Abstract
MICs of tetracyclines were determined for 86 human Bifidobacterium isolates and three environmental strains. The tet(O) gene was found to be absent in these isolates. tet(W) and tet(M) were found in 26 and 7%, respectively, of the Bifidobacterium isolates, and one isolate contained both genes. Chromosomal DNA hybridization showed that there was one chromosomal copy of tet(W) and/or tet(M).
Bifidobacteria are gram-positive anaerobic bacteria found in the gastrointestinal tracts of humans and animals. Strains belonging to the genus Bifidobacterium have been reported to have several health-promoting effects (15, 16, 21), explaining why they are increasingly used as probiotics in a wide range of functional foods (9). For probiotic safety (20), guidelines have recently recommended that probiotic bacteria should not harbor transmissible genes encoding resistance to antibiotics that are used clinically (17). For bifidobacteria two molecular antibiotic resistance determinants have been described, the bbmr gene of Bifidobacterium breve, which confers moderate resistance to macrolides (10), and tet genes coding for ribosomal protection proteins involved in resistance to tetracyclines (3). Although different types of acquired tetracycline resistance genes have been found in anaerobes (3, 18), only the tet(M) and tet(W) genes encoding ribosomal protection proteins have been selectively found in bifidobacteria (6, 8, 11, 12, 22). In order to better understand resistance mechanisms in human bifidobacterial strains, we investigated the prevalence and distribution of the tet(M), tet(W), and tet(O) genes encoding ribosomal protection proteins involved in acquired tetracycline resistance in this human commensal genus.
Eighty-nine strains of bifidobacteria belonging to nine species were included in our study (Table 1). Eighty-six of these strains were isolated, as described previously (2), from feces of healthy humans (adults and newborns), and three were environmental strains (laboratory collection). Bacteria were assigned to the genus Bifidobacterium on the basis of their anaerobic requirement, cellular morphology, Gram staining, and fructose-6-phosphate phosphoketolase activity and by PCR (7). Species were identified using a validated multiplex PCR (13) which included control strains. Isolates whose identities were not clear-cut were not included in the study.
TABLE 1.
MICs of tetracycline, minocycline, and doxycycline for Bifidobacterium isolates
| Antibiotic | Bifidobacterium sp. | No. of isolates | No. of isolates inhibited at a concn (mg/liter) of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | |||
| Tetracycline | B. longum longum type | 30 | 1 | 2 | 11 | 4 | 2 | 2 | 2 | 2 | 4 | |
| B. longum infantis type | 4 | 1 | 2 | 1 | ||||||||
| B. pseudocatenulatum | 12 | 5 | 2 | 5 | ||||||||
| B. breve | 14 | 1 | 1 | 9 | 1 | 1 | 1 | |||||
| B. angulatum | 2 | 2 | ||||||||||
| B. bifidum | 12 | 4 | 4 | 1 | 1 | 2 | ||||||
| B. adolescentis | 3 | 1 | 2 | |||||||||
| B. dentium | 5 | 3 | 1 | 1 | ||||||||
| B. animalis subsp. animalis | 3 | 1 | 1 | 1 | ||||||||
| B. animalis subsp. lactis | 4 | 1 | 3 | |||||||||
| Minocycline | B. longum longum type | 30 | 1 | 7 | 10 | 2 | 2 | 2 | 1 | 5 | ||
| B. longum infantis type | 4 | 1 | 2 | 1 | ||||||||
| B. pseudocatenulatum | 12 | 4 | 2 | 1 | 4 | 1 | ||||||
| B. breve | 14 | 1 | 4 | 7 | 1 | 1 | ||||||
| B. angulatum | 2 | 2 | ||||||||||
| B. bifidum | 12 | 4 | 3 | 1 | 1 | 3 | ||||||
| B. adolescentis | 3 | 2 | 1 | |||||||||
| B. dentium | 5 | 1 | 2 | 2 | ||||||||
| B. animalis subsp. animalis | 3 | 1 | 1 | 1 | ||||||||
| B. animalis subsp. lactis | 4 | 3 | 1 | |||||||||
| Doxycycline | B. longum longum type | 30 | 1 | 8 | 10 | 1 | 1 | 3 | 5 | 1 | ||
| B. longum infantis type | 4 | 1 | 2 | 1 | ||||||||
| B. pseudocatenulatum | 12 | 4 | 2 | 1 | 2 | 3 | ||||||
| B. breve | 14 | 2 | 7 | 3 | 2 | |||||||
| B. angulatum | 2 | 2 | ||||||||||
| B. bifidum | 12 | 5 | 3 | 1 | 2 | 1 | ||||||
| B. adolescentis | 3 | 2 | 1 | |||||||||
| B. dentium | 5 | 2 | 3 | 1 | 1 | |||||||
| B. animalis subsp. animalis | 3 | 1 | 1 | 1 | ||||||||
| B. animalis subsp. lactis | 4 | 2 | 2 | |||||||||
Most of the anaerobe efflux proteins confer resistance to tetracycline but not to minocycline (3). Therefore, the phenotypic patterns of resistance to tetracyclines of Bifidobacterium isolates were determined using three tetracyclines: tetracycline, minocycline, and doxycycline (Sigma-Aldrich, Saint Quentin Fallavier, France). MICs were determined by the agar dilution method, as described previously (12), following the CLSI (formerly NCCLS) recommendations (14). In general, the MICs of tetracycline were one- to twofold higher than those of the two other molecules tested (Table 1).
