Abstract
Its metabolic characteristics suggest that Zymobacter palmae gen. nov., sp. nov. could serve as a useful new ethanol-fermenting bacterium, but its biotechnological exploitation will require certain genetic modifications. We therefore engineered Z. palmae so as to broaden the range of its fermentable sugar substrates to include the pentose sugar xylose. The Escherichia coli genes encoding the xylose catabolic enzymes xylose isomerase, xylulokinase, transaldolase, and transketolase were introduced into Z. palmae, where their expression was driven by the Zymomonas mobilis glyceraldehyde-3-phosphate dehydrogenase promoter. When cultured with 40 g/liter xylose, the recombinant Z. palmae strain was able to ferment 16.4 g/liter xylose within 5 days, producing 91% of the theoretical yield of ethanol with no accumulation of organic acids as metabolic by-products. Notably, xylose acclimation enhanced both the expression of xylose catabolic enzymes and the rate of xylose uptake into recombinant Z. palmae, which enabled the acclimated organism to completely and simultaneously ferment a mixture of 40 g/liter glucose and 40 g/liter xylose within 8 h, producing 95% of the theoretical yield of ethanol. Thus, efficient fermentation of a mixture of glucose and xylose to ethanol can be accomplished by using Z. palmae expressing E. coli xylose catabolic enzymes.
Ethanol-producing bacteria have attracted much attention in recent years because their growth rate is substantially higher than that of the Saccharomyces presently used for practical production of fuel alcohol, and with the recent advances in biotechnology, they have the potential to play a key role in making production of ethanol much more economical. Among these ethanol-producing bacteria is the well-known organism Zymomonas mobilis, which has been used historically in tropical areas to make alcoholic beverages from plant sap (19). The advantages of Z. mobilis are its high growth rate and high specific ethanol production; unfortunately, its fermentable carbohydrate substrates are limited to glucose, fructose, and sucrose. On the other hand, the gram-negative bacterium Zymobacter palmae, which was isolated by Okamoto et al. (16) by using a broad range of carbohydrate substrates, is a facultative anaerobe that ferments hexoses, α-linked di- and trisaccharides, and sugar alcohols (glucose, fructose, galactose, maltose, melibiose, sucrose, raffinose, mannitol, and sorbitol). This organism produces approximately 2 mol of ethanol per mol of glucose without accumulation of by-products and shows productivity similar to that of Z. mobilis (16). Moreover, we previously described a host-vector system for Z. palmae and showed that by using the system we could breed a strain capable of producing ethanol from cellobiose (22).
In recent years, it has been suggested that, instead of traditional feedstocks (starch crops), cellulosic biomass (cellulose and hemicellulose), including agricultural and forestry residues, waste paper, and industrial waste, could be used as an ideally inexpensive and abundant source of sugars for fermentation into transportation fuel ethanol. One of those sugars, xylose, is the second most abundant carbohydrate in nature, and its commercial fermentation to ethanol could represent a practical alternative fuel source for the future.
Microbes such as yeasts and bacteria are essential for fermentation of xylose. For instance, Eliasson et al. and others have described the fermentation of xylose by a genetically engineered strain of Saccharomyces cerevisiae that expressed xylose reductase, xylitol dehydrogenase, and xylulokinase (XK) from Pichia stipitis (7, 10, 11, 12). In bacteria, there have been two approaches to breeding a strain capable of fermenting xylose (5). One approach was to genetically engineer a strain capable of utilizing a broad range of sugar substrates. Using that approach, Ingram et al. demonstrated xylose fermentation by strains of Escherichia coli and Klebsiella oxytoca harboring the pet operon (1, 15, 20). The second approach was to genetically engineer the ethanol-producing bacterium Z. mobilis by introducing E. coli genes encoding the xylose catabolic enzymes xylose isomerase (XI), XK, transaldolase (TA), and transketolase (TK). Using that approach, Zhang et al. demonstrated ethanol production from xylose that nearly reached the theoretical yield limit (23). For a process to be economically viable, however, there must be complete bioconversion of lignocellulosic biomass to ethanol, which requires the simultaneous fermentation of both glucose and xylose. For that reason, we have been focusing on Z. palmae, given its broad range of carbohydrate substrates and its ability to efficiently produce ethanol. But wild-type Z. palmae does not ferment xylose. We therefore investigated the possibility of conferring that ability on Z. palmae by introducing the E. coli genes encoding XI, XK, TA, and TK by using our previously described host-vector system (22). Here we report the simultaneous fermentation of xylose and glucose to ethanol by a recombinant Z. palmae strain expressing E. coli xylose catabolic enzymes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli JM 109 [recA supE endA hsdR gyrA relA thi Δ(lac-proAB) F′ tra ΔproAB lacIq lacZdM15] was used as the host strain for recombinant DNA manipulations. Z. palmae ATCC 51623 was used as the breeding host strain for the fermentation of xylose. E. coli K-12 was used as the source of the XI (xylA), XK (xylB), TA (tal), and TK (tktA) genes (2). Z. mobilis ATCC 29191 was used as the source of promoter genes of glycelaldehyde-3-phosphate dehdyrogenase (Pgap) and enolase (Peno) (3, 4). E. coli was grown in Luria-Bertani medium (10 g/liter Bacto tryptone, 5 g/liter Bacto yeast extract, 5 g/liter sodium chloride, pH 7.2) containing 100 μg/ml ampicillin. Z. mobilis was cultured statically in RM medium (20 g/liter glucose, 10 g/liter Bacto yeast extract, 2 g/liter potassium phosphate, pH 6.0) at 30°C. Z. palmae was cultured statically in T medium [20 g/liter glucose, 10 g/liter Bacto yeast extract, 10 g/liter potassium phosphate, 2 g/liter (NH4)2SO4, 0.5 g/liter MgSO4 · 7H2O, pH 6.0] at 30°C.
pUC118 was used to construct recombinant plasmids in E. coli. The broad-host-range vector plasmid used was pMFY31 harboring an RSF1010 replication origin (13.7 kbp; tetracycline, chloramphenicol, and ampicillin resistance) (9).
Transformation of Z. palmae.
