Abstract
For quantification of bacterial adherence to biomaterial surfaces or to other surfaces prone to biofouling, there is a need for methods that allow a comparative analysis of small material specimens. A new method for quantification of surface-attached biotinylated bacteria was established by in situ detection with fluorescence-labeled avidin-D. This method was evaluated utilizing a silicon wafer model system to monitor the influences of surface wettability and roughness on bacterial adhesion. Furthermore, the effects of protein preadsorption from serum, saliva, human serum albumin, and fibronectin were investigated. Streptococcus gordonii, Streptococcus mitis, and Staphylococcus aureus were chosen as model organisms because of their differing adhesion properties and their clinical relevance. To verify the results obtained by this new technique, scanning electron microscopy and agar replica plating were employed. Oxidized and poly(ethylene glycol)-modified silicon wafers were found to be more resistant to bacterial adhesion than wafers coated with hydrocarbon and fluorocarbon moieties. Roughening of the chemically modified surfaces resulted in an overall increase in bacterial attachment. Preadsorption of proteins affected bacterial adherence but did not fully abolish the influence of the original surface chemistry. However, in certain instances, mostly with saliva or serum, masking of the underlying surface chemistry became evident. The new bacterial overlay method allowed a reliable quantification of surface-attached bacteria and could hence be employed for measuring bacterial adherence on material specimens in a variety of applications.
Colonization of material surfaces by bacteria is an unwanted event in many medical applications and also in situations where materials become subject to biofouling (16). Therefore, the evaluation of bacterial adherence to existent materials and to newly developed experimental materials or material coatings is of great importance. A number of techniques for the quantification of attached bacteria have been described, including in situ visualization by epifluorescence microscopy, confocal laser scanning microscopy, scanning electron microscopy (SEM), and atomic force microscopy. Also, indirect techniques for quantification of bacteria after removal from the surface, such as scintillation counting of radiolabeled organisms, quantification by metabolites (5-cyano-2,3-ditolyltetrazolium chloride), and enumeration by use of a Coulter Counter, by flow cytometry (fluorescence-activated cell sorting), or by spectrophotometry (1), are available. A simple method for estimation of the extent of bacterial colonization is provided by imprinting the colonized material surface onto agar growth media (replica plating technique), followed by visual comparison of colony densities (10). Although microscopic methods provide the most direct determination of attached bacteria, counting of organisms is a meticulous task due to the small sections that can be observed at once by the required high-magnifying optics. On the other hand, detachment of bacteria, which is a prerequisite for indirect counting methods, is difficult to control, because the number of bacteria that cannot be removed from the surface or become damaged remains unknown. In addition, most of these methods are not well suited for analyzing many parallel small samples, a feature that would be desirable for testing different biomaterials for their repulsive capacities on microbial colonization.
Bacterial adhesion is commonly assumed to depend on a variety of material-intrinsic parameters, including surface chemistry and topography. But in most in vivo situations, particularly in the case of medical biomaterials, bacteria encounter a surface that has already been covered by a layer of adsorbed proteins from the surrounding body fluids. This layer by itself is also assumed to be influenced in terms of quantity, composition, and structure by the physicochemical properties of the underlying material surfaces (37). Extensive investigation of salivary proteins adsorbed on hydroxyapatite (19) or on prosthetic materials (4) suggests that such a proteinaceous film may serve as a substratum for bacterial attachment by providing specific receptor sites for bacterial adhesins (33). Once the early colonizing bacteria are firmly attached, the formation of a biofilm is governed by subsequent cell division, bacterial interspecies coadhesion, coaggregation, and the production of a slimy extracellular matrix (29). Although formation of a bacterial biofilm that fosters increased resistance to antibiotics may be the ultimate reason for biomaterial-related infections (16, 34), it is still the very early events of bacterial colonization that are the decisive determinants of the fate of the biomaterial in the body.
Because initial bacterial adhesion in vivo is influenced by a multitude of factors that cannot be well controlled experimentally, in vitro model systems have been established in the past to better observe the isolated effects of certain parameters on the very early events of bacterial adhesion in a controlled environment. Glass surfaces have been widely utilized as model surfaces to study bacterial adhesion in vitro under stationary or flow chamber conditions (3, 5, 30). Glass offers good possibilities for alteration of the surface properties through chemical modification by silanization techniques, but most real biomaterials differ from glass in that they are not translucent, thereby preventing enumeration of attached bacteria by optical techniques. Silicon wafers are nontranslucent and are equally well suited as a model surface because they provide a simple and highly smooth model surface that can be modified in a controlled manner by established silane chemistry and can be roughened by sandblasting (21, 36, 42).
The aim of this investigation was to evaluate a new technique for in situ detection and quantification of surface-attached bacteria that can be performed simultaneously on many parallel small, nontranslucent material samples. This technique, which is based on a modification of the bacterial overlay method (24, 32), was utilized in the silicon wafer model system and was validated by monitoring the influences of surface wettability, roughness, and protein preadsorption on bacterial adhesion. The results were verified by comparison with SEM and agar replica plating. Three clinically relevant bacterial strains exhibiting quite different adhesion properties were chosen for this model system, namely, Streptococcus mitis 49456 and Streptococcus gordonii DL1, which are known as early colonizers on oral surfaces (12, 20), and Staphylococcus aureus 25923 as an example of an ubiquitous bacterial species that is known to be a common cause of implant infections (43). Since most implants are exposed to serum or serum-like fluids (28), and dental biomaterials are exposed to saliva (9, 13), these two complex body fluids were exemplarily chosen to include the influence of protein preadsorption on subsequent bacterial adhesion.
MATERIALS AND METHODS
Modification of silicon wafers.
