Abstract
Bacteria in the phylum Acidobacteria are widely distributed and abundant in soils, but their ecological roles are poorly understood, owing in part to a paucity of cultured representatives. In a molecular survey of acidobacterial diversity at the Michigan State University Kellogg Biological Station Long-Term Ecological Research site, 27% of acidobacterial 16S rRNA gene clones in a never-tilled, successional plant community belonged to subdivision 1, whose relative abundance varied inversely with soil pH. Strains of subdivision 1 were isolated from these never-tilled soils using low-nutrient medium incubated for 3 to 4 weeks under elevated levels of carbon dioxide, which resulted in a slightly acidified medium that matched the pH optima of the strains (between 5 and 6). Colonies were approximately 1 mm in diameter and either white or pink, the latter due to a carotenoid(s) that was synthesized preferentially under 20% instead of 2% oxygen. Strains were gram-negative, aerobic, chemo-organotrophic, nonmotile rods that produced an extracellular matrix. All strains contained either one or two copies of the 16S rRNA encoding gene, which along with a relatively slow doubling time (10 to 15 h at ca. 23°C) is suggestive of an oligotrophic lifestyle. Six of the strains are sufficiently similar to one another, but distinct from previously named Acidobacteria, to warrant creation of a new genus, Terriglobus, with Terriglobus roseus defined as the type species. The physiological and nutritional characteristics of Terriglobus are consistent with its potential widespread distribution in soil.
Soils typically contain 109 to 1010 microorganisms per gram (dry weight), which may represent more than a million bacterial species (21). However, characterization of the small fraction of microbes that has been cultivated provides only a glimpse of their potential physiological capacity and influence on soil ecosystems. The absence of pure cultures or genome sequences makes it difficult to ascertain the roles of specific microbes in soil environments: this is particularly true for bacteria in the phylum Acidobacteria, which are broadly distributed in soils but poorly represented in culture.
The phylum Acidobacteria is defined by a large collection of 16S rRNA gene sequences (>1,500 in the Ribosome Database Project) (10) retrieved from diverse environments including soils and sediments (3, 17), soil crusts of sand dunes (69), wastewater (13, 41), water distribution systems (49), peat bogs (15), acid mine drainage (33), hot springs (26), shallow submarine hydrothermal vents (67), and the surfaces of Paleolithic cave paintings and catacombs (62-64, 75, 76). In situ hybridization with Acidobacteria-specific probes has also confirmed the presence of intact acidobacteria in many environments and revealed multiple cellular morphotypes, including cocci, short rods, and thin filaments (46).
Acidobacterium capsulatum was the first described member of the phylum Acidobacteria. It was among eight strains isolated from acidic mine drainage and acidic muds in the early 1990s (33, 34). This was the first report of the isolation of acidophilic, heterotrophic bacteria other than Acidiphilium from an acidic mineral environment (33). By the mid-1990s, the growing collection of rRNA gene sequences from molecular surveys of diverse environments resulted in the recognition of A. capsulatum as a member of a large, deeply branching, monophyletic lineage within the Bacteria (25). The phylum Acidobacteria was named after the only validly described species at the time (46).
Shortly after the characterization of A. capsulatum, Holophaga foetida was isolated and described as a novel, homoacetogenic bacterium capable of degrading methoxylated aromatic compounds (43). Based in part on the 81.6% divergence in the 16S rRNA gene sequences of these two cultivars, the phylum was sometimes referred to as Holophaga/Acidobacterium. Geothrix fermentans, a strain capable of Fe(III) reduction, was later placed in the Holophaga/Acidobacterium phylum based on the similarity of its 16S rRNA gene sequence to that of H. foetida (ca. 94%) (9).
The phylum Acidobacteria is now officially recognized in Bergey's Manual of Systematic Bacteriology (22) and includes three genera with cultured representatives: Acidobacterium (33), Geothrix (9), and Holophaga (43). The genus Solibacter was recently proposed as the fourth genus in this phylum (http://jgi.doe.gov). There are currently eight recognized monophyletic subdivisions within this phylum (28) that encompasses the molecular diversity first recognized as Acidobacteria (38) and additional unnamed and mostly uncharacterized cultivars in subdivisions 1, 2, 3, and 4 (14, 29-31, 60, 61, 70). A recent survey of 23S rRNA genes in microbial communities associated with Paleolithic paintings uncovered additional acidobacteria, expanding the number of subdivisions to as many as eleven (76).
Acidobacteria are oftentimes the most abundant bacteria represented in molecular surveys of soil environments: as many as half of all clones from Arizona soil samples clustered in the phylum Acidobacteria (16), as did more than 40% of PCR-amplified and cloned rRNA-encoding genes in soils of alpine ecosystems (44). In a comprehensive review of acidobacterial abundance in soil communities (29), acidobacteria averaged ca. 20% of the total bacterial community. The breadth of divergence of 16S rRNA gene sequences in the phylum Acidobacteria (ca. 77% based on >1,000 nearly full-length sequences [this study]) is similar to that within the metabolically diverse phylum Proteobacteria (28), suggesting the capacity for extensive metabolic diversity. Although the metabolic potential of acidobacteria is poorly described, their abundance suggests a major impact on nutrient cycling in soil environments.