To transform Z. palmae, the strain was initially precultured statically in 5 ml of T medium for 15 h at 30°C, after which the cells were added to 50 ml of fresh T medium and incubated for 90 min at 30°C. The culture broth was then chilled on ice and centrifuged at 3,000 rpm for 10 min at 4°C. The harvested cells were washed with 25 ml of ice-cold 10% glycerol and then resuspended in 1 ml of ice-cold glycerol. An aliquot (0.2 ml) of cell suspension was then transferred to a 0.2-cm electroporation cuvette, and 10 μl of plasmid DNA solution was added. Immediately after pulsing of the cell suspension with a Bio-Rad gene pulser apparatus (1.8 kV, 200 Ω, 25 μF; Bio-Rad Laboratories, Inc.), 1 ml of prewarmed (30°C) T medium was added and the cells were statically incubated for 1 h at 30°C. Finally, the cells were spread on selection plates and incubated for 1 to 3 days at 30°C.
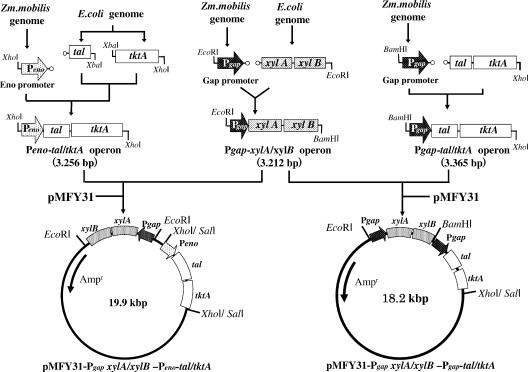
Construction of pMFY31-Pgap-xylA/xylB-Peno-tal/tktA and pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA.
An expression vector encoding a fused gene consisting of E. coli xylA and xylB under the control of the strong, constitutive Z. mobilis glyceraldehyde-3-phosphate dehydrogenase promoter was constructed by recombinant PCR (Fig. 1). Initially, a DNA fragment containing Pgap was amplified from Z. mobilis chromosomal DNA with the primer pair 5′-CGGAATTCGTTCGATCAACAACCCGAATCCTATCG (XYL1; 5′ primer; EcoRI site is underlined) and 5′-ATCGACTGGTCAAAATAGGCTTGCATAACGAAGACCATTTATTCCAGTA (XYL3; 3′ primer), while xylA and xylB with the Pgap flanking region were amplified from E. coli K-12 chromosomal DNA with the primer pair 5′-TACTGGAATAAATGGTCTTCGTTATGCAAGCCTATTTTGACCAGCCTCGAT (XYL2; 5′ primer) and 5′-ATAGCCCTCGAGGCGAATTATCCCCCACCCGGTCAGGCAG (XYL5; 3′ primer; BglI site is underlined). The two DNA fragments were then mixed, heated at 94°C for 20 min, and then incubated for an additional 45 min at 37°C to form a heteroduplex having an EcoRI site at its 5′ end and a BglI site at its 3′ end. Thereafter, xylA and xylB were amplified from the heteroduplex with primers XYL1 and XYL5. The 3,212-bp fused xylA/xylB fragment was then blunted and inserted into the HincII site within the multicloning site of pUC118, yielding pUC118-Pgap-xylA/xylB.
FIG. 1.
Construction of pMFY31-Pgap-xylA/xylB-Peno-tal/tktA and pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA. Arrows indicate the transcriptional direction. The sparsely dotted arrow indicates the promoter of the Z. mobilis enolase gene (eno), while the densely dotted bar indicates the promoter of the Z. mobilis glycelaldehyde-3-phosphate dehydrogenase gene (gap). The blank bars indicate the E. coli TA (tal) and TK (tktA) genes, respectively. The dotted bars indicate the E. coli XI (xylA) and XK (xylB) genes, respectively. The solid line indicates vector plasmid pMFY31.
In analogous fashion, a fused gene encoding E. coli tal and tktA under the control of the strong, constitutive Z. mobilis enolase (eno) promoter was constructed. A DNA fragment containing Peno was amplified from Z. mobilis chromosomal DNA with the primer pair 5′-TATGCCTCGAGGGCCAGTTACTCAATACGTAACAATAA (TAL9; 5′ primer; BglI site is underlined) and 5′-ACGAAGGGAGGTCAATTTGTCCGTCATATCGAAACCTTTCTTAAAATCTT (TAL3; 3′ primer). E. coli tal with the Peno flanking region was amplified from E. coli K-12 chromosomal DNA with the primer pair 5′-AAGATTTTAAGAAAGGTTTCGATATGACGGACAAATTGACCTCCCTTCGT (TAL2; 5′ primer) and 5′-CATTTTGACTACCAGATCTAGATTACAGCAGATCGCCGATCATTTTTTCC (TAL4; 3′ primer; XbaI site is underlined). With primers TAL9 (5′ primer) and TAL3 (3′ primer), the fused tal gene was amplified from the heteroduplex and BglI and XbaI sites were added at its 5′ and 3′ ends, respectively. The 1,290-bp fused tal fragment was then blunted and inserted into the HincII site within the multicloning site of pUC118, yielding pUC118-Peno-tal. The tktA fragment was then amplified from E. coli K-12 chromosomal DNA with the primer pair 5′-CGGAATTCTCGAGCTCCAGTTACTCAATACGTAACAATAA (TKT1; 5′ primer; XbaI site is underlined) and 5′-CGGCATGCCTCGAGGCAAACGGACATTATCAAGGTAATAAAAAAGGTCGC (TKT2; 3′ primer; XhoI site is underlined), which added XbaI and XhoI sites at the 5′ and 3′ ends, respectively. The 1,200-bp tktA fragment was then blunted and inserted into the HincII site of pUC118, yielding pUC118-tktA. To construct the operon consisting of tal and tktA under the control by Peno, the 1,200-bp tktA fragment was excised with XbaI and SphI, after which the SphI site was blunted and the fragment was inserted into the XbaI and SmaI sites of pUC118-Peno-tal downstream of Peno-tal, yielding pUC118-Peno-tal/tktA. In addition, to assess the effect of a promoter on the expression of tal and tktA, in some cases Z. mobilis Pgap was inserted upstream of tal. A DNA fragment containing Pgap was amplified from Z. mobilis chromosomal DNA with the primer pair 5′-TATGCCTCGAGGGCGTTCGATCAACAACCCGAATCCTATC (TAL8; 5′ primer; BglI site is underlined) and 5′-ACGAAGGGAGGTCAATTTGTCCGTCATGTTTATTCTCCTAACTTATTAAG (TAL7; 3′ primer). tal with the Pgap flanking region was then amplified from E. coli chromosomal DNA with primers 5′-CTTAATAAGTTAGGAGAATAAACATGACGGACAAATTGACCTCCCTTCGT (TAL6; 5′ primer) and TAL4, which added BglI and XbaI sites at the 5′ and 3′ ends, respectively. The 12,906-bp fused fragment was then blunted and inserted into the HincII site of pUC118, yielding pUC118-Pgap-tal. In the same way, tktA was then inserted downstream of tal in pUC118-Pgap-tal, yielding pUC118-Pgap-tal/tktA.