Single-sided, polished silicon wafers (n type; phosphor doped; 1- to 10-Ωcm resistivity; 575- to 675-μm thickness) were purchased from Wafer World Inc. The wafers were cut into specimens of 10 mm by 10 mm by Disco GmbH (Germany). Wafer roughening was performed by abrasive blasting of the surfaces with corundum particles with an average size of 50 μm (Harnisch & Rieth, Germany). The abrasive was applied with a working pressure of 2 × 105 Pa by a PG 360/3 sand-blasting machine from Harnisch & Rieth. Here, the nozzle was moved by hand at a constant distance of about 3 cm under an angle of about 30° to the wafer surface for about 30 s per wafer. Measurement of surface roughness using a S6P perthometer from Mahr GmbH (Germany) revealed an arithmetic average peak-to-valley value of 1.72 (range, 1.35 to 1.99) μm for the sandblasted surfaces, compared with 0.04 (range, 0.04 to 0.04) μm for the smooth surfaces.
Wafer surfaces were cleansed by sonicating the specimens successively in acetone, toluene, acetone, ethanol, and water for 5 min each. The organic solvents (analysis grade) were purchased from VWR (Germany) and Riedel-de-Haën (Germany), and the water was purified by an ultrafiltration apparatus from Millipore Corp. For the creation of hydroxyl groups, the specimens were sonicated in 32.5% nitric acid solution (VWR) for 30 min at room temperature. Excess acid was removed by repeated water rinsing, and the oxidized wafers were stored in 0.1% sodium azide solution to prevent adsorption of air contaminants and growth of microorganisms.
Oxidized wafer surfaces were further modified to obtain different degrees of surface wettability. For this purpose, dried wafers were silanized by being boiled for 3 h in toluene solutions containing 15 mg/ml of either N-(triethoxysilylpropyl)-O-poly(ethylene glycol)urethane carrying five ethylene glycol units to modify wafers with poly(ethylene glycol) groups (PEG), n-octyltriethoxysilane to introduce surface-bound octyl groups (OTS), or (heptadecafluoro-1,1,2,2-tetrahydrodecyl)trichlorosilane to coat surfaces with heptadecafluoro-tetrahydrodecyl groups (HFS). All silanes were obtained from ABCR GmbH (Germany). After silanization, the specimens were sonicated successively in pure toluene, chloroform, and methanol and then dried in a nitrogen stream and stored in a vacuum desiccator. Water contact angles on the modified wafer surfaces were measured using the sessile drop method with a P1 goniometer from Erna Inc. (Japan). Two-microliter droplets were advanced toward the samples by a syringe tip until the droplets made contact with the sample surfaces. Measurements were performed in an ambient atmosphere. X-ray photoelectron spectroscopy demonstrated successful surface coating (data not shown).
Protein preadsorption.
Unstimulated human whole saliva was collected by expectoration into a 50-ml polypropylene vial (Falcon), followed by filtration with a low protein-binding membrane filter with a 0.45-μm pore size (Sartorius, Germany) fitted to a syringe. Saliva was kept on ice for no more than 30 min before use. Fetal bovine serum (0.1 μm; sterile filtered) was purchased from PAN Biotech GmbH, Germany (catalog no. P30-3402). Human serum albumin (HSA; fraction V; catalog no. A-1653; Sigma, Germany) was dissolved in 10 mM phosphate-buffered saline (PBS; pH 7.0) to a final concentration of 10 mg/ml. Fibronectin from human plasma was purchased from Sigma (F-0635) and dissolved in PBS to a final concentration of 5 μg/ml (14). Wafer specimens were mounted onto 10-cm by 7-cm polystyrene plates and incubated in the protein solutions for 1 h at 20°C. For measuring wettability after protein adsorption, mounted wafers were washed three times for 10 min with 60 ml PBS in a 12.5-cm by 8.5-cm jar on an orbital shaker (type KM2; Bühler, Germany) set at 50 rpm to remove excess protein and with water to remove salt. After the wafers were dried in a vacuum, advancing water contact angles were measured as described above.
Amounts of adsorbed proteins were determined directly on the wafer surfaces by a chemiluminescence-based method involving labeling of amino groups with biotin, followed by binding of horseradish peroxidase-conjugated avidin-D (22). Briefly, amino groups were biotinylated with sulfo-NHS-LC-biotin (Pierce, Rockford, IL), followed by binding of horseradish peroxidase-conjugated avidin-D. For detection of chemiluminescence, each wafer surface was overlaid with a luminol-containing solution and immediately exposed to a light-sensitive film. For establishment of standard curves, serial twofold dilutions of the protein solutions were prepared and applied to an SRC96 Minifold I dot blot system (Whatman Schleicher & Schuell, Dassel, Germany) to immobilize the proteins onto nitrocellulose membranes (Whatman Schleicher & Schuell). Densitometric analysis of signals on the X-ray films, recorded as inverse gray levels, was performed using Optimas software (version 6.2; Optimas Corporation, Bothell, WA) after scanning the films utilizing an image scanner.
Bacterial culture.
The bacterial strains used in this study were Streptococcus gordonii strain DL1 (Challis), S. mitis ATCC 49456, and Staphylococcus aureus ATCC 25923, which was kindly provided by the Department of Medical Microbiology and Hygiene at the University of Regensburg. Streptococci were grown as stationary suspension cultures in brain heart infusion (VWR) in a microaerophilic atmosphere (5% CO2), and staphylococci were grown under constant motion and aerobic conditions in caso-bouillon (VWR) for 24 h at 37°C. For use in the adhesion assays, bacteria were washed in PBS, and suspensions of 108 organisms per ml PBS containing 1 mM CaCl2 and 1 mM MgCl2 were prepared. The bacterial densities were determined by measuring the optical densities at 600 nm.