To learn more about the metabolic properties and potential ecological roles of members of this poorly explored phylum, we sought to cultivate and characterize new strains from terrestrial habitats. By using incubation conditions and media designed to mimic natural environments, eight strains of the phylum Acidobacteria were isolated from soil as well as from the hindguts of soil-dwelling termites. These strains were characterized, with an emphasis on properties that may bear on their ecological roles in soil. Results from the physiological and phylogenetic characterization warrant creation of a new genus, Terriglobus.
MATERIALS AND METHODS
Phylogenetic survey and isolation of strains.
Soil samples were collected between August 2001 and August 2005 from the Michigan State University W. K. Kellogg Biological Station Long-Term Ecological Research (KBS LTER) site. The KBS LTER is a 48-hectare research site established in 1989 to study ecological processes in agroecosystems. The dominant soil types are of the Kalamazoo (fine-loamy, mixed, mesic Typic Hapludalfs) and Oshtemo (coarse-loamy, mixed, mesic Typic Hapludalfs) series. A detailed description of this site can be accessed at http://www.kbs.msu.edu.
For the survey of acidobacterial diversity, soil cores (2.5-cm diameter by 20-cm depth, divided into 0 to 7 cm and 13 to 20 cm) were collected from four different treatments at the KBS LTER: treatment 1 is managed as a conventional agriculture site with a rotation of corn, soybean, and wheat; treatment 7 is a successional plant community on historically tilled soil (tillage was abandoned after spring plowing in 1989); treatment 8 (T8) is a successional plant community on never-tilled soil; and the fourth treatment is a native deciduous forest. Five soil cores per plot were pooled and homogenized using a 2-mm-pore-size sieve; portions of the sieved soil were used to determine soil pH and soil moisture (58), and the remaining soil was frozen immediately in liquid nitrogen and stored at −80°C until use. DNA was extracted from soil samples using an UltraClean Fecal DNA MoBio DNA Extraction Kit (MoBio, Carlsbad, CA). The 16S rRNA genes were amplified from the DNA using PCR with an Acidobacteria-specific forward primer (ACD31F, 5′-GATCCTGGCTCAGAATC-3′) (3) and a broadly inclusive bacterial reverse primer (1492R, 5′-GGTTACCTTGTTACGACTT-3′) (39, 40). Each 25-μl PCR mixture contained 1× PCR buffer, 1 mM MgCl2, a 0.03 mM concentration of each deoxynucleoside triphosphate, a 0.2 μM concentration of each primer, and 5 U of Taq DNA polymerase (Invitrogen, Carlsbad, CA). Thermal cycling consisted of the following steps: 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 45 s, and a final step at 72°C for 10 min. Genomic DNA purified from A. capsulatum (ATCC 51196) was used as a positive control. PCR products were electrophoresed through a 1% agarose gel in 0.5× Tris-borate-EDTA buffer and visualized with GelStar Nucleic Acid Stain (BioWhittaker Molecular Applications, Rockland, MA).
PCR products were cloned with an Invitrogen TOPO TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA), and the inserts were reamplified using the primers modified-M13F (F2; 5′-CAGTCACGACGTTGTAAAACGACGGC-3′) and modifed-M13R (F4; 5′-CAGGAAACAGCTATGACCATG-3′) (32) under the PCR conditions described above. The PCR products for sequencing were treated with ExoSAP-IT (USB, Cleveland, OH) using a modification of the manufacturer's protocol (ExoSAP enzyme was diluted 1:8, and the reaction mixture was incubated at 37°C for 30 min, followed by incubation at 80°C for 15 min) and submitted for sequencing with primer 531R (5′-TACCGCGGCTGCTGGCAC-3′). Sequencing was performed at the Michigan State University Research Technology Support Facility (http://genomics.msu.edu). Sequences were aligned with other sequences downloaded from the GenBank database using an integrated aligner (ARB Software) (47) as well as manual corrections based on secondary structure models of the 16S rRNA gene.