To introduce these genes into Z. palmae, the operons described above were inserted into broad-host-range vector plasmid pMFY31. The 3.2-kb Pgap-xylA/xylB fragment was excised from pUC118-Pgap-xylA/xylB with EcoRI and BglI, and the 3.4-kb Peno-tal/tktA fragment was excised from pUC118-Peno-tal/tktA with BglI and XhoI, after which the two were ligated into pMFY31 with EcoRI and SalI, yielding pMFY31-Pgap-xylA/xylB-Peno-tal/tktA. In addition, the 3.4-kb Pgap-tal/tktA fragment was excised from pUC118-Pgap-tal/tktA with BglI and XhoI and ligated, along with the Pgap-xylA/xylB fragment, into pMFY31 with EcoRI and SalI to construct pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA.
Acclimation of the recombinant Z. palmae strain with rapid growth in xylose.
A transformant on a 20-g/liter xylose agar plate supplemented with 100 μg/ml ampicillin was inoculated into a medium containing 40 g/liter xylose as a sole source of carbon and cultivated statically at 30°C. An aliquot of the culture medium in the early exponential growth phase was inoculated into the above medium and subcultured to enrich the strain capable of growing rapidly in xylose as a sole carbon source. After 10 passages, an aliquot of the culture medium was streaked onto an agar plate containing 40 g/liter xylose and 100 μg/ml ampicillin. Large colonies that formed were picked up and inoculated into 40 g/liter xylose medium, and then a strain that exhibited rapid growth in xylose medium was selected as an acclimated recombinant strain.
Preparation of crude extracts and enzyme assays.
Cells were harvested by centrifugation for 10 min at 5,000 × g and then washed with buffer (20 mM potassium phosphate buffer, pH 7.0). After sonication (2 × 30 s at 35 to 40 W) in an ice bath with an INSONICATOR (Kubota), the resultant lysate was cleared by centrifugation for 15 min at 35,000 × g and the supernatant was used as cell extract. The protein content of the extract was determined with a protein assay kit (Bio-Rad Laboratories, Inc.); bovine serum albumin served as the standard.
XI activity was assayed by determination of xylulose formed from xylose. The assay mixture contained 14 mM potassium phosphate (pH 7.0), 0.7 mM MnCl2, and cell extract in a total volume of 650 μl. The reaction was started by adding 8 mM d-xylose and subsequently stopped by adding 50 μl of 50% trichloroacetic acid. The amount of xylulose formed was determined colorimetrically by the cysteine-carbazole method (21). One unit of XI was defined as the amount of enzyme needed to produce 1 μmol of d-xylulose in 1 min.
XK was assayed by measuring xylulose-5-phosphate formation in a coupled reaction catalyzed by pyruvate kinase and lactate dehydrogenase (8). The assay mixture contained 56 mM Tris-HCl, 56 mM KCl (pH 7.8), 1 mM EDTA, 5.6 mM MgCl2, 1 mM phosphoenol pyruvate, 0.5 mM ATP, 0.3 mM NADH, 0.3 U lactate dehydrogenase, 0.3 U pyruvate kinase, and cell extract in a total volume of 900 μl. The reaction was started by adding 100 μl of 10 mM d-xylulose, after which NADH oxidation was determined spectroscopically as absorbance at 340 nm recorded at 30°C. One unit of XK was defined as the amount of enzyme needed to oxidize 1 μmol of NADH in 1 min.
TA was assayed in a reaction mixture containing 84 mM triethanolamine, 17 mM EDTA (pH 7.8), 33 mM fructose-6-phosphate, 0.15 mM NADH, 0.34 U glycerol 3-phosphate-dehydrogenase, 1 U triosephosphate isomerase, and cell extract (8a) in a total volume of 900 μl. The reaction was started by adding 100 μl of 6 mM erythrose 4-phosphate. NADH oxidation was determined spectroscopically as absorbance at 340 nm recorded at 30°C. One unit of TA was defined as the amount of enzyme needed to oxidize 1 μmol of NADH in 1 min.
TK was assayed in a reaction mixture containing 84 mM triethanolamine, 17 mM EDTA (pH 7.8), 0.56 mM ribose-5-phosphate, 0.154 mM thiamine pyrophosphate, 0.33 mM NADH, 0.34 U glycerol 3-phosphate-dehydrogenase, 1 U triosephosphate isomerase, and cell extract in a total volume of 900 μl (8a). The reaction was started by adding 100 μl of 5 mM xylulose 5-phosphate. NADH oxidation was determined spectroscopically as absorbance at 340 nm recorded at 30°C. One unit of TK was defined as the amount of enzyme needed to oxidize 1 μmol of NADH in 1 min.
Uptake of labeled xylose.