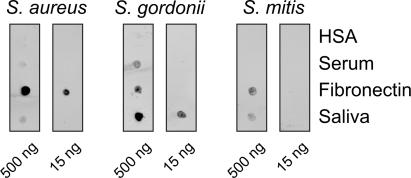
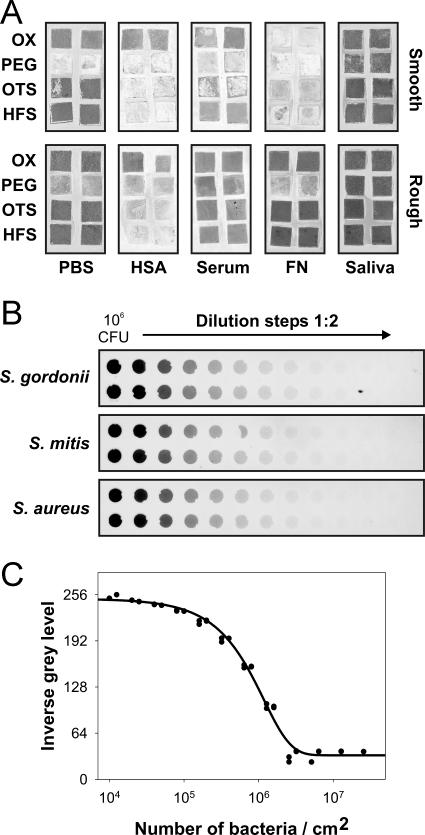
The adhesion properties of the bacterial strains employed were tested utilizing a dot blot array technique that has recently been described (44). Briefly, nitrocellulose membranes (Invitrogen, Carlsbad, CA) were spotted with 1-μl volumes containing 500 ng or 15 ng of HSA, serum, fibronectin, or saliva and were overlaid with fluorescein isothiocyanate (FITC)-labeled S. aureus 25923, S. gordonii DL1, and S. mitis 49456. The fluorescence of adherent bacteria was detected by a Typhoon 9200 imaging system (Amersham Biosciences, Freiburg, Germany). Dark spots are indicative of strong binding activities. As shown in Fig. 1, no bacterial binding to purified HSA could be observed. Weak binding to serum was found for S. aureus and S. gordonii. Purified fibronectin mediated strong binding of S. aureus 25923 and weaker binding of S. gordonii and S. mitis. Saliva mediated strong binding of S. gordonii DL1 and weak binding of S. aureus and S. mitis.
FIG. 1.
Properties of binding of the bacterial strains employed in this study to the proteins and biological fluids used for surface precoating. Proteins were immobilized as dots on nitrocellulose membranes. Binding of fluorescence-labeled bacteria to the immobilized proteins was measured by a fluorescence scanner and is visualized by dark spots.
Detection of surface-attached bacteria by imprint of wafers onto agar (replica plating).
Wafer specimens, reversibly mounted onto polystyrene plates and precoated with protein, were washed as described above to remove excess protein and were then incubated with 50 ml of bacterial suspensions for 1 h at 4°C. After loosely bound bacteria were removed by washing the mounted wafers in a 12.5-cm by 8.5-cm jar three times for 5 min with 50 ml PBS on the orbital shaker set to 50 rpm and draining excess liquid, single wafers were imprinted with the coated side down onto the surface of Columbia agar (VWR) containing 8% sterile defibrinated sheep blood. For each bacterial strain, imprints from all surface modifications were arranged in pairs on one large petri dish (150 mm by 20 mm; catalog no. 82.1184; Sarstedt, Germany) for better comparison. Streptococcal colonies were grown for 24 h in a microaerophilic atmosphere, and staphylococcal colonies were grown for 8 h under aerobic conditions. Photographs from the culture plates were taken with a digital camera (Canon Powershot S45).
Visualization of surface-attached bacteria by SEM.
Precoating with proteins and incubation with bacteria were performed as described above. Wafers were washed as described above, fixed for 30 min by the addition of glutaraldehyde to a final concentration of 2.5%, washed again two times with PBS and two times with H2O, and air dried. Specimens were mounted on stubs, and attached bacteria were visualized by a Quanta 400 FEG scanning electron microscope (FEI, Germany) in low vacuum mode (0.08 torr) using a large-field detector.
Detection and quantification of surface-attached bacteria by fluorescence-based overlay.
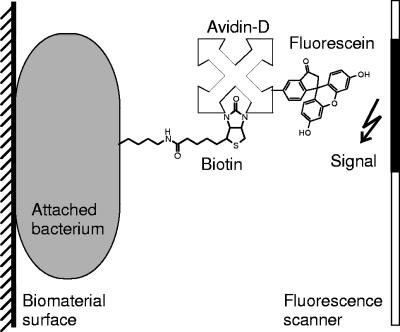
Bacteria were labeled with biotinamidohexanoic acid 3-sulfo-N-hydroxysuccinimide ester (sulfo-NHS-LC-biotin; Pierce) as previously described (33). Wafer specimens, reversibly mounted onto polystyrene plates and precoated with protein, were incubated for 1 h at 4°C with biotinylated bacteria to allow binding. After being washed with PBS to remove loosely bound bacteria as described above, wafers were incubated for 30 min in the dark with a solution of 0.5 μg/ml FITC-conjugated avidin-D (Vector Laboratories Inc.) in Tris-buffered saline-Tween 20 containing 1 mM CaCl2 and 1 mM MgCl2. After four 5-min washings with PBS on the orbital shaker to remove excess avidin-D-FITC, the fluorescence of attached bacteria was detected by the Typhoon imaging system and recorded as a TIFF file (schematic diagram shown in Fig. 2). Densitometric analysis of signals was performed using Optimas software (version 6.2; Optimas Corporation).
FIG. 2.
Schematic drawing of the bacterial overlay method modified for detection of surface-adsorbed microorganisms. Attached biotinylated bacterial organisms were detected by fluorescein-tagged avidin-D. The fluorescence signal emitted was recorded by a fluorescence scanner.