For isolation attempts, five cores (2.5-cm diameter by 10-cm depth) from the never-tilled, successional treatment (T8) were collected and homogenized with a sterile spatula in a 500-ml beaker under a hypoxic atmosphere ([vol/vol] 2% O2, 5% CO2, 93% N2) within a flexible vinyl chamber fitted with an oxygen sensor/controller (Coy Laboratory Products, Grass Lake, MI). Portions of the homogenized soil were used to determine soil moisture (58). Approximately 30 g of soil was added to 100 ml of a phosphate-buffered salts solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4); pH was adjusted to 7.0, and the solution was supplemented with 2.24 mM Na4P2O7·10H2O as a dispersal agent and dithiothreitol (1 mM) as a reducing agent (73). The soil suspension was stirred vigorously with a magnetic stir bar for 30 min. Denser soil aggregates were then allowed to settle for 30 min, after which an aliquot of the supernatant fraction was used for total direct cell counts after staining with 5-(4,6-dichlorotriazine-2-yl) aminofluorescein (4). Another aliquot was used to prepare 10-fold serial dilutions in the same buffer for subsequent inoculation of the isolation medium consisting of vitamins (excluding thiamine), inorganic salts, and trace elements, as described previously (70); hereafter, this composition will be referred to as vitamins and salts base (VSB; numbers in VSB designations indicate pH values). VSB was buffered to a pH 7.0 (VSB-7) with 10 mM HEPES for isolation attempts and amended with one or more of the following additives (prepared in distilled water unless otherwise noted): the humic acid analog anthraquinone-2,6-disulfonic acid disodium salt (AQDS; final concentration, 25 mM) as a potential electron acceptor; a mixture of N-acylhomoserine lactones prepared in ethyl-acetate acidified with 0.1% (vol/vol) acetic acid at a final concentration in the medium of 1 μM (each) N-(butyryl, heptanoyl, hexanoyl, β-ketocaproyl, octanoyl, and tetradecanoyl)-dl-homoserine lactones (Sigma-Aldrich Co., St. Louis, MO) as possible growth-promoting signals; catalase, added directly to plates at a concentration of 65 U/ml or to cooled, molten agar just prior to pouring plates at 130 U/ml to protect cells from peroxides; and a mixture of yeast extract, Bacto protease peptone 3, casamino acids, and dextrose at 0.05 g/liter each. Cultivation conditions for samples from the hindguts of soil-dwelling termites, Reticulitermes flavipes, have been described previously (70).
Plates were incubated for 20 to 30 days at 12°C or 23°C under one of the following atmospheres: CO2-enriched hypoxia ([vol/vol] 2% O2, 5% CO2, and N2 for the balance); CO2-enriched air ([vol/vol] 95% air, 5% CO2), or air. Plates were screened for the presence of acidobacteria using plate wash PCR (70). A chi-square test was used to assess the significance of treatments on the frequency with which acidobacteria colonies were detected on the plates. Analysis of variance (ANOVA) was used to assess the significance of any treatments on the total number of colonies formed.
Optimization of growth rate.
VSB was amended with a mixture of organic compounds (yeast extract, protease peptone, casamino acids, and glucose at a concentration of 0.15 g/liter each) to assess capacity for growth under a range of conditions. This complex medium (designated VSM; numbers in VSM designations indicate pH values) was used to determine the effect of pH on growth rate. Strains were grown at room temperature (23°C) between pH 2.0 and 7.5 at intervals of 0.5 pH units: morpholineethanesulfonic acid was used to buffer at pH ranges between 5.0 to 6.0, 4-morpholinepropanesulfonic acid was used at pH ranges between 6.5 to 7.5, and citric acid was used as a buffer below pH 5.0, all at a final concentration of 10 mM. Culture tubes (Bellco catalog no. 2048-18150) containing 5 to 10 ml of medium were stoppered under an air atmosphere and held on a reciprocating shaker operating at ca. 190 strokes/min to disrupt cellular flocs. Optical density at 600 nm was monitored periodically with a Thermo Spectronic model 20D+ spectrophotometer. Inocula consisted of a 1:100 dilution of cultures in mid- or late-log-phase growth (ca. 1 × 108 cells/ml) in R2B medium, which is a complex medium containing yeast extract, amino acids, peptone, carbohydrates, pyruvate, and inorganic salts (56). After the optimal pH for growth was identified, the range of growth-permissive temperatures was determined by assessing the growth rate at 4°C, 12°C, 23°C, or 37°C in VSM at a pH of 6 (VSM-6) for all strains except KBS 89, which was grown in VSM-5.
To determine if the growth rate was influenced by the elevated concentration of carbon dioxide used during isolation, strains were grown on VSM-6 in triplicate culture tubes and were incubated with either 0.05% CO2 (vol/vol) or 5% CO2 (vol/vol) with air. Sodium bicarbonate (7 mM) was used as an additional buffer under elevated levels of carbon dioxide. To control for the possible influence of an increased concentration of sodium when sodium bicarbonate was added, growth was also monitored in a medium containing 7 mM sodium chloride without a CO2-enriched atmosphere. ANOVA was used to assess if culture conditions resulted in significant differences in growth rates (SAS System, version 8e; SAS Institute Inc., Cary, NC).
Colony and cell morphology.
Bacterial colonies on plates of R2A medium (BD, Franklin Lakes, NJ) were examined under a Nikon SMZ-2T dissecting microscope at a magnification of ×10 to ×15 for size, pigmentation, form, elevation, and margin (68). Gram staining reactions were performed as described previously (65). Gross cell morphology and motility were assessed by using phase-contrast microscopy with a Zeiss Axioskop microscope (Carl Zeiss Inc., Thornwood, NY). Cell shape, length, and width were measured with the CMEIAS image analysis software (45). Transmission and scanning and electron microscopy, including whole-cell negative staining (using 2% uranyl acetate in water), were completed at the Michigan State University Center for Advanced Microscopy (http://ceo.msu.edu).