Cells in the late logarithmic phase were harvested by centrifugation and suspended to an optical density at 610 nm (OD610) of 20 (8.2 mg [dry weight] cells/ml) in 2× T medium without a carbon source. Thereafter, 2-ml aliquots of cell suspension were incubated in a shaking water bath for 10 min at 30°C to allow temperature equilibration. For analysis of xylose uptake, 80 g/liter xylose containing 1.5 μCi [U-14C]xylose (Amersham-Japan Corp.) was then added to the cell suspensions, after which samples (100 μl) were removed at 2-min intervals and filtered through membrane filters (0.45-μm pore size; Millipore Corp.) that were then washed with 3 ml of cold saline. The filters with cells were then transferred to 10 ml of scintillation cocktail (Organic Counting Scintillant; Amersham) for radioassay in a Packard liquid scintillation spectrometer. Results are expressed as nmoles of xylose taken up per milligram (dry weight) of cells.
Analysis.
The growth of strains was measured as the turbidity of culture medium at 610 nm, and a reading of 1.0 at 610 nm was equivalent to 0.41 mg (dry weight) of cells/ml. Level of xylose and glucose in culture broth were determined by high-performance liquid chromatography with a Shodex Ionpak KS801 (4.6 by 300 mm; Soko Co. Ltd.) at a column temperature of 75°C. The mobile phase was degassed water, the flow rate was 0.7 ml/min, and a refractive-index detector (model RID-10A; Shimadzu Corp.) was used. For metabolites such as lactate and acetate, an Aminex 8 (Bio-Rad Laboratories, Inc.) was used with 0.1 M hydrogen sulfate as the mobile phase at a column temperature of 40°C. Ethanol was assayed by gas chromatography with a glass column (0.26 by 200 cm) filled with Porapak type QS (80 to 100 mesh; Waters Corp.) at 180°C; nitrogen was the carrier gas (40 ml/min), and a flame ionization detector was used. The theoretical yield of ethanol was defined as 0.511 g of ethanol per g of xylose (5 mol of ethanol/3 mol of xylose). DNA sequencing was carried out by the dideoxy-chain termination method in an automated DNA sequencer (Applied Biosystems model 373A). The sequencing reaction was carried out with a BigDye Terminator v.3.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions.
RESULTS
Expression of xylose catabolic genes in Z. palmae.
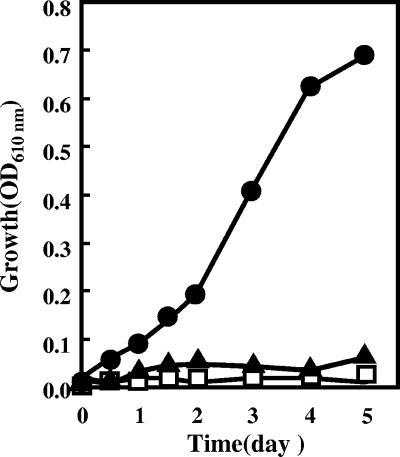
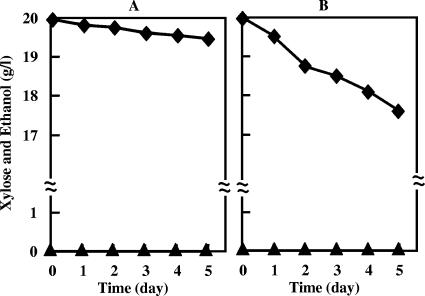
First, to enable Z. palmae fermentation of xylose to ethanol, the organism was transformed with recombinant plasmid pMFY31-Pgap-xylA/xylB-Peno-tal/tktA. Following transformation, the expression of XI, XK, TA, and TK activities driven by the Z. mobilis promoters was examined in cells grown in glucose under standard conditions (Table 1). Z. palmae harboring the vector pMFY31 showed no detectable XI or XK activity, which catalyzes the initial steps of xylose utilization. Some TA and TK activity was observed in the pentose phosphate pathway. Although the level of that activity was low, the primary reason Z. palmae cannot ferment xylose appears to be the absence of both XI and XK. Z. palmae harboring pMFY31-Pgap-xylA/xylB-Peno-tal/tktA efficiently expressed both XI and XK, but levels of TA and TK activity were comparatively low, indicating that the Z. mobilis Peno promoter did not drive TA and TK expression efficiently. Neither the wild-type strain nor the recombinant strain harboring pMFY31-Pgap-xylA/xylB-Peno-tal/tktA was able to grow when xylose was the sole source of carbon (Fig. 2). To clarify whether Z. palmae could transport xylose or not, resting cells of both the wild-type strain and the recombinant strain harboring pMFY31-Pgap-xylA/xylB-Peno-tal/tktA and grown in glucose were suspended in medium supplemented with 20 g/liter xylose and cultured at 30°C. Assays of xylose degradation by resting cells showed that although the wild-type strain could not degrade xylose at all (Fig. 3A), the strain harboring pMFY31-Pgap-xylA/xylB-Peno-tal/tktA gradually degraded xylose, there was no accumulation of ethanol (Fig. 3B). Apparently, this recombinant strain was only able to take up xylose and metabolize it to xylulose-5-phosphate via xylulose with XI and XK.
TABLE 1.
Expression of E. coli xylA, xylB, tal, and tktA in Z. palmae
| Plasmid or strain | Activitya of:
|
|||
|---|---|---|---|---|
| XIc (mU/mg) | XKd (U/mg) | TAe (U/mg) | TKf (U/mg) | |
| pMFY31 | <0.01 | <0.01 | 0.01 | 0.01 |
| pMFY31-Pgap-xylA/xylB-Peno-tal/tktA | 9.78 | 0.64 | 0.05 | 0.06 |
| pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA | 9.33 | 0.81 | 0.25 | 0.26 |
| E. coli K-12b | 3.23 | 0.28 | 0.19 | 0.14 |
Cells were cultured in T medium containing 20 g/liter glucose and 100 μg/ml of ampicillin for 48 h at 30°C, and harvested cells were disrupted with an ultrasonic oscillator. The resulting supernatant was used as cell extract.
E. coli K-12 was cultured in Luria-Bertani medium for 24 h at 37°C with shaking, and cell extract was prepared from the harvested cells as described in footnote a.
One unit of XI produces 1 μmol of xylulose/min.
One unit of XK produces 1 μmol of NADH/min.
One unit of TA decreases 1 μmol of NADH/min.
One unit of TK decreases 1 μmol of NADH/min.
FIG. 2.