For establishment of standard curves, serial twofold dilutions of the bacterial suspensions were prepared and applied to an SRC96 Minifold I dot blot system (Schleicher & Schuell, Germany) to immobilize the FITC-labeled bacteria onto nitrocellulose membranes (Schleicher & Schuell). Fluorescence was detected and densitometric analysis was performed analogously to the method described for the wafers.
Statistical analysis.
All results are shown as medians, including 25 to 75% quartiles, which were calculated from the values for at least three independent experiments, each performed with two wafer specimens. Statistically significant differences were calculated by the Mann-Whitney test using SPSS for Windows, version 13.0, from SPSS Inc. Significance was accepted at P values of <0.05. In the standards, inverse gray level values with corresponding amounts of adsorbed proteins (μg/cm2) and numbers of bacteria/cm2 were put into a two-dimensional general fit (TableCurve 2D, V5.01; SYSTAT Software Inc.).
RESULTS
Properties of modified silicon wafers before and after protein adsorption.
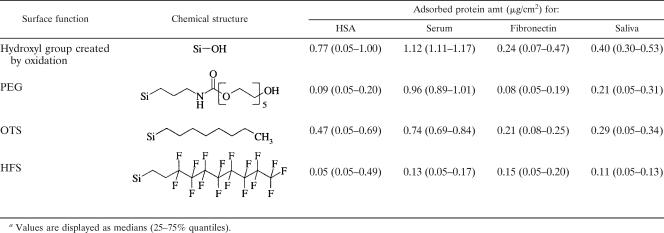
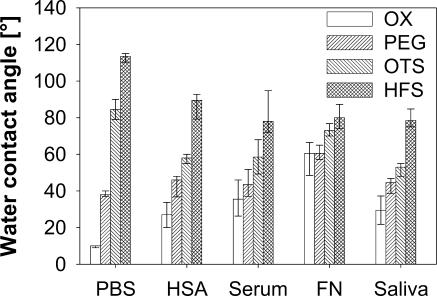
Table 1 summarizes the adsorption behaviors of albumin, serum, fibronectin, and saliva on silicon wafers, depending on their chemical surface modifications. The changes in advancing water contact angle resulting from surface modification before and after protein adsorption are displayed in Fig. 3. Water contact angle measurements confirmed the high wettability of oxidized and PEG-modified surfaces and the low wettability of OTS- and HFS-coated wafers. As a general result, it was observed that on hydrophilic surfaces the contact angles were increased after protein adsorption, whereas on hydrophobic surfaces, the adsorbed proteins caused decreases in the water contact angles. In particular, the increases in the contact angles on oxidized surfaces were significant (P < 0.001) for all proteins, for which the significantly highest value, 60° (P < 0.001), was detected after fibronectin adsorption. On PEG-modified surfaces, only the adsorption of fibronectin led to significant increases in the water contact angles, from 38° to 60° (P < 0.001), whereas the adsorption of all other proteins enhanced the contact angles only slightly. On OTS-modified surfaces, all proteins induced significant decreases in the water contact angles (P < 0.001), and the significantly highest value, 73° (P < 0.001), was found again after fibronectin adsorption. After adsorption to the HFS-modified wafers, all four types of proteins significantly (P < 0.001) reduced the water contact angles to similar extents. Thus, the adsorbed proteins moderated but did not fully mask the original surface properties.
TABLE 1.
Values are displayed as medians (25-75% quantiles).
FIG. 3.
Effect of protein precoating on advancing water contact angles of the chemically modified silicon wafers. FN, fibronectin; OX, hydroxyl group created by oxidation.
Detection of surface-attached bacteria by fluorescence-based overlay in comparison with that by SEM and agar replica plating.
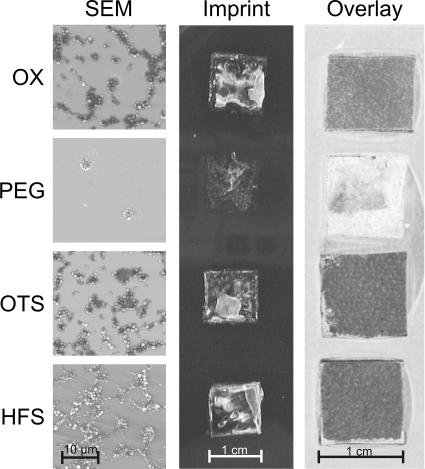
For detection of attached organisms, bacteria were biotinylated prior to incubation with the wafer surfaces. Attached biotinylated bacteria were labeled by the avidin-D-FITC coupling, followed by exposure to a fluorescence scanner. Figure 4 shows the scanned images of the four chemical surface modifications on smooth wafers after colonization by S. gordonii DL1. The intensities of the signals reflect the numbers of microorganisms attached to the wafer surfaces, with high fluorescence intensities (dark staining) corresponding to high numbers of attached bacterial organisms. High fluorescence intensities were detected on oxidized and OTS- and HFS-modified surfaces. On wafers covered with PEG, very weak signals, corresponding to low numbers of attached bacteria, were observed. Scanning electron micrographs of identically modified wafer surfaces colonized by S. gordonii DL1 revealed large and interconnecting aggregates of bacteria on surfaces that had been oxidized or modified with hydrocarbon and fluorocarbon moieties. On the contrary, only a few small, isolated groups of bacteria could be detected on PEG-modified surfaces. Imprints of colonized wafers onto agar also displayed dense colonization on oxidized and OTS- and HFS-modified wafer surfaces, whereas few colonies were recovered from PEG-modified specimens. The qualitative agreement of the findings obtained by SEM and agar replica plating with the results obtained by the fluorescence-based overlay technique adds support to the validity of the newly developed method for detection of surface-attached bacteria.