Carbon source utilization.
The ability of strains to use various organic compounds for aerobic growth was tested in replicate 25-ml Erlenmeyer flasks containing 5 ml of VSB-6 for all strains except KBS 89 (VSB-5), amended with the following carbon sources at a final concentration of 10 mM (unless indicated otherwise): d-glucose, d-fructose, d-galactose, d-mannose, d-ribose, d-xylose, l-arabinose, d-mannitol, d-sorbitol, sucrose, d-maltose, d-raffinose, d-cellobiose, methyl cellulose (0.1% [vol/vol] final concentration), carboxymethyl cellulose (0.1% [vol/vol] final concentration), sodium acetate, sodium pyruvate, sodium formate, sodium succinate, a mixture of organic acids (sodium citrate, sodium pyruvate, sodium fumarate, sodium dl-lactate, and malic acid; 0.02% each, final concentration), a mixture of purines and pyrimidines (adenine, guanine, thymine, cytosine, and uracil; 10 μg/ml each final concentration), d-glucuronate, d-galacturonate, d-gluconic acid, trimethoxybenzoate, syringate, ferulate, vanillate, sodium benzoate, resorcinol, and tannic acid (0.1% [vol/vol] final concentration). Flasks were incubated at room temperature (ca. 23°C) and shaken on an orbital shaker (ca. 190 rotations/min). A positive result was defined as visible turbidity after incubation for ca. 14 days.
Strains were also tested for their ability to grow anaerobically by fermentation of glucose, lysine, or ornithine (BBL Enterotube II, Sparks, MD) or by anaerobic respiration with nitrate (10 mM), iron citrate (10 mM), or AQDS (8 mM) as the electron acceptor with glucose (10 mM) as the sole carbon and energy source in VSB-6 for all strains except KBS 89 (VSB-5) with the addition of 7 mM bicarbonate. The headspace gas for these tests was 85% nitrogen, 10% hydrogen, and 5% carbon dioxide, except for samples with nitrate, which had a headspace of helium.
Carotenoid characterization.
Replicate 50-ml cultures of the pigmented strains in 250-ml side-arm flasks were grown in R2B medium to late log phase under elevated carbon dioxide (5% carbon dioxide) and either 20% oxygen or 2% oxygen in the dark. Cultures were normalized to the same optical density at 600 nm, and then cells were harvested by centrifugation at 5,000 × g for 25 min. The cell pellet was resuspended in a mixture of acetone:methanol (7:2, vol/vol), incubated overnight at 4°C in the dark (18), and centrifuged to remove cell debris, and the absorption spectra of the extracts were determined between 220 and 800 nm with a Perkin Elmer model Lambda 14 scanning UV-visible light spectrometer. Carotenoids were redissolved in chloroform for comparison of their spectra with previously characterized carotenoids (7).
16S rRNA gene phylogeny.
Nearly full-length sequences of the 16S rRNA gene (ca. 1,500 nucleotides) were generated. Briefly, the 16S rRNA gene was amplified using the Acidobacteria-specific forward primer ACD31F and the broadly inclusive bacterial reverse primer 1492R under the conditions described above. PCR products were cloned and prepared for sequencing as described above. Ten sequencing primers were used to obtain an average coverage of 6× for the 16S rRNA gene using the following primers: ACD31F, 1492R, 338F (5′-ACTCCTACGGGAGGCAGC-3′), 338R (5′-GCTGCCTCCCGTAGGAGT-3′), 531R, 810R (5′-GGCGTGGACTTCCAGGGTATCT-3′), 776F (5′-AGCAAACAGGATTAGATACCCTGG-3′), 1087F (5′-GGTTAAGTCCCGCAACGA-3′), modified-M13F, and the modified-M13R. Each primer was used in duplicate sequencing reactions. Sequences were assembled using DNA Star LaserGene software (Madison, WI), aligned using ARB software (47), and compared to acidobacterial 16S rRNA gene sequences downloaded from the GenBank database. The phylogeny algorithms in ARB were used for the generation of the phylogenetic trees; PAUP*, version 4.0b10, was used for bootstrapping analysis (72).
Characterization of genomic DNA.
The mole percent G+C content of genomic DNA from strains KBS 63, KBS 89, TAA 43, and TAA 166 was determined as described previously (51). Briefly, genomic DNA was extracted using a QIAGEN Genomic DNA Extraction Kit (QIAGEN, Valencia, CA), and approximately 5 μg of DNA was digested with P1 nuclease and alkaline phosphatase. The nucleosides were separated and quantified using a Shimadzu high-pressure liquid chromatograph fitted with a UV detector and a VP series Alltima C18 column (250 by 4.6 mm; particle size, 5 μm) (Alltech Associates, Inc., Deerfield, IL). Genomic DNA purified from A. capsulatum (ATCC 51196) was used as a positive control.