Growth of recombinant Z. palmae in xylose-containing medium. Transformants were inoculated into T medium containing 20 g/liter xylose and 100 μg/ml ampicillin and cultured statically at 30°C. Closed circles indicate the strain harboring pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA, closed triangles indicate the strain harboring pMFY31-Pgap-xylA/xylB-Peno-tal/tktA, and open squares indicate the strain harboring pMFY31.
FIG. 3.
Degradation of xylose by recombinant Z. palmae under resting conditions. Strains harboring pMFY31 (A) or pMFY31-Pgap-xylA/xylB-Peno-tal/tktA (B) were cultured in T medium containing 20 g/liter glucose and 100 μg/ml ampicillin for 48 h at 30°C, after which harvested cells were resuspended in T medium containing 20 g/liter xylose to give a concentration of 8.2 mg (dry weight) of cells/ml. The time course of xylose degradation was determined by submitting aliquots of culture medium to high-performance liquid chromatography as described in Materials and Methods. Diamonds indicate residual xylose, and triangles indicate accumulated ethanol.
On the basis of the above results, to express both TA and TK efficiently, recombinant plasmid pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA was constructed and then introduced into Z. palmae. The Z. mobilis Pgap promoter drove efficient expression of all four enzyme genes in Z. palmae, where the levels of all of the enzymes' activities were two- to threefold higher than in E. coli (Table 1). Z. palmae harboring pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA was able to grow when xylose was the sole carbon source (Fig. 2), which reflects both the functionality of all four of the enzymes involved in xylose metabolism and the importance for cell growth of converting xylulose phosphate to glyceraldehyde-3-phosphate and fructose-6-phosphate via the pentose phosphate pathway.
Fermentation of xylose by Z. palmae harboring pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA.
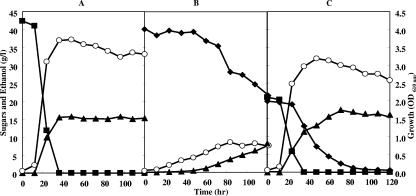
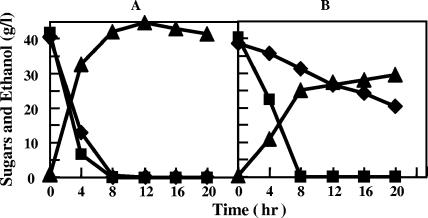
After preculturing of recombinant Z. palmae for 48 h in T medium supplemented with 20 g/liter xylose, the cells were harvested and washed with T medium lacking a carbon source. The washed cells were then inoculated into T medium supplemented with 40 g/liter xylose and cultivated statically. The culture medium initially had an OD610 of 0.1 (about 106 cells/ml). The strain harboring pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA grew in xylose, reaching an OD610 of about 0.8 after 5 days of cultivation, which was lower than that obtained when the cells were cultured in 40 g/liter glucose (Fig. 4A and B). During that 5-day period, 16.4 g/liter xylose was consumed to produce 7.4 g/liter ethanol, which is 91% of the theoretical yield. Although no organic acids (succinate, lactate, or acetate) formed as by-products were detected during the cultivation, a small amount of an unknown metabolite was detected (data not shown). In the presence of 20 g/liter glucose, the rate of xylose consumption was increased about twofold of that in 40 g/liter xylose alone, and 20 g/liter xylose was utilized perfectly to produce 16.5 g/liter ethanol within 96 h of cultivation (Fig. 4C). However, the fermentation profile of a mixture of glucose and xylose by the recombinant strain exhibited a diauxic shift from glucose to xylose, indicating that xylose was fermented by a large amount of cells grown preferentially in glucose.
FIG. 4.
Cell growth and fermentation of glucose and xylose in Z. palmae/pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA. The recombinant strain was cultured statically in T medium containing 40 g/liter glucose (A), 40 g/liter xylose (B), or a mixture of 20 g/liter glucose and 20 g/liter xylose (C) at 30°C. Open circles indicate the growth of the strain, closed squares indicate residual glucose, closed diamonds indicate residual xylose, and closed triangles indicate the accumulation of ethanol.
Acclimation of Z. palmae harboring pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA on xylose.
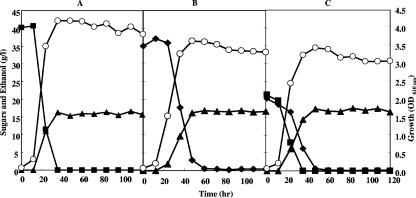
To select for Z. palmae harboring pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA and capable of higher ethanol production from xylose with rapid growth, the strain was repetitively grown and subcultured in medium supplemented with 40 g/liter xylose. After 10 passages, an aliquot of the culture broth was streaked onto a plate containing xylose and the large colony that formed was collected. This acclimated strain grew well with xylose as the sole carbon source, exhibiting a growth rate similar to that seen in glucose (Fig. 5A and B), and after only 2 days of cultivation had fermented the 40 g/liter xylose completely to produce ethanol at 93% of the theoretical yield (Fig. 5B). Moreover, in the presence of a mixture of 20 g/liter glucose and 20 g/liter xylose, the strain completely fermented both sugars to ethanol with growth of the organism (Fig. 5C). Although glucose was preferentially utilized and fermented at a faster rate than xylose, simultaneous xylose fermentation was ongoing in the presence of glucose.
FIG. 5.
Cell growth and fermentation of glucose and xylose in acclimated Z. palmae/pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA. The acclimated strain was cultured statically in T medium containing 40 g/liter glucose (A), 40 g/liter xylose (B), or a mixture of 20 g/liter glucose and 20 g/liter xylose (C) at 30°C. Open circles indicate the growth of the strain, closed squares indicate residual glucose, closed diamonds indicate residual xylose, and closed triangles indicate the accumulation of ethanol.
Simultaneous fermentation of a mixture of glucose and xylose by acclimated Z. palmae precultured in either glucose or xylose.