FIG. 4.
Bacterial attachment detected by SEM and agar replica plating (imprint) in comparison to that detected by the fluorescence-based bacterial overlay technique. The comparison of the three techniques is exemplarily shown for S. gordonii DL1 on differently modified smooth wafers without protein precoating. In the images of the agar imprint, whitish areas are indicative of colony formation. In the images of the bacterial overlay, dark areas are indicative of fluorescence from attached bacteria. OX, hydroxyl group created by oxidation.
Quantification of attached bacteria by the fluorescence-based overlay technique.
Figure 5A shows the scanned images of all surface conditions after colonization by S. gordonii DL1. For the establishment of standard curves, defined numbers of the respective bacterial strains were immobilized in twofold serial dilutions on nitrocellulose (Fig. 5B). The signal intensities correlated with the numbers of bacteria immobilized on the membranes. Signals from these standards were analyzed by densitometry, the inverse gray levels were fitted against the known numbers of bacteria/cm2, and a four-parameter sigmoidal curve fit was found best. An exemplary standard curve is shown for S. gordonii DL1 (Fig. 5C).
FIG. 5.
Quantification of attached bacteria by bacterial overlay on differently modified silicon wafers. (A) Image of fluorescent signals (dark areas) after attachment of S. gordonii DL1 to surfaces of various morphologies and surface chemistries. (The upper left panel, right column, is identical to the overlay image shown in Fig. 4.) (B) Signals obtained after fluorescence labeling of serial twofold dilutions of the three different bacterial strains immobilized on nitrocellulose membranes. (C) Standard curve for quantification of S. gordonii DL1 after densitometric analysis of signals obtained from the standard dilutions. FN, fibronectin; OX, hydroxyl group created by oxidation.
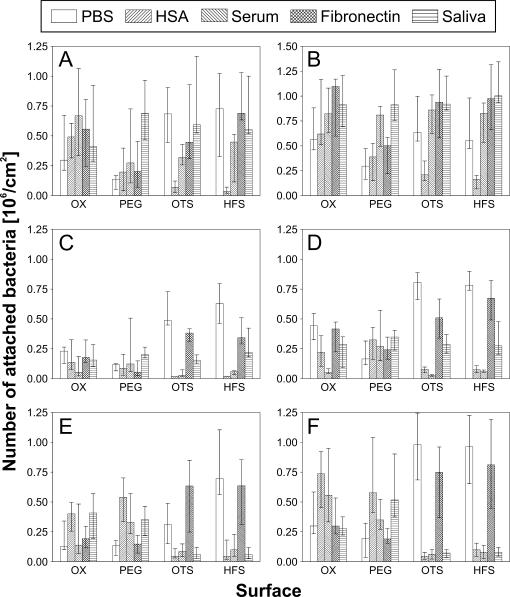
Standard curves analogous to the one shown in Fig. 5C were prepared for all bacterial strains and were subsequently used to transform gray levels obtained on the different wafer surfaces into numbers of attached bacteria per area. Figure 6 shows the transformed data compiled from all experiments. Consistently higher numbers of attached bacteria were detected on roughened surfaces (Fig. 6B, D, and F), for which the influence of surface chemistry on the adhesion pattern was similar to that found on smooth surfaces (Fig. 6A, C, and E). The adhesion patterns differed between different bacterial strains and displayed clear dependencies on surface chemistry and preadsorbed protein layers. Only on surfaces which had not been precoated with protein were similar adhesion patterns for all three microbial strains observed. The highest numbers were detected on OTS- and HFS-modified surfaces, and the lowest numbers were found on PEG-modified wafers.
FIG. 6.
Numbers of bacteria attached on silicon wafers, depending on surface roughness, chemical surface modification, and preadsorbed protein layers. Numbers were calculated by comparison of the signal intensities from wafer surfaces with the standard curves for each bacterium. (A) S. gordonii on smooth surfaces; (B) S. gordonii on rough surfaces; (C) S. mitis on smooth surfaces; (D) S. mitis on rough surfaces; (E) S. aureus on smooth surfaces; (F) S. aureus on rough surfaces. OX, hydroxyl group created by oxidation.
HSA precoating enhanced attachment of S. gordonii DL1 and to a higher extent that of S. aureus on oxidized and PEG-modified surfaces, whereas on hydrophobic surfaces, the attachment of these two strains was nearly suppressed (P = 0.002) by HSA. In the case of S. mitis, a trend of reduced adhesion after HSA precoating was observed on all surfaces. Precoating hydrophobic wafers with serum reduced the attachment of S. mitis and S. aureus significantly (P = 0.01), while the attachment of S. gordonii DL1 was not altered in a noticeable manner. On hydrophilic surfaces, highly different responses of bacterial attachment after serum precoating were observed. Adhesion was enhanced for S. gordonii DL1 and significantly reduced for S. mitis (P = 0.028), while no clear influence was found for S. aureus. On PEG-modified wafers precoated with serum, slight increases in microbial attachment were observed for all strains employed.
Precoating of wafers with fibronectin did not appear to qualitatively change the adhesion behavior of any of the three bacterial strains, regardless of surface modification. High numbers of attached organisms were found for S. mitis and S. aureus on hydrophobic surfaces, while rather low numbers were observed on oxidized and PEG-modified wafers. S. gordonii DL1 adhered well to all fibronectin precoated surfaces, with the exception of PEG-modified wafers. Adhesion of this oral strain was enhanced by precoating the wafer surfaces with saliva. High numbers of attached cells were detected on all types of surfaces independently of chemical modification. For S. mitis and S. aureus, significantly reduced adhesion was noted on hydrophobic surfaces (P = 0.002), whereas on oxidized and PEG-modified wafers, no noticeable change in adhesion could be recognized.