The number of 16S rRNA-encoding genes was determined by nonradioactive Southern hybridization after restriction endonuclease digestion of genomic DNA, as described previously (35, 36) (http://rrndb.cme.msu.edu/rrndb/servlet/controller), using a digoxigenin-labeled, Acidobacteria-specific 16S rRNA gene probe targeting regions between residues 31 and 531 (Escherichia coli numbering). Genomic DNA purified from A. capsulatum was used as a positive control.
Other physiological tests.
Catalase and oxidase tests were performed using standard methods (65, 68). E. coli strain REL 607, a derivative of E. coli B/r (42), was used as a positive control for the catalase test and as a negative control for the oxidase test. Pseudomonas aeruginosa ATCC 10145 was used as a positive control for the oxidase test. Fatty acid profiles were generated and analyzed by Microbial ID (Newark, DE; http://www.microbialid.com) after all strains, including A. capsulatum, were grown on glucose-yeast extract medium adjusted to within their optimal pH range.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the Acidobacteria strains described in this study have been deposited in the GenBank database under accession numbers AY587227 through AY587230 and DQ660892 through DQ660895.
RESULTS AND DISCUSSION
Occurrence and isolation of acidobacteria.
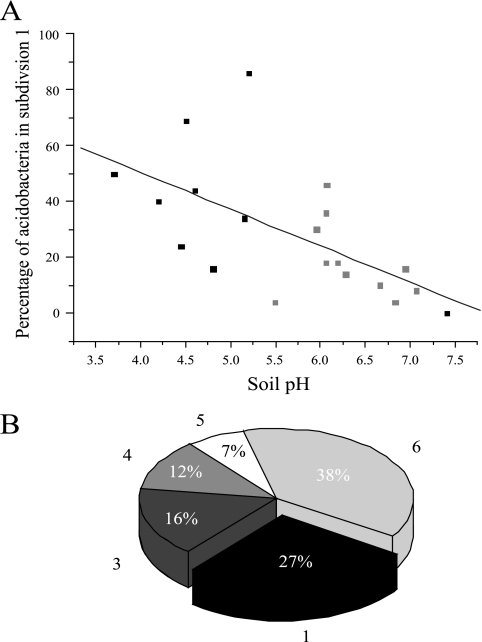
Acidobacteria are abundant members of the microbial community in soils at the KBS LTER; their rRNAs account for ca. 1 to 6% of the total bacterial rRNA (8). The specific phylogenetic affiliations of acidobacteria in these soils were determined from samples collected during August 2005 from different management treatments and soil depths at the KBS LTER. Partial sequences (ca. 500 nucleotides) of 50 cloned 16S rRNA genes from each depth and treatment (total number of clones, 550) revealed the presence of acidobacteria from subdivisions 1, 3, 4, 5, and 6. The relative abundance of clones of each subdivision varied with depth and treatment, suggesting that physical and chemical properties of the soil influence the distribution of the various subdivisions. In particular, the percentage of acidobacteria in subdivision 1 correlated with soil pH, generally increasing in mildly acidic soils (Fig. 1A, gray boxes). To determine if the relationship between soil pH and the relative abundance of subdivision 1 acidobacteria was significant for other terrestrial environments as well, previously published sequences from a variety of soil environments (15, 23, 38, 44, 50, 53, 54, 59, 69) were analyzed. The combined data sets reveal a significant correlation (P < 0.004) (Fig. 1A) between pH and the percentage of acidobacteria that cluster within subdivision 1. This result is consistent with a previously determined relationship revealing that as soil pH decreased, there was an increased abundance of subdivision 1 acidobacteria in relation to the total bacterial community (60).
FIG. 1.
Composition of acidobacterial communities in soil based on 16S rRNA gene surveys. (A) Percent abundance of Acidobacteria subdivision 1 out of the total acidobacterial community at various soil pH values. Data from the KBS LTER are depicted as gray boxes, whereas data from published soil surveys are depicted as black boxes. If a range in pH was given in these studies, the average of that range was used for this analysis. The equation of the line is y = −13.1x + 103 with an R2 value of 0.38 and a P value of <0.004. (B) Average percent abundance of Acidobacteria subdivisions 1, 3, 4, 5, and 6 (subdivisions are indicated next to each pie wedge) from replicate plots of the never-tilled, successional community with herbaceous plants at the KBS LTER, depth of 0 to 7 cm.