To verify simultaneous fermentation of a mixture of glucose and xylose by the acclimated strain experimentally, an initial consumption of sugars in a mixture of glucose and xylose by the resting cells of the acclimated strain was measured with the passage of time. Acclimated Z. palmae harboring pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA was precultured for 48 h in T medium supplemented with either xylose or glucose as the sole carbon source, after which the cells were harvested and suspended at 8.2 mg (dry weight) cells/ml in T medium containing mixtures of 40 g/liter glucose and 40 g/liter xylose. As shown in Fig. 6A, cells precultured in xylose rapidly fermented all of the glucose and xylose available within 8 h. In addition, because the rates of consumption of glucose and xylose were similar, both glucose and xylose were consumed synchronously and ethanol was produced to a level approximating the theoretical yield. By contrast, cells precultured in glucose fermented 20 g/liter xylose during a 20-h incubation in medium containing 40 g/liter glucose and 40 g/liter xylose (Fig. 6 B). The cells fermented glucose and xylose simultaneously, but the rate of xylose fermentation was substantially slower than that of glucose fermentation. Although the rate of xylose fermentation appeared to be dependent on a carbohydrate with precultivation of the acclimated strain, the difference in the rate of xylose fermentation was not observed in the usual batch fermentation test accompanying cell growth by inoculation of the acclimated cells precultured in either glucose or xylose.
FIG. 6.
Simultaneous fermentation of xylose by acclimated Z. palmae/pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA under resting conditions. The acclimated strain was precultured statically in T medium containing 40 g/liter xylose (A) or 40 g/liter glucose (B) for 48 h at 30°C. Harvested cells were resuspended in T medium containing a mixture of 40 g/liter glucose and 40 g/liter xylose to give a concentration of 8.2 mg (dry weight) of cells/ml and incubated statically at 30°C. Closed squares indicate residual glucose, closed diamonds indicate residual xylose, and closed triangles indicate the accumulation of ethanol.
Two possible explanations for the difference in the rate of xylose fermentation by the acclimated cells precultured in glucose or xylose are that cultivating the cells in xylose (i) upregulates expression of XI, XK, TA, and TK or (ii) enhances xylose uptake into cells. To test the effect of the carbon source on enzyme expression, xylose-acclimated Z. palmae was cultivated for 48 h with glucose or xylose as the sole carbon source, after which the cells were harvested and disrupted by ultrasonication to prepare a cell extract. As shown in Table 2, the activities of XI, XK, TA, and TK were two to eightfold higher in acclimated Z. palmae grown in xylose than in the unacclimated strain (compare Tables 1 and 2). However, the levels of enzyme expression in acclimated Z. palmae grown in glucose or xylose were similar (Table 2). It thus does not appear that the carbon source significantly affects enzyme expression, which means that the enhanced xylose fermentation by cells grown in xylose was not caused by increased expression of enzyme genes in acclimated Z. palmae.
TABLE 2.
Effects of carbon sources on expression of E. coli enzyme genes in acclimated Z. palmae/pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA
| Plasmid | Carbon source | Activiya of:
|
|||
|---|---|---|---|---|---|
| XIb (mU/mg) | XKc (U/mg) | TAd (U/mg) | TKe (U/mg) | ||
| pMFY31 | Glucose | <0.01 | <0.01 | 0.01 | 0.02 |
| pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA | Glucose | 45.5 | 2.65 | 0.85 | 0.47 |
| pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA | Xylose | 65.3 | 3.75 | 1.94 | 1.14 |
Cells were cultured in T medium containing 40 g/liter glucose or xylose and 100 μg/ml of ampicillin for 48 h at 30°C, and harvested cells were disrupted with an ultrasonic oscillator. The resulting supernatant was used as cell extract.
One unit of XI produces 1 μmol of xylulose/min.
One unit of XK produces 1 μmol of NADH/min.
One unit of TA decreases 1 μmol of NADH/min.
One unit of TK decreases 1 μmol of NADH/min.
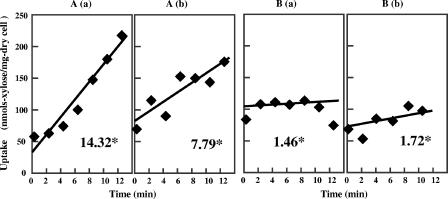
We next assessed the effect of the carbon source on the uptake of isotopic d-xylose by acclimated Z. palmae harboring pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA and precultured in xylose or glucose. Harvested cells were resuspended in medium without a carbon source, after which xylose uptake was initiated by addition of an equal volume of 80 g/liter xylose with 1.5 mCi/ml [U-C14]xylose to the cell suspensions. As shown in Fig. 7, the rate of incorporation of isotopic xylose was dependent on the carbon source. The rate of xylose uptake in xylose-grown cells was 10-fold higher than that in glucose-grown cells (Fig. 7A, part a, and B, part a), and although the presence of glucose reduced the rate of xylose uptake by xylose-precultured cells by nearly half (Fig. 7A, parts a and b), the rate remained significantly greater than that seen in glucose-precultured cells (Fig. 7B, part b).
FIG. 7.
Xylose uptake into Z. palmae/pMFY31-Pgap-xylA/xylB-Pgap-tal/tktA. The acclimated strain was cultured statically under the conditions described in the legend to Fig. 6. Washed cells precultured in xylose (A) or glucose (B) were suspended to give a concentration of 8.2 mg (dry weight) of cells/ml in 2 ml of 2× T medium without a carbon source. Uptake was then initiated by adding to the cell suspension 2 ml of 80 g/liter xylose containing 1.5 μCi [U-14C]xylose (a) or 2 ml of a mixture of 80 g/liter glucose and 80 g/liter xylose containing 1.5 μCi [U-14C]xylose (b). Samples (100 μl) were removed at 2-min intervals and passed through membrane filters, after which the filters were washed with 3 ml of cold saline and transferred to 10 ml of scintillation cocktail for radioassay. The diamonds indicate the xylose uptake, which is expressed as nanomoles of xylose per milligram (dry weight) of cells. The asterisks indicate the rate of xylose uptake (nmol/min · mg of cells).