To differentiate resistant strains from susceptible strains, we use the wild-type/nonwild-type definition from EUCAST (http://www.escmid.org/sites/index.aspx), which allows strain differentiation based on the presence or absence of resistance genes. Therefore, purified genomic DNA (23) of all 89 strains was used as a template for PCR amplification of the tet(M), tet(W), and tet(O) genes using sense and antisense primers as described previously (4, 19, 22). Our results showed that the prevalence of tetracycline-resistant Bifidobacterium strains was 33% (Table 2). PCR results showed that the tet(O) determinant was not present in any of the Bifidobacterium isolates tested, while 29 of the 89 isolates carried either tet(W) or tet(M) or both. tet(W) was the most widely distributed gene among Bifidobacterium species and was found in 83% of the tetracycline-resistant isolates, while the prevalence of tet(M) was lower (21%).
TABLE 2.
Distribution of tet genes in Bifidobacterium isolates
| Bacteria | No. of isolates | No. (%) of isolates with the following resistance gene(s):
|
|||
|---|---|---|---|---|---|
| tet(W) | tet(M) | tet(O) | tet(W) and tet(M) | ||
| All Bifidobacterium isolates | 89 | 23 (26) | 5 (6) | 0 | 1 (1) |
| Human Bifidobacterium isolates | |||||
| B. longum longum type | 30 | 6 (20) | 2 (6) | 0 | 0 |
| B. longum infantis type | 4 | 1 (25) | 0 | 0 | 0 |
| B. pseudocatenulatum | 12 | 5 (41) | 0 | 0 | 0 |
| B. breve | 14 | 1 (7) | 2 (14) | 0 | 1 (7) |
| B. angulatum | 2 | 0 | 0 | 0 | 0 |
| B. bifidum | 12 | 4 (33) | 1 (8) | 0 | 0 |
| B. adolescentis | 3 | 0 | 0 | 0 | 0 |
| B. dentium | 5 | 0 | 0 | 0 | 0 |
| B. animalis subsp. lactis | 4 | 4 (100) | 0 | 0 | 0 |
| Environmental Bifidobacterium isolates | |||||
| B. animalis subsp. animalis | 3 | 2 (67) | 0 | 0 | 0 |
Based on the CLSI anaerobic bacterium tetracycline breakpoints (14), two isolates carrying a tet(M) gene were clinically susceptible to tetracylines (MICs, ≤4 mg/liter) and four isolates carrying tet(W) were determined to be intermediate strains (MICs, 8 mg/liter). These results suggest that when bifidobacteria are categorized as clinically intermediate for tetracycline resistance, they should be screened genetically for the presence of tet genes.
We report here for the first time the presence of the tet(M) gene in the human species Bifidobacterium bifidum, Bifidobacterium longum, and Bifidobacterium breve. The fact that tet(W) has a G+C content (50 to 55%) closer to the average G+C content of the Bifidobacterium chromosome (58% G+C) is a possible explanation for the spread of tet(W) in this genus at the expense of tet(M) (32 to 40% G+C). Interestingly, one B. breve tetracycline-resistant isolate contained both the tet(W) and tet(M) genes (MIC of tetracycline, 64 mg/liter), an uncommon feature that has not been described previously for the genus Bifidobacterium. The presence of both these tet genes was not associated with an MIC that was higher than the MICs for all strains that contained only tet(W) or tet(M), for which the MIC of tetracycline was 64 mg/liter. This suggests that a need for an increased level of tetracycline resistance is not the selective pressure for the presence of more than one gene and is consistent with genetic events in the dissemination of tet resistance genes that are independent of antibiotic pressure.