The strains characterized in this study were isolated from never-tilled soils (T8) with an average pH of 6.1 and a high percentage of subdivision 1 acidobacteria (27%) (Fig. 1B). Media and incubation conditions were designed to mimic the microenvironments thought to be experienced by microbes in these soils. In particular, relatively low concentrations of organic carbon in conjunction with extended incubation periods were used to accommodate oligotrophic, slow-growing acidobacteria (35). The concentrations of oxygen and carbon dioxide during incubation were also manipulated to better simulate conditions in soil aggregates. Although upland soils like those at the KBS LTER are generally well aerated, they include pockets of anoxia within soil aggregates and also include transition zones between oxic and anoxic conditions (27, 66). Moreover, metabolism of heterotrophic microbes in soil leads to increased concentrations of carbon dioxide (20), which has the potential to influence the pH of microenvironments. Accordingly, hypoxic and CO2-enriched incubation atmospheres were included, as was the incorporation of catalase in the medium to minimize the damaging effects of hydrogen peroxide produced during aerobic respiration.
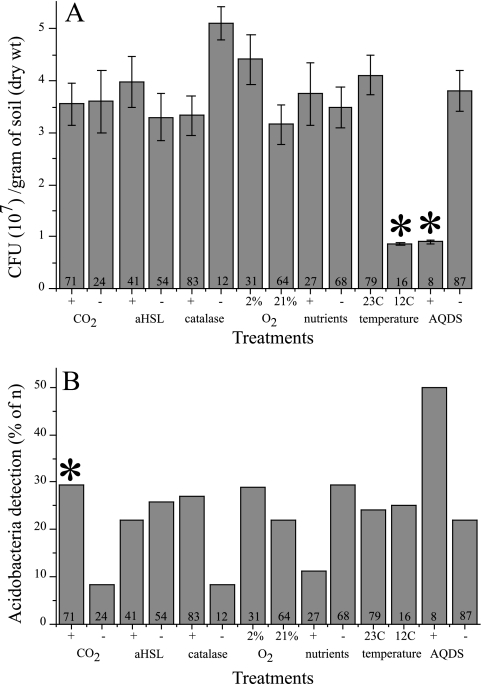
With the exception of decreased recoveries at lower temperature and in the presence of AQDS, the recovery of soil microorganisms under the various conditions tested was between 3.5 × 107 and 5 × 107 CFU/gram of soil (dry weight), which was 2.5 to 6% of the direct cell count (Fig. 2A). Previous experiments with three different types of solidifying agents (Bacto Agar, washed Bacto agar in VSB-7, and agarose in distilled water) indicated a decrease in total CFU recovery when samples were incubated under oxic conditions as the purity of the solidifying agent increased: agar, ca. 5.2 × 107 CFU/gram of soil (dry weight); washed agar, ca. 4.4 × 107 CFU/gram of soil (dry weight); and agarose, ca. 0.3 × 107 CFU/gram of soil (dry weight). Agar was chosen as a solidifying agent to increase recovery of soil microorganisms, presumably increasing our chances of isolating members of the phylum Acidobacteria. Across treatments used for isolation, acidobacteria were detected more frequently under atmosphere(s) containing elevated levels of carbon dioxide (0.05< P < 0.10) (Fig. 2B). This was apparently due to a mild acidification of the medium under 5% carbon dioxide (plates incubated under 5% CO2 had a pH of ca. 6.5), which favored growth of these acidobacteria (see below).
FIG. 2.
Summary of cultivation experiments for total recovery of colonies per gram of soil (dry weight) (A) and acidobacteria detection (B). Panel A presents the average CFU recovery from soil ± the standard error; the number of times a particular treatment was employed (n) is listed at the base of each bar. Panel B shows the percentage of treatments (where n is the number of treatments) that yielded a positive test for acidobacteria using plate wash PCR from the plates of panel A. The asterisks indicate treatments that yielded a significant difference (panel A, ANOVA α = 0.05; panel B, chi-square goodness of fit, α = 0.10).
Although overall recoveries of bacteria here were similar to results from other studies using low-nutrient concentrations and extended incubation times (12, 14), the specific conditions used in this study resulted in the isolation of several novel, pink-pigmented strains of acidobacteria (strains KBS 62, KBS 63, KBS 68, and KBS 112). All were isolated from plates that contained a mixture of organic carbon substrates and that were incubated for 20 to 30 days under CO2-enriched air.
Previous isolation efforts with homogenized hindguts of the soil-dwelling termites R. flavipes, under similar cultivation conditions, resulted in the isolation of three acidobacteria strains (strains TAA 43, TAA 48, and TAA 166), which appeared to be allocthonous contaminants from nearby soil (74a), and strain KBS 89 from soils at the KBS LTER. These strains are also characterized further in the current study.
Colonial and cell morphology and pigmentation.
Colonies of all acidobacteria were small, approximately 1 mm in diameter (after a 7-day incubation, except for KBS 89 which took about 14 days), and had a circular form with a convex elevation and entire margin when grown on VSM-6, VSM-5 (KBS 89), or R2A medium. With the exception of KBS 89, all isolates took approximately 3 to 5 days before there were visible, smooth, butyrous colonies, while KBS 89 took approximately 14 to 16 days to form visible colonies. All strains achieved maximal growth rates in complex medium, such as R2B, at room temperature (ca. 23°C) with a doubling time between 10 and 15 h.