DISCUSSION
We have demonstrated simultaneous fermentation by genetically engineered Z. palmae of glucose and xylose, which is the second most abundant sugar in nature. There have been reported several microorganisms capable of producing ethanol from xylose by genetic engineering. Eliasson et al. (7) and Karhumaa et al. (12) have constructed a xylose-fermenting S. cerevisiae strain by introduction and coexpression of genes encoding P. stipitis xylose reductase (xyl1), xylitol dehydrogenase (xyl2), and XK (xks1) and/or Thermus thermophilus XI (xylA). They have reported that the ethanol yield from xylose by the recombinant S. cerevisiae was 0.21 to 0.29 g ethanol/g xylose with accumulation of xylitol (0.23 g/g xylose) as a by-product. Recombinant Z. mobilis (23), E. coli (20), and K. oxytoca (15) have been reported as xylose-fermenting bacterial strains. Although recombinant E. coli and K. oxytoca could ferment xylose with almost the theoretical yield of ethanol, these bacteria appeared to be unable to ferment xylose at high ethanol concentrations. Meanwhile, the recombinant Z. mobilis strain that was constructed to coexpress E. coli genes xylA, xylB, tal, and tktA could ferment xylose with almost the theoretical yield of ethanol (23). However, the recombinant Z. mobilis strain could not ferment a mixture of glucose and xylose synchronously. In contrast, the acclimated Z. palmae strain could ferment a mixture of glucose and xylose synchronously with almost the theoretical yield of ethanol. This peculiar property of the acclimated Z. palmae strain appears to be suitable for the continuous fermentation of acid hydrolysates prepared from cellulosic feedstocks.
Little is known about the glycolytic and pentose phosphate pathways in Z. palmae; however, we observed here that wild-type Z. palmae does not express either XI or XK, which catalyze the first steps of xylose catabolism in bacteria (Table 1), while levels of TA and TK activity were lower in Z. palmae than in E. coli. When we then tried to express the E. coli genes encoding XI, XK, TA, and TK in Z. palmae, we found that cells harboring pMFY31-Pgap-xylA/B-Peno-tal/tktA, which expressed only little TA and TK activity, could not grow in xylose, although both XI and XK were expressed at higher levels than in E. coli. This suggests that the level of TA and TK expression was not sufficient to convert xylulose phosphate to glyceraldehyde-3-phosphate and fructose-6-phosphate via the pentose phosphate pathway, raising the possibility that an accumulated metabolite might have inhibited the growth of the strain. Consistent with that idea, Feldmann et al. reported that Z. mobilis harboring xylA and xylB from Klebsiella pneumoniae could not grow in xylose because of growth inhibition caused by the accumulation of xylitol phosphate due to the activities of endogenous aldose reductase and XK (8a).
When Lawford and Rousseau tested the performance of Z. mobilis metabolically engineered for cofermentation of glucose, xylose and arabinose (13), they found that the order of sugar exhaustion from the medium was glucose followed by xylose and arabinose. Similarly, Mohagheghi et al. observed the same order of sugar preference in a genomic-DNA-integrated xylose-arabinose-fermenting Z. mobilis strain, AX101 (14). The efficient expression of the genes encoding all four xylose catabolic enzymes under the control of the Pgap promoter provided Z. palmae with the abilities to grow in xylose and produce ethanol at a level that was 91% of the theoretical yield. Moreover, there was no accumulation of organic acids (succinate, lactate, and acetate) as metabolic end products. Thus, comparison with the rate of glucose fermentation revealed that the recombinant strain fermented xylose relatively slowly. Interestingly, in a mixture of 20 g/liter glucose and 20 g/liter xylose, the xylose consumption rate was increased markedly compared to that in the presence of xylose alone (Fig. 4B and C). However, xylose fermentation was observed after glucose consumption. In this case, the increased numbers of cells grown by utilizing glucose reflected the increased rate of xylose fermentation. To improve the rate of xylose fermentation by recombinant Z. palmae, we used xylose acclimation to select a recombinant strain that would exhibit significant growth when xylose was the sole carbon source. The acclimated strain was able to ferment xylose at a rate similar to that of glucose fermentation, and the fermentation of xylose was synchronized with that of glucose.
It is noteworthy that the rate of fermentation by xylose-acclimated cells was dependent on the carbon source used for precultivation of the strain. Although resting xylose-acclimated cells precultivated in glucose fermented glucose and xylose simultaneously, the rate of xylose fermentation was markedly slower than that of cells precultivated in xylose. We initially considered the possibility that the different rates of xylose and glucose fermentation reflected different levels of expression of xylose catabolic enzymes. We found, however, that although the activities of XI, XK, TA, and TK were all increased in the xylose-acclimated strain, the expression levels in acclimated cells grown in glucose or xylose did not significantly differ. Apparently, incorporated xylose was converted to fructose-6-phosphate and glyceraldehyde-3-phosphate at similar rates, irrespective of whether the acclimated cells were precultivated in glucose or xylose, which means that a difference in enzyme expression cannot account for the difference in the rates of xylose fermentation between cells grown in glucose or xylose.
Instead, we found that while all xylose-acclimated cells exhibited high rates of uptake compared to unacclimated cells, whether they were precultivated in glucose or xylose, the rate of xylose uptake by cells grown in xylose was almost 10-fold the rate of uptake by cells grown in glucose (Fig. 7). In Z. mobilis, xylose is transported into cells via glucose facilitator (Glf), a carrier-mediated facilitated diffusion system (6, 17). In that system, uptake of xylose is inhibited in the presence of glucose, which appears to behave as a competitive inhibitor. Our finding that the rate of xylose uptake by acclimated Z. palmae was slower in the presence of glucose (Fig. 7A, part b) is consistent with that idea, although to our knowledge there have been no reports on the sugar transport system of Z. palmae. It is also noteworthy that the rate of xylose fermentation by acclimated Z. palmae precultivated in glucose did not increase after the glucose was exhausted (Fig. 6B), which suggests that glucose does not act solely as a competitive inhibitor and/or that glucose is not the only factor exerting an effect. It may be that in Z. palmae xylose is incorporated via a different transport system with a broad range of substrate sugars other than glucose. In addition, a certain sugar transport system in the xylose-acclimated strain might be upregulated by xylose as a source of carbon. Further work is in progress to clarify the mechanism of the acclimation by analysis of the expression level of genes encoding carbohydrate metabolism and its transport system during cultivation in xylose or glucose.