Partial sequencing (495 nucleotides) of the tet(W) genes of 12 tetracycline-resistant isolates revealed that the nucleotide sequences exhibited 98 to 100% identity to an internal fragment (nucleotides 330 to 825) of the tet(W) genes of Butyrivibrio fibrisolvens (1) and B. longum (5). The partial sequences (500 nucleotides) of the tet(M) genes of two tetracycline-resistant isolates exhibited 97% identity with Enterococcus faecalis and Streptococcus pneumoniae tet(M) genes (GenBank accession no. AY466395 and AJ585081, respectively). The high level of sequence identity between the tet(W) genes of bifidobacteria and the rumen anaerobe B. fibrisolvens or between the tet(M) genes of bifidobacteria and E. faecalis suggests that horizontal gene transfer occurred.
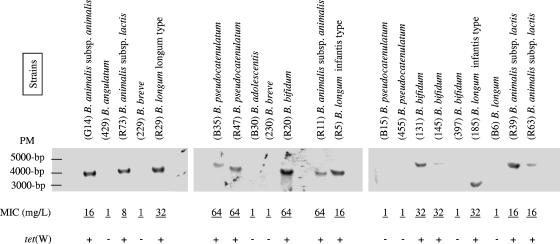
The tet(W) or tet(M) locus is thought to be located on the bacterial chromosome since when strains were found to harbor plasmids, no tet genes could be amplified by PCR (data not shown). For all strains, chromosomal localization of tet(W) or tet(M) was assessed under standard conditions by hybridization of PvuII-digested total DNA using a 1,200-bp PCR fragment of tet(M) or tet(W) as a chemiluminescently labeled probe (ECL kit; Amersham, Sacley, France). Southern blots contained single hybridization bands at 3,000 to 5,000 bp for tetracycline-resistant Bifidobacterium strains carrying tet(W) (Fig. 1) or tet(M) (data not shown), which was consistent with the MICs and the PCR results. The fact that the tet(W) hybridization signal appeared at different positions suggests that there is variability in the DNA region containing the tet genes and/or in the genetic transfer mechanism(s).
FIG. 1.
MICs of tetracycline and tet(W)-specific hybridization patterns of PvuII-restricted chromosomal DNA of Bifidobacterium isolates. Not all strains analyzed in this study are included. PM, molecular weight.
This study is the first study showing a high prevalence and wide distribution of acquired resistance to tetracyclines due to ribosomal protection proteins in human Bifidobacterium isolates and three strains from the environment. The findings suggest that bifidobacteria in the human gastrointestinal tract have access to tetracycline resistance genes and may be involved in their dissemination. However, when we investigated the possible transfer of tet(W) among Bifidobacterium isolates by conducting conjugations experiments, preliminary results showed that there were no transconjugants (data not shown). How tet genes are maintained and disseminate through bifidobacteria needs to be addressed. Indeed, Bifidobacterium is of special interest because several Bifidobacterium strains are used as probiotics and because of general concern concerning the safety of probiotics (i.e., the potential transferability of antibiotic resistance determinants).
Nucleotide sequence accession numbers.