DISCUSSION
For direct in situ quantification of attached bacteria on various biomaterial surfaces or on other surfaces exposed to biofouling, a novel fluorescence-based bacterial overlay technique was developed. The detection of bacteria by this method, which was based on biotinylation of bacteria and subsequent detection by fluorescence-tagged avidin, was methodically simple to perform and allowed the simultaneous measurement of a large number of different small material specimens. This method was tested in an established model system based on silanized silicon wafers with and without prior protein coating by using exemplary bacterial strains exhibiting different adhesion properties. With this novel method, bacterial attachment was found to depend on a variety of parameters, including surface wettability, surface roughness, strain of bacteria employed, and prior protein adsorption.
Bacterial overlay methods are well-established tools for examination of bacterial adhesin-mediated binding to immobilized host glycolipid (18) or glycoprotein (24, 32, 44) receptors separated by either thin-layer chromatography or gel electrophoresis and subsequent blotting but have so far not been employed for quantification of bacteria attached to material surfaces. When the newly established overlay technique was compared to the very simple and direct agar replica technique or to visualization by SEM, similar qualitative adhesion patterns on the differently modified wafer surfaces could be verified. Certain quantitative deviations between these methods might be caused by inherent technical shortcomings. The agar imprint method requires viable bacteria and reflects only the number of bacteria that are transferable to the agar medium, whereas an uncertainty regarding the number of firmly adherent residual bacteria on the material surface remains. SEM, on the other hand, shows only the firmly attached bacteria remaining on the material surface, whereas uncertainty regarding organisms lost during sample preparation exists. Alternative methods for quantification, such as radiolabeling, crystal violet-staining, quantification by metabolites, and enumeration by use of a Coulter Counter or flow cytometry, require large surface areas to obtain reliable results. Such large surface areas are sometimes difficult to realize with certain materials. Similar limitations might apply also to other techniques that have been described in the past to quantify surface-attached bacteria (1). Thus, the technique of choice depends on the particular problem that one is interested in as well as on the type of material examined. The most obvious advantage of the fluorescence-based overlay method described in the present study is the possibility for screening of in situ bacterial attachment on many parallel small samples within one experimental setting.
Despite the inherent limitations of the different techniques for enumeration of bacteria attached to material surfaces, certain common principles regarding the influence of surface properties on bacterial adhesion can be deduced from the literature. There is a general tendency for hydrophilic materials to be more resistant to bacterial or fungal adhesion than hydrophobic ones (2, 6). This is reflected in the present investigation by the low numbers of bacteria found on oxidized wafers compared to those on OTS or HFS-modified surfaces. This is also in agreement with previous reports investigating the adhesion of a variety of bacteria, including S. aureus, to self-assembled monolayers (8, 40) and with SEM studies using various materials within a wide range of surface free energy (41). In the present investigation, also in agreement with these reports, the lowest numbers of attached organisms for each of the three bacterial strains tested were recorded for PEG-modified surfaces. This may be explained by the fact that PEG chains, depending on their lengths (31), provide a template for water nucleation resulting in the formation of an interfacial water layer, whereby these PEG brushes prevent direct contact between bacteria and the surface (17).
Higher roughness on the chemically modified wafer surfaces was found in the present study to increase bacterial attachment but did not alter the relative adhesion patterns found for smooth wafer surfaces. Other studies also found that roughening of, e.g., polymeric surfaces, promoted bacterial adhesion and biofilm deposition, whereas smooth surfaces did not (27, 39). An accepted explanation for this behavior is that rough surfaces have greater surface areas and that the depressions in the roughened surfaces provide more favorable sites for colonization (2), an observation that was also observed by SEM visualization (images not shown).
Exposure of the modified wafers to protein solutions before incubation with the bacterial strains resulted in significantly altered adhesion patterns that were to a great degree dependent on the type of protein or protein mixture and on the particular bacterial strain examined. Differences in wettability between the chemically modified wafer surfaces were moderated to a certain extent by adsorption of a protein layer, whereby hydrophobic surfaces became more hydrophilic and vice versa. However, preadsorption of the proteins did not completely mask the influence of the original surface chemistry. Preadsorption of wafers with HSA reduced bacterial attachment in most cases, which is in agreement with prior studies using different material surfaces (23, 25). The hypothesis that this reduction of bacterial adhesion by albumin might be caused by changing substratum surface hydrophobicity (2) was supported by the combined results for the water contact angle measurements and bacterial adhesion experiments of the present study. Preadsorption of wafers with serum, similar to that with HSA, reduced bacterial attachment in most instances, which might in fact be due to the high albumin content in serum, as suggested previously (23). However, for S. gordonii DL1, an increase in bacterial numbers attached to serum-precoated surfaces was observed. This exceptional adhesion behavior of S. gordonii DL1 could be explained by the expression of an adhesin activity recognizing putative receptor molecules in this complex body fluid. This was further supported by the present dot blot experiments, showing preferential binding of this strain to serum proteins immobilized on nitrocellulose (Fig. 1). Precoating of wafers with fibronectin, in certain instances, resulted in greater numbers of attached bacteria. This could be explained by expression of fibronectin-binding adhesin activities in these bacteria (7, 35) and was also reflected by bacterial binding to fibronectin in the dot blot experiment (Fig. 1). For saliva precoating, it is not surprising that particularly the adhesion of S. gordonii DL1 was enhanced in all situations because multiple adhesin activities that interact with salivary proteins have been described for this bacterium (33), including a lectin-like sialic acid binding adhesin (38) that might in part also be responsible for the observed binding to serum proteins. This was supported in the present investigation by strong binding of S. gordonii DL1 to mixed whole saliva in the dot blot experiment. S. mitis, although considered one of the earliest colonizers of the tooth surface (12, 20), did not exhibit stronger binding to saliva-precoated wafers and was found to bind only weakly to whole saliva immobilized on nitrocellulose. Thus, it appears that adsorbed proteins do not mask the original surface properties but rather decorate the surface in a fashion whereby their presence adds additional biochemical properties to the already present physicochemical properties. It is, however, still the original surface chemistry that determines the amount, the composition, the conformation, and ultimately the biological activity of these adsorbed proteins.
