model organisms with which to analyze the metabolic pathways and biochemistry of DMS oxidation and to develop functional gene markers for studying the microbial ecology of marine DMS oxidation.
Acknowledgments
I thank Willie Wilson and Declan Schroeder (Plymouth Marine Laboratory and Marine Biological Association) for provision of Emiliania huxleyi cultures and Mike Cox for culturing them at Warwick. Ian Joint and Martin Mühling from Plymouth Marine Laboratory are thanked for providing samples from L4, Sanjeev Kumar is thanked for technical assistance, and Sue Slade is thanked for mass spectrometry analysis. I am grateful to Colin Murrell and Don Kelly for support and advice and to Josh Neufeld and Rich Boden for comments on the manuscript.
This work was supported by the Natural Environment Research Council with a postdoctoral fellowship (NE/B501404/1) and a project grant (NE/C001109/1).
Footnotes
Published ahead of print on 23 February 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andreae, M. O. 1990. Ocean-atmosphere interactions in the global biogeochemical sulfur cycle. Mar. Chem. 30:1-29. [Google Scholar]
- 3.Bates, T. S., R. P. Kiene, G. V. Wolfe, P. A. Matrai, F. P. Chavez, K. R. Buck, B. W. Blomquist, and R. L. Cuhel. 1994. The cycling of sulfur in surface water of the northeast Pacific. J. Geophys. Res. 99:7835-7843. [Google Scholar]
- 4.Borodina, E., D. P. Kelly, F. A. Rainey, N. L. Ward-Rainey, and A. P. Wood. 2000. Dimethylsulfone as a growth substrate for novel methylotrophic species of Hyphomicrobium and Arthrobacter. Arch. Microbiol. 173:425-437. [DOI] [PubMed] [Google Scholar]
- 5.Broadbent, A. D., and G. B. Jones. 2004. DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Mar. Freshw. Res. 55:849-855. [Google Scholar]
- 6.Buchan, A., J. M. González, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlson, R. J., J. E. Lovelock, M. O. Andreae, and S. G. Warren. 1987. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326:655-661. [Google Scholar]
- 8.Chistoserdova, L., and M. E. Lidstrom. 1997. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology 153:1729-1736. [DOI] [PubMed] [Google Scholar]
- 9.De Bont, J. A. M., J. P. van Dijken, and W. Harder. 1981. Dimethyl sulphoxide and dimethyl sulphide as a carbon, sulphur and energy source for growth of Hyphomicrobium S. J. Gen. Microbiol. 127:315-323. [Google Scholar]
- 10.deZwart, J. M. M., P. N. Nelisse, and J. G. Kuenen. 1996. Isolation and characterization of Methylophaga sulfidovorans sp. nov.: an obligately methylotrophic, aerobic, dimethylsulfide oxidizing bacterium from a microbial mat. FEMS Microbiol. Ecol. 20:261-270. [Google Scholar]
- 11.Doronina, N. V., T. D. Darmaeva, and Y. A. Trotsenko. 2003. Methylophaga alcalica sp. nov., a novel alkaliphilic and moderately halophilic, obligately methylotrophic bacterium from an East Mongolian saline soda lake. Int. J. Syst. Evol. Microbiol. 53:223-229. [DOI] [PubMed] [Google Scholar]
- 12.Doronina, N. V., T. D. Darmaeva, and Y. A. Trotsenko. 2003. Methylophaga natronica sp. nov., a new alkaliphilic and moderately halophilic, restricted-facultatively methylotrophic bacterium from soda lake of the Southern Transbaikal region. Syst. Appl. Microbiol. 26:382-389. [DOI] [PubMed] [Google Scholar]
- 13.Doronina, N. V., T. D. Lee, E. G. Ivanova, and Y. A. Trotsenko. 2005. Methylophaga murata sp. nov.: a haloalkaliphilic aerobic methylotroph from deteriorating marble. Microbiology 74:440-447. [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. University of Washington, Seattle, WA.
- 15.Fuse, H., O. Takimura, K. Murakami, Y. Yamoaka, and T. Omori. 2000. Utilization of dimethyl sulfide as a sulfur source with the aid of light by Marinobacterium sp. strain DMS-S1. Appl. Environ. Microbiol. 66:5527-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanlon, S. P., R. A. Holt, G. R. Moore, and A. G. McEwan. 1994. Isolation and characterization of a strain of Rhodobacter sulfidophilus: a bacterium which grows autotrophically with dimethylsulfide as electron-donor. Microbiology 140:1953-1958. [Google Scholar]
- 18.Hansen, T. A., P. Quist, M. J. E. C. van der Maarel, and L. Dijkhuizen. 1993. Isolation of marine dimethylsulfide-oxidizing bacteria, p. 37-41. In G. Restelli and G. Angeletti (ed.), Dimethylsulfide: oceans, atmosphere, and climate. ECSE-EEC-EAEC, Brussels, Belgium.