The accession numbers for the partial nucleotide sequences of the tet(W) genes that have been deposited in the GenBank database are as follows: DQ988358 and DQ988363 for Bifidobacterium animalis subsp. animalis; DQ988360, DQ988361, and DQ988362 for B. animalis subsp. lactis; DQ988353 and DQ988359 for B. longum longum type; DQ988357 for B. longum infantis type; DQ988352 for B. breve; and DQ988354, DQ988355, and DQ988356 for B. bifidum.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 2.Butel, M. J., N. Roland, A. Hibert, F. Popot, A. Favre, A. C. Tessèdre, M. Bensaada, A. Rimbault, and O. Szylit. 1998. Clostridial pathogenicity in experimental necrotising enterocolitis in gnotobiotic quails and protective role of bifidobacteria. J. Med. Microbiol. 47:391-399. [DOI] [PubMed] [Google Scholar]
- 3.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, W. O., K. Young, Z. Leng, and M. C. Roberts. 1999. Mobile elements carrying ermF and tetQ genes in gram-positive and gram-negative bacteria. J. Antimicrob. Chemother. 44:329-335. [DOI] [PubMed] [Google Scholar]
- 5.Florez, A. B., M. S. Ammor, P. Álvarez-Martín, A. Margolles, and B. Mayo. 2006. Molecular analysis of tet(W) gene-mediated tetracycline resistance in dominant intestinal Bifidobacterium species from healthy humans. Appl. Environ. Microbiol. 72:7377-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kastner, S., V. Perreten, H. Bleuler, G. Hugenschmidt, C. Lacroix, and L. Meile. 2006. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 29:145-155. [DOI] [PubMed] [Google Scholar]
- 7.Kok, R. G., A. de Waal, F. Schut, G. W. Welling, G. Weenk, and K. J. Hellingwerf. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 62:3668-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacroix, J. M., and C. B. Walker. 1995. Detection and incidence of the tetracycline resistance determinant tet(M) in the microflora associated with adult periodontitis. J. Periodontol. 66:102-108. [DOI] [PubMed] [Google Scholar]
- 9.Lin, D. C. 2003. Probiotics as functional foods. Nutr. Clin. Pract. 18:497-506. [DOI] [PubMed] [Google Scholar]
- 10.Margolles, A., J. A. Moreno, D. van Sinderen, and C. G. de los Reyes-Gavilan. 2005. Macrolide resistance mediated by a Bifidobacterium breve membrane protein. Antimicrob. Agents Chemother. 49:4379-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masco, L., K. Van Hoorde, E. De Brandt, J. Swings, and G. Huys. 2006. Antimicrobial susceptibility of Bifidobacterium strains from humans, animals and probiotic products. J. Antimicrob. Chemother. 58:85-94. [DOI] [PubMed] [Google Scholar]
- 12.Moubareck, C., F. Gavini, L. Vaugien, M. J. Butel, and F. Doucet-Populaire. 2005. Antimicrobial susceptibility of bifidobacteria. J. Antimicrob. Chemother. 55:38-44. [DOI] [PubMed] [Google Scholar]
- 13.Mullie, C., M. F. Odou, E. Singer, M. B. Romond, and D. Izard. 2003. Multiplex PCR using 16S rRNA gene-targeted primers for the identification of bifidobacteria from human origin. FEMS Microbiol. Lett. 222:129-136. [DOI] [PubMed] [Google Scholar]
- 14.NCCLS. 2004. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 6th ed. Approved Standard M11-A6. NCCLS, Wayne, PA.
- 15.Parvez, S., K. A. Malik, K. S. Ah, and H. Y. Kim. 2006. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 100:1171-1185. [DOI] [PubMed] [Google Scholar]
- 16.Picard, C., J. Fioramonti, A. Francois, T. Robinson, F. Neant, and C. Matuchansky. 2005. Review article: bifidobacteria as probiotic agents—physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 22:495-512. [DOI] [PubMed] [Google Scholar]
- 17.Reid, G. 2005. The importance of guidelines in the development and application of probiotics. Curr. Pharm. Des. 11:11-16. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 19.Roberts, M. C., Y. Pang, D. E. Riley, S. L. Hillier, R. C. Berger, and J. N. Krieger. 1993. Detection of tet(M) and tet(O) tetracycline resistance genes by polymerase chain reaction. Mol. Cell. Probes 7:387-393. [DOI] [PubMed] [Google Scholar]
- 20.Salminen, S., A. von Wright, L. Morelli, P. Marteau, D. Brassart, W. M. De Vos, R. Fonden, M. Saxelin, K. Collins, G. Mogensen, S. E. Birkeland, and T. Mattila-Sandholm. 1998. Demonstration of safety of probiotics—a review. Int. J. Food Microbiol. 44:93-106. [DOI] [PubMed] [Google Scholar]
- 21.Santosa, S., E. Farnworth, and P. J. Jones. 2006. Probiotics and their potential health claims. Nutr. Rev. 64:265-274. [DOI] [PubMed] [Google Scholar]
- 22.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu, H., F. Qu, and L. H. Zhu. 1993. Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res. 21:5279-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]