For many applications, there is a need for materials that resist bacterial colonization (15, 16). The new method described in the present study not only allows quantification of surface-attached bacteria but also more closely imitates the real situation by considering the conditioning influence of the surrounding macromolecule-containing fluid environment. For each particular application, e.g., when evaluating implant materials, blood-contacting transcutaneous or extracorporal devices, contact lenses, dental restorative materials, or even surfaces exposed to a marine environment (9, 11, 16), it is crucial to select those bacteria that are of relevance in each given particular situation. It would be advantageous to establish model strains for each test system together with a set of adhesin-deficient mutants (26, 38, 44) for discrimination between general physicochemical influences and the more specific biochemical sterical recognition mechanisms.
Acknowledgments
We are grateful to Katja Bomertl, Katharina Schindler, Martina Kreuzer, and Wei-Qi Zhou for excellent technical assistance and to Brigitte Bey and Helga Ebensberger for performing SEM. We further thank Norbert Lehn, Ulrike Thalmaier, and Wulf Schneider (Department of Medical Microbiology and Hygiene, University of Regensburg) for helpful advice as well as Reinhard Rachel (Department of Microbiology, University of Regensburg) and Christof von Eiff (Institute of Medical Microbiology and Immunology, University of Münster) for fruitful discussions.
This work was supported by a grant (ReForM-C) from the University of Regensburg.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.An, Y. H., and R. J. Friedman. 1997. Laboratory methods for studies of bacterial adhesion. J. Microbiol. Methods 30:141-152. [Google Scholar]
- 2.An, Y. H., and R. J. Friedman. 1998. Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J. Biomed. Mater. Res. 43:338-348. [DOI] [PubMed] [Google Scholar]
- 3.An, Y. H., J. B. McGlohorn, B. K. Bednarski, K. L. Martin, and R. J. Friedman. 2001. An open channel flow chamber for characterizing biofilm formation on biomaterial surfaces. Methods Enzymol. 337:79-88. [DOI] [PubMed] [Google Scholar]
- 4.Busscher, H. J., G. I. Geertsema-Doornbusch, and H. C. van der Mei. 1997. Adhesion to silicone rubber of yeasts and bacteria isolated from voice prostheses: influence of salivary conditioning films. J. Biomed. Mater. Res. 34:201-209. [DOI] [PubMed] [Google Scholar]
- 5.Busscher, H. J., and H. C. van der Mei. 1995. Use of flow chamber devices and image analysis methods to study microbial adhesion. Methods Enzymol. 253:455-477. [DOI] [PubMed] [Google Scholar]
- 6.Chandra, J., J. D. Patel, J. Li, G. Zhou, P. K. Mukherjee, T. S. McCormick, J. M. Anderson, and M. A. Ghannoum. 2005. Modification of surface properties of biomaterials influences the ability of Candida albicans to form biofilms. Appl. Environ. Microbiol. 71:8795-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie, J., R. McNab, and H. F. Jenkinson. 2002. Expression of fibronectin-binding protein FbpA modulates adhesion in Streptococcus gordonii. Microbiology 148:1615-1625. [DOI] [PubMed] [Google Scholar]
- 8.Cunliffe, D., C. A. Smart, C. Alexander, and E. N. Vulfson. 1999. Bacterial adhesion at synthetic surfaces. Appl. Environ. Microbiol. 65:4995-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgerton, M., and M. J. Levine. 1993. Biocompatibility: its future in prosthodontic research. J. Prosthet. Dent. 69:406-415. [DOI] [PubMed] [Google Scholar]
- 10.Fijan, S., S. Sostar-Turk, and A. Cencic. 2005. Implementing hygiene monitoring systems in hospital laundries in order to reduce microbial contamination of hospital textiles. J. Hosp. Infect. 61:30-38. [DOI] [PubMed] [Google Scholar]
- 11.Flemming, H. C. 2002. Biofouling in water systems—cases, causes and countermeasures. Appl. Microbiol. Biotechnol. 59:629-640. [DOI] [PubMed] [Google Scholar]
- 12.Frandsen, E. V., V. Pedrazzoli, and M. Kilian. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol. Immunol. 6:129-133. [DOI] [PubMed] [Google Scholar]
- 13.Glantz, P. O., T. Arnebrant, T. Nylander, and R. E. Baier. 1999. Bioadhesion—a phenomenon with multiple dimensions. Acta Odontol. Scand. 57:238-241. [DOI] [PubMed] [Google Scholar]
- 14.Grinnell, F. 1987. Fibronectin adsorption on material surfaces. Ann. N. Y. Acad. Sci. 516:280-290. [DOI] [PubMed] [Google Scholar]
- 15.Gristina, A. G. 1987. Biomaterial-centered infection: microbial adhesion versus tissue integration. Science 237:1588-1595. [DOI] [PubMed] [Google Scholar]
- 16.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 17.Heuberger, M., T. Drobek, and N. D. Spencer. 2005. Interaction forces and morphology of a protein-resistant poly(ethylene glycol) layer. Biophys. J. 88:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson, K. A., and N. Strömberg. 1987. Overlay and solid-phase analysis of glycolipid receptors for bacteria and viruses. Methods Enzymol. 138:220-232. [DOI] [PubMed] [Google Scholar]