Pyruvate decarboxylase is reportedly involved in the decarboxylation of pyruvate in Z. palmae (18), which is known to be a key enzyme involved in ethanol production in the traditional brewing microorganisms S. cerevisiae and Z. mobilis. However, no reports have yet appeared in which members of the glycolysis and pentose phosphate pathways were examined in Z. palmae. In this study, we demonstrated that Z. palmae is a potentially useful strain with which to produce ethanol from the component sugars present in lignocellulosic biomass. We anticipate that further characterization of sugar transport and metabolism in Z. palmae will facilitate genetic engineering of strains capable of producing ethanol from a variety of sugars.
Acknowledgments
This work was financed by the New Energy and Industrial Technology Development Organization (NEDO), Tokyo, Japan.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Alterthum, F., and L. O. Ingram. 1989. Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl. Environ. Microbiol. 55:1943-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Colladovides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Burnett, M. E., J. Liu, and T. Conway. 1992. Molecular characterization of the Zymomonas mobilis enolase (eno) gene. J. Bacteriol. 174:6548-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conway, T., G. W. Sewell, and L. O. Ingram. 1987. Glyceraldehyde-3-phosphate dehydrogenase gene from Zymomonas mobilis: cloning, sequencing, and identification of promoter region. J. Bacteriol. 169:5653-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dien, B. S., M. A. Cotta, and T. W. Jeffries. 2003. Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63:258-266. [DOI] [PubMed] [Google Scholar]
- 6.Dimarco, A. A., and A. H. Romano. 1985. d-Glucose transport system of Zymomonas mobilis. Appl. Environ. Microbiol. 49:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eliasson, A., C. Christensson, C. F. Wahlbom, and B. Hahn-Hagerdal. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66:3381-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann, S. D., H. Sahm, and G. A. Sprenger. 1992. Cloning and expression of the genes for xylose isomerase and xylulokinase from Klebsiella pneumoniae 1033 in Escherichia coli K12. Mol. Gen. Genet. 234:201-210. [DOI] [PubMed] [Google Scholar]
- 8a.Feldmann, S. D., H. Sahm, and G. A. Sprenger. 1992. Pentose metabolism in Zymomonas mobilis wild-type and recombinant strains. Appl. Microbiol. Biotechnol. 38:354-361. [Google Scholar]
- 9.Fukuda, M., and K. Yano. 1985. Construction of broad host range cloning vectors for gram-negative bacteria. Agric. Biol. Chem. 49:2719-2724. [Google Scholar]
- 10.Jeffries, T. W. 2006. Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 17:320-326. [DOI] [PubMed] [Google Scholar]
- 11.Jeppsson, M., B. Johansson, B. Hahn-Hagerdal, and M. F. Gorwa-Grauslund. 2002. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl. Environ. Microbiol. 68:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karhumaa, K., B. Hahn-Hagerdal, and M. F. Gorwa-Grauslund. 2005. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22:359-368. [DOI] [PubMed] [Google Scholar]
- 13.Lawford, H. G., and J. D. Rousseau. 2002. Performance testing of Zymomonas mobilis metabolically engineered for cofermentation of glucose, xylose, and arabinose. Appl. Biochem. Biotechnol. 98-100:429-448. [DOI] [PubMed] [Google Scholar]
- 14.Mohagheghi, A., K. Evans, Y. C. Chou, and M. Zhang. 2002. Cofermentation of glucose, xylose, and arabinose by genomic DNA-integrated xylose/arabinose fermenting strain of Zymomonas mobilis AX101. Appl. Biochem. Biotechnol. 98-100:885-898. [PubMed] [Google Scholar]
- 15.Ohta, K., D. S. Beall, J. P. Mejia, K. T. Shanmugam, and L. O. Ingram. 1991. Metabolic engineering of Klebsiella oxytoca M5A1 for ethanol production from xylose and glucose. Appl. Environ. Microbiol. 57:2810-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamoto, T., H. Taguchi, K. Nakamura, H. Ikenaga, H. Kuraishi, and K. Yamasato. 1993. Zymobacter palmae gen. nov., sp. nov., a new ethanol-fermenting peritrichous bacterium isolated from palm sap. Arch. Microbiol. 160:333-337. [DOI] [PubMed] [Google Scholar]
- 17.Parker, C., W. O. Barnell, J. L. Snoep, L. O. Ingram, and T. Conway. 1995. Characterization of the Zymomonas mobilis glucose facilitator gene product (glf) in recombinant Escherichia coli: examination of transport mechanism, kinetics and the role of glucokinase in glucose transport. Mol. Microbiol. 15:795-802. [DOI] [PubMed] [Google Scholar]
- 18.Raj, K. C., L. A. Talarico, L. O. Ingram, and J. A. Maupin-Furlow. 2002. Cloning and characterization of the Zymobacter palmae pyruvate decarboxylase gene (pdc) and comparison to bacterial homologues. Appl. Environ. Microbiol. 68:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skotnicki, M. L., R. G. Warr, A. E. Goodman, K. J. Lee, and P. L. Rogers. 1983. High-productivity alcohol fermentations using Zymomonas mobilis. Biochem. Soc. Symp. 48:53-86. [PubMed] [Google Scholar]
- 20.Underwood, S. A., M. L. Buszko, K. T. Shanmugam, and L. O. Ingram. 2004. Lack of protective osmolytes limits final cell density and volumetric productivity of ethanologenic Escherichia coli KO11 during xylose fermentation. Appl. Environ. Microbiol. 70:2734-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamanaka, K. 1975. Xylose isomerase. Methods Enzymol. 41:588-593. [DOI] [PubMed] [Google Scholar]
- 22.Yanase, H., K. Yamamoto, D. Sato, and K. Okamoto. 2005. Ethanol production from cellobiose by Zymobacter palmae carrying the Ruminococcus albus β-glucosidase gene. J. Biotechnol. 118:35-43. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, M., C. Eddy, K. Deanda, M. Finkelstein, and S. Picataggio. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240-243. [DOI] [PubMed] [Google Scholar]