- 19.Hatton, A. D., L. Darroch, and G. Malin. 2004. The role of dimethylsulphoxide in the marine biogeochemical cycle of dimethylsulphide, p. 29-55. In R. N. Gibson et al. (ed.), Oceanography and marine biology: an annual review, vol. 42. Taylor and Francis, London, United Kingdom. [Google Scholar]
- 20.Hoeft, S. E., D. R. Rogers, and P. T. Visscher. 2000. Metabolism of methyl bromide and dimethyl sulfide by marine bacteria isolated from coastal and open waters. Aquat. Microb. Ecol. 21:221-230. [Google Scholar]
- 21.Janvier, M., and P. A. Grimont. 1995. The genus Methylophaga, a new line of descent within phylogenetic branch gamma of Proteobacteria. Res. Microbiol. 146:543-550. [DOI] [PubMed] [Google Scholar]
- 22.Janvier, M., B. Regnault, and P. Grimont. 2003. Development and use of fluorescent 16S rRNA-targeted probes for the specific detection of Methylophaga species by in situ hybridization in marine sediments. Res. Microbiol. 154:483-490. [DOI] [PubMed] [Google Scholar]
- 23.Jonkers, H. M., M. Jansen, M. J. E. C. van der Maarel, and H. van Gemerden. 1999. Aerobic turnover of dimethyl sulfide by the anoxygenic phototrophic bacterium Thiocapsa roseopersicina. Arch. Microbiol. 172:150-156. [DOI] [PubMed] [Google Scholar]
- 24.Kanagawa, T., and D. P. Kelly. 1986. Breakdown of dimethyl sulfide by mixed cultures and by Thiobacillus thioparus. FEMS Microbiol. Lett. 34:13-19. [Google Scholar]
- 25.Kettle, A. J., M. O. Andreae, D. Amouroux, T. W. Andreae, T. S. Bates, H. Berresheim, H. Bingemer, R. Boniforti, M. A. J. Curran, G. R. DiTullio, G. Helas, G. B. Jones, M. D. Keller, R. P. Kiene, C. Leck, M. Levasseur, G. Malin, M. Maspero, P. Matrai, A. R. McTaggart, N. Mihalopoulos, B. C. Nguyen, A. Novo, J. P. Putaud, S. Rapsomanikis, G. Roberts, G. Schebeske, S. Sharma, R. Simo, R. Staubes, S. Turner, and G. Uher. 1999. A global database of sea surface dimethylsulfide (DMS) measurements and a procedure to predict sea surface DMS as a function of latitude, longitude, and month. Global Biogeochem. Cycles 13:399-444. [Google Scholar]
- 26.Kiene, R. P., and T. S. Bates. 1990. Biological removal of dimethyl sulfide from sea-water. Nature 345:702-705. [Google Scholar]
- 27.Kiene, R. P., L. J. Linn, and J. A. Bruton. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43:209-224. [Google Scholar]
- 28.Kiene, R. P., R. S. Oremland, A. Catena, L. G. Miller, and D. G. Capone. 1986. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl. Environ. Microbiol. 52:1037-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labbé, N., P. Juteau, S. Parent, and R. Villemur. 2003. Bacterial diversity in a marine methanol-fed denitrification reactor at the Montreal Biodome, Canada. Microb. Ecol. 46:12-21. [DOI] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 31.Lovelock, J. E., R. J. Maggs, and R. A. Rasmussen. 1972. Atmospheric dimethyl sulphide and the natural sulphur cycle. Nature 237:452-453. [Google Scholar]
- 32.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maidak, B. L., J. R. Cole, C. T. J. Parker, G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens, T., T. Heidorn, R. Pukall, M. Simon, B. J. Tindall, and T. Brinkhoff. 2006. Reclassification of Roseobacter gallaeciensis Ruiz-Ponte et al. 1998 as Phaeobacter gallaeciensis gen. nov., comb. nov., description of Phaeobacter inhibens sp. nov., reclassification of Ruegeria algicola (Lafay et al. 1995) Uchino et al. 1999 as Marinovum algicola gen. nov., comb. nov., and emended descriptions of the genera Roseobacter, Ruegeria and Leisingera. Int. J. Syst. Evol. Microbiol. 56:1293-1304. [DOI] [PubMed] [Google Scholar]
- 35.Martin, B., O. Humbert, M. Camara, E. Guenzi, J. Walker, T. Mitchell, P. Andrew, M. Prudhomme, G. Alloing, and R. Hakenbeck. 1992. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 20:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDevitt, C. A., P. Hugenholtz, G. R. Hanson, and A. G. McEwan. 2002. Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes. Mol. Microbiol. 44:1575-1587. [DOI] [PubMed] [Google Scholar]
- 37.Moosvi, S. A., I. R. McDonald, D. A. Pearce, D. P. Kelly, and A. P. Wood. 2005. Molecular detection and isolation from Antarctica of methylotrophic bacteria able to grow with methylated sulfur compounds. Syst. Appl. Microbiol. 28:541-554. [DOI] [PubMed] [Google Scholar]