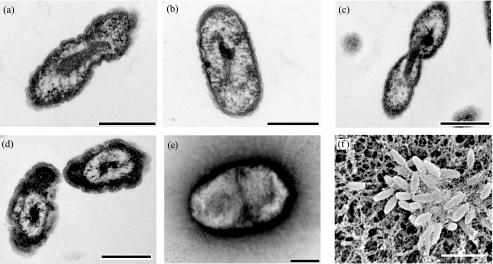
Cells of all strains were short, plump, gram-negative, nonmotile rods measuring 1 μm by 0.6 μm when grown on R2A medium (Fig. 3). Cells possessed slightly convoluted outer membranes when viewed by transmission electron microscopy, as is typical of a gram-negative-type cell wall. All strains produced an extracellular matrix of as yet unknown chemical composition but which apparently caused cells to stick together tightly (Fig. 3f) in colonies and form visible clumps in liquid culture, qualitatively to the same extent under microoxic and oxic conditions. The formation of such extracellular material in soils may serve as a form of protection from predation or as a web to trap water or nutrients (57) and may be involved in the formation of soil aggregates.
FIG. 3.
Electron micrographs of thin sections of strains TAA 43 (a), TAA 48 (b), TAA 166 (c), and KBS 63 (d) grown in R2A medium. Also shown are a negatively stained cell of strain KBS 63 (e) and a scanning electron micrograph of strain KBS 89 (f). Bars, 500 nm (a to e) and 2 μm (f).
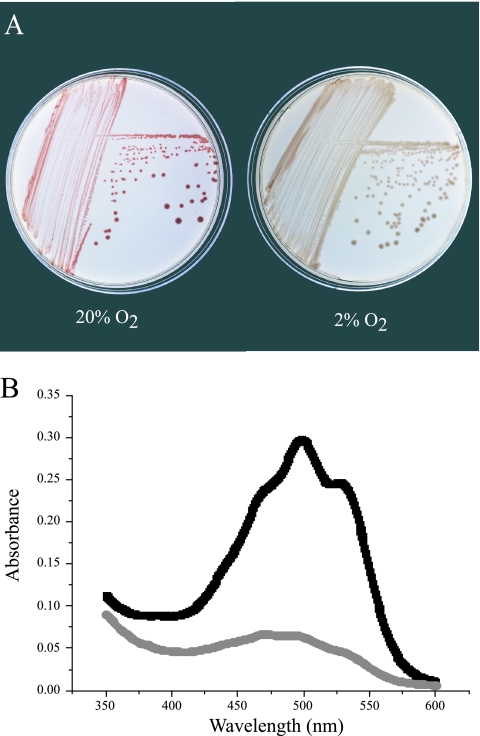
Four strains (KBS 62, 63, 68, and 112) were pink, owing to the presence of a pigment(s), whose solvent extractability and absorption spectrum were typical of carotenoids. The pigmentation was more pronounced in cells grown under 20% O2 than under 2% O2 (Fig. 4). The ratio of the absorbance maxima of the two dominant peaks in acetone:methanol (A498 and A528) were nearly identical, 1.2 ± 0.0 and 1.3 ± 0.1, respectively, suggesting that the same carotenoid(s) was produced under each oxygen regime. The visible absorption spectrum of strain KBS 63 pigment(s) in chloroform had maxima at 473, 505, and 539 nm, which are most similar to those of spirilloxanthin (475, 505, and 543 nm) (7).
FIG. 4.
(A) Strain KBS 63 grown on R2A medium under 20% and 2% oxygen (vol/vol) mixed with nitrogen and 5% CO2. (B) Absorption spectra of strain KBS 63 cells extracted in acetone:methanol (7:2) after growth in 20% (black line) and 2% (gray line) oxygen on R2B medium.
Carotenoids are naturally occurring accessory pigments that typically absorb visible light maximally between 450 and 550 nm (48) and are known to play roles in light-harvesting for photosynthesis and protection from photooxidative damage (6), including the quenching of singlet oxygen (11). Carotenoids are found not only in phototrophic bacteria but also in numerous, phylogenetically diverse genera of heterotrophic soil bacteria, including Streptomyces, Myxococcus, Flavobacterium, and Erwinia (2). Carotenoid synthesis has long been known to be regulated by two key environmental factors, oxygen and light (2). While light had no measurable influence on the carotenoid(s) content of the Terriglobus strains, the concentration of carotenoid(s) in T. roseus increased in response to oxygen. We suggest that increased production of carotenoid(s) under 20% oxygen compared to 2% oxygen concentration represents a response to oxidative stress. The potential antioxidant property of carotenoids is well known: β-carotene is effective in quenching singlet oxygen (11), and the addition of carotenoid genes to E. coli inhibits peroxidation (1). Carotenoids can influence the growth rate and survival of heterotrophic bacteria (52), presumably through the inactivation of reactive oxygen species generated during respiration. Differential expression of carotenoids might also be advantageous in the soil environment, where oxygen gradients change in response to soil moisture and depth. Higher concentrations of oxygen occur in the upper layers of soil that are also subject to more intense solar radiation, where carotenoids would provide protection against the damaging effects of UV radiation.