- 19.Lendenmann, U., J. Grogan, and F. G. Oppenheim. 2000. Saliva and dental pellicle—a review. Adv. Dent. Res. 14:22-28. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., E. J. Helmerhorst, C. W. Leone, R. F. Troxler, T. Yaskell, A. D. Haffajee, S. S. Socransky, and F. G. Oppenheim. 2004. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97:1311-1318. [DOI] [PubMed] [Google Scholar]
- 21.Mrksich, M., and G. M. Whitesides. 1996. Using self-assembled monolayers to understand the interactions of man-made surfaces with proteins and cells. Annu. Rev. Biophys. Biomol. Struct. 25:55-78. [DOI] [PubMed] [Google Scholar]
- 22.Müller, R., K.-A. Hiller, G. Schmalz, and S. Ruhl. 2006. Chemiluminescence-based detection and comparison of adsorbed protein amounts on differently modified silica surfaces. Anal. Biochem. 359:194-202. [DOI] [PubMed] [Google Scholar]
- 23.Paulsson, M., M. Kober, C. Freij-Larsson, M. Stollenwerk, B. Wesslen, and A. Ljungh. 1993. Adhesion of staphylococci to chemically modified and native polymers, and the influence of preadsorbed fibronectin, vitronectin and fibrinogen. Biomaterials 14:845-853. [DOI] [PubMed] [Google Scholar]
- 24.Prakobphol, A., P. A. Murray, and S. J. Fisher. 1987. Bacterial adherence on replicas of sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 164:5-11. [DOI] [PubMed] [Google Scholar]
- 25.Pringle, J. H., and M. Fletcher. 1986. Influence of substratum hydration and adsorbed macromolecules on bacterial attachment to surfaces. Appl. Environ. Microbiol. 51:1321-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proctor, R. A., C. von Eiff, B. C. Kahl, K. Becker, P. McNamara, M. Herrmann, and G. Peters. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295-305. [DOI] [PubMed] [Google Scholar]
- 27.Quirynen, M., H. C. van der Mei, C. M. Bollen, A. Schotte, M. Marechal, G. I. Doornbusch, I. Naert, H. J. Busscher, and D. van Steenberghe. 1993. An in vivo study of the influence of the surface roughness of implants on the microbiology of supra- and subgingival plaque. J. Dent. Res. 72:1304-1309. [DOI] [PubMed] [Google Scholar]
- 28.Ratner, B. D., and S. J. Bryant. 2004. Biomaterials: where we have been and where we are going. Annu. Rev. Biomed. Eng. 6:41-75. [DOI] [PubMed] [Google Scholar]
- 29.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 30.Roosjen, A., N. P. Boks, H. C. van der Mei, H. J. Busscher, and W. Norde. 2005. Influence of shear on microbial adhesion to PEO-brushes and glass by convective-diffusion and sedimentation in a parallel plate flow chamber. Colloids Surf. B 46:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Roosjen, A., H. C. van der Mei, H. J. Busscher, and W. Norde. 2004. Microbial adhesion to poly(ethylene oxide) brushes: influence of polymer chain length and temperature. Langmuir 20:10949-10955. [DOI] [PubMed] [Google Scholar]
- 32.Ruhl, S., J. O. Cisar, and A. L. Sandberg. 2000. Identification of polymorphonuclear leukocyte and HL-60 cell receptors for adhesins of Streptococcus gordonii and Actinomyces naeslundii. Infect. Immun. 68:6346-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruhl, S., A. L. Sandberg, and J. O. Cisar. 2004. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral actinomyces and streptococci. J. Dent. Res. 83:505-510. [DOI] [PubMed] [Google Scholar]
- 34.Sbordone, L., and C. Bortolaia. 2003. Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease. Clin. Oral Investig. 7:181-188. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52:631-641. [DOI] [PubMed] [Google Scholar]
- 36.Silberzan, P., L. Leger, D. Ausserre, and J. J. Benattar. 1991. Silanation of silica surfaces. A new method of constructing pure or mixed monolayers. Langmuir 7:1647-1651. [Google Scholar]
- 37.Slack, S. M., and T. A. Horbett. 1995. The Vroman effect: a critical review, p. 112-128. In T. A. Horbett and J. Brash (ed.), Proteins at interfaces II: fundamentals and applications. American Chemical Society, Washington, DC.
- 38.Takahashi, Y., K. Konishi, J. O. Cisar, and M. Yoshikawa. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, R. L., J. Verran, G. C. Lees, and A. J. Ward. 1998. The influence of substratum topography on bacterial adhesion to polymethyl methacrylate. J. Mater. Sci. Mater. Med. 9:17-22. [DOI] [PubMed] [Google Scholar]
- 40.Tegoulia, V. A., and S. L. Cooper. 2002. Staphylococcus aureus adhesion to self-assembled monolayers: effect of surface chemistry and fibrinogen presence. Colloids Surf. B 24:217-228. [Google Scholar]
- 41.van Pelt, A. W., A. H. Weerkamp, M. H. Uyen, H. J. Busscher, H. P. de Jong, and J. Arends. 1985. Adhesion of Streptococcus sanguis CH3 to polymers with different surface free energies. Appl. Environ. Microbiol. 49:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vergnat, M., N. H. Zoubir, and A. Burneau. 1995. Homogenous chemical etching of sand-blasted silicon substrates. Thin Solid Films 255:231-233. [Google Scholar]
- 43.von Eiff, C., B. Jansen, W. Kohnen, and K. Becker. 2005. Infections associated with medical devices: pathogenesis, management and prophylaxis. Drugs 65:179-214. [DOI] [PubMed] [Google Scholar]
- 44.Walz, A., S. Odenbreit, J. Mahdavi, T. Borén, and S. Ruhl. 2005. Identification and characterization of binding properties of Helicobacter pylori by glycoconjugate arrays. Glycobiology 15:700-708. [DOI] [PubMed] [Google Scholar]