- 38.Moran, M. A., J. M. González, and R. P. Kiene. 2003. Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol. J. 20:375-388. [Google Scholar]
- 39.Neufeld, J. D., and W. W. Mohn. 2005. Fluorophore-labeled primers improve the sensitivity, versatility, and normalization of denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 71:4893-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinhassi, J., M. M. Sala, H. Havskum, F. Peters, O. Guadayol, A. Malits, and C. L. Marrase. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Pol, A., H. J. M. Op den Camp, S. G. M. Mees, M. A. S. H. Kersten, and C. van der Drift. 1994. Isolation of a dimethylsulfide-utilizing Hyphomicrobium species and its application in biofiltration of polluted air. Biodegradation 5:105-112. [DOI] [PubMed] [Google Scholar]
- 41.Rademaker, J. L. W. 2003. Rep-PCR genomic fingerprint typing, p. 57-70. In A. van Belkum, B. Duim, and J. P. Hays (ed.), Experimental approaches for assessing genetic diversity among microbial pathogens. Werkgroep Epidemiologische Typering, Wageningen, The Netherlands.
- 42.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1998. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting, p. 1-26. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, supplement 3. Kluwer, Dordrecht, The Netherlands.
- 43.Schaefer, J. K., K. D. Goodwin, I. R. McDonald, J. C. Murrell, and R. S. Oremland. 2002. Leisingera methylohalidivorans gen. nov., sp. nov., a marine methylotroph that grows on methyl bromide. Int. J. Syst. Evol. Microbiol. 52:851-859. [DOI] [PubMed] [Google Scholar]
- 44.Schäfer, H., I. R. McDonald, P. D. Nightingale, and J. C. Murrell. 2005. Evidence for the presence of a CmuA methyltransferase pathway in novel marine methyl halide-oxidising bacteria. Environ. Microbiol. 7:839-852. [DOI] [PubMed] [Google Scholar]
- 45.Schäfer, H., and G. Muyzer. 2001. Denaturing gradient gel electrophoresis in marine microbial ecology, p. 425-468. In J. H. Paul (ed.), Methods in microbiology, vol. 30. Academic Press, San Diego, CA. [Google Scholar]
- 46.Sunda, W., D. J. Kieber, R. P. Kiene, and S. Huntsman. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418:317-320. [DOI] [PubMed] [Google Scholar]
- 47.Suylen, G. M. H., and J. G. Kuenen. 1986. Chemostat enrichment and isolation of Hyphomicrobium EG a dimethyl sulfide oxidizing methylotroph and reevaluation of Thiobacillus MS1. Antonie Leeuwenhoek 52:281-293. [DOI] [PubMed] [Google Scholar]
- 48.Tanimoto, Y., and F. Bak. 1994. Anaerobic degradation of methylmercaptan and dimethyl sulfide by newly isolated thermophilic sulfate-reducing bacteria. Appl. Environ. Microbiol. 60:2450-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vila-Costa, M., D. A. Del Valle, J. M. González, D. Slezak, R. P. Kiene, O. Sánchez, and R. Símo. 2006. Phylogenetic identification and metabolism of dimethylsulfide degrading marine bacteria. Environ. Microbiol. 8:2189-2200. [DOI] [PubMed] [Google Scholar]
- 50.Visscher, P. T., P. Quist, and H. van Gemerden. 1991. Methylated sulfur compounds in microbial mats: in situ concentrations and metabolism by a colorless sulfur bacterium. Appl. Environ. Microbiol. 57:1758-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visscher, P. T., and B. F. Taylor. 1993. Aerobic and anaerobic degradation of a range of alkyl sulfides by a denitrifying marine bacterium. Appl. Environ. Microbiol. 59:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visscher, P. T., and B. F. Taylor. 1994. Demethylation of dimethylsulfoniopropionate to 3-mercaptopropionate by an aerobic marine bacterium. Appl. Environ. Microbiol. 60:4617-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visscher, P. T., and B. F. Taylor. 1993. A new mechanism for the aerobic catabolism of dimethyl sulfide. Appl. Environ. Microbiol. 59:3784-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoch, D. C. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68:5804-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeyer, J., P. Eicher, S. G. Wakeham, and R. P. Schwarzenbach. 1987. Oxidation of dimethyl sulfide to dimethyl sulfoxide by phototrophic bacteria. Appl. Environ. Microbiol. 53:2026-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2002. Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep-Sea Res. Part II 49:3017-3038. [Google Scholar]