The frequency of acidobacteria detection was notably higher in the presence of catalase (Fig. 2B), suggesting that acidobacteria are sensitive to damage by reactive oxygen species. All strains, except for KBS 89, tested positive for the presence of catalase (Table 1), and some strains produced a carotenoid(s) which was preferentially expressed under higher oxygen concentrations (Fig. 4). It would be beneficial for a soil microorganism to have mechanisms to deal with the damaging effects of reactive oxygen species, since there are oxygen gradients in the three-dimensional soil structure that change with soil moisture and in response to the metabolism of nearby microbes.
TABLE 1.
Phenotypic characteristics of Terriglobus and Acidobacteriaceae strains and A. capsulatum
| Characteristic |
Terriglobus strain
|
Acidobacteriaceae strain
|
A. capsulatuma | ||
|---|---|---|---|---|---|
| KBS 63b | TAA 43c | KBS 89 | TAA 166 | ||
| Origin | Soil | Termite hindgut | Soil | Termite hindgut | Acidic mine drainage |
| Cell shaped | Rod | Rod | Rod | Rod | Rod |
| Length (μm) | 1.1 ± 0.2 | 1.2 ± 0.2 | ca. 1 | 1.1 ± 0.3 | 1.1-2.3 |
| Width (μm) | 0.6 ± 0.1 | 0.6 ± 0.1 | ca. 0.5 | 0.6 ± 0.0 | 0.3-0.8 |
| Pigment | Pink | White | White | White | Orange |
| rrs copy no. | 2 | 1 | 1 | 1 | 1 |
| G+C content (mol %)e | 59.8 ± 0.5 | 58.1 ± 0.04 | 59.7 ± 1.8 | 54.7 ± 0.7 | 60.8 |
| Catalase | + | + | − | + | + |
| Oxidase | − | − | − | − | − |
| Motility | − | − | − | − | + |
| pH range (optimum) | 5-7 (6) | 5-6.5 (5) | NDf | 5-7 (6) | 3-6 |
| Growth at 4°C | − | − | − | − | NDf |
| Growth at 12°C | + | + | + | + | ND |
| Growth at 23°C | + | + | + | + | ND |
| Growth at 37°C | − | − | − | − | + |
| Differences in carbon utilization | |||||
| d-Ribose | − | − | + | − | ND |
| d-Sorbitol | − | − | + | − | ND |
| Purine and pyrimidine mixg | − | + | − | − | ND |
Characteristics are as described from a previous study (33). The G+C content determined in the present study is 62.7 ± 0.1.
Data are identical for strains KBS 62, KBS 63, KBS 68, and KBS 112.
Data are identical for strains TAA 43 and TAA 48.
Cell length and width were determined from a culture in mid-log phase on R2B medium.
The mole percent G+C content was determined only for strains KBS 63, TAA 43, KBS 89, and TAA 166.
ND, not determined.
The mix of adenine, guanine, thymine, cytosine, and uracil had a final concentration of 10 μg/ml each.
Physiological properties of strains.
Physiological properties of all strains are listed in Table 1. The growth rates of strains grown in VSM-5 (KBS 89) or VSM-6 (all others) were highest (μ ≈ 0.15) at room temperature under air. Growth was observed over a pH range of 4.5 to 7.0, with optima between pH 5 and 6. Mildly acidic pH optima are not surprising, given that soils are slightly acidic due to the presence of humic substances, which make up 70 to 80% of the soil organic matter (24). Sait et al. recently suggested that pH influences the isolation and distribution of subdivision 1 acidobacteria in soil, with subdivision 1 acidobacteria forming colonies more frequently in medium at pH 5.5 than at pH 7.0 after a 4-month incubation (60). Furthermore, the results of our molecular-based survey reveals that subdivision 1 acidobacteria are found in a higher abundance in mildly acidic to acidic soil environments (Fig. 1A). Taken together, these results suggest that members of subdivision 1 of the Acidobacteria have a preference for mildly acidic pH conditions, which is consistent with their ubiquitous distribution in soil.
We examined the effect of CO2 on the growth of isolated strains to explore the greater frequency at which acidobacteria formed colonies on plates incubated under 5% CO2 (Fig. 2B). There was no significant difference in growth rate of strains KBS 63, TAA 43, and TAA 166 incubated under an atmosphere of air (ca. 0.05% CO2, vol/vol) or 5% CO2 (balance, air) (P value of 0.83, 0.98, and 1.0, respectively). Therefore, the increased frequency of appearance of acidobacteria on plates incubated under 5% CO2 appears to be the result of mild acidification of the isolation medium, as originally suggested (70). The degree of acidification that can occur on plates of medium without added bicarbonate and incubated under elevated levels of carbon dioxide (pH ca. 6.5) is consistent with the optimal pH of the strains (Table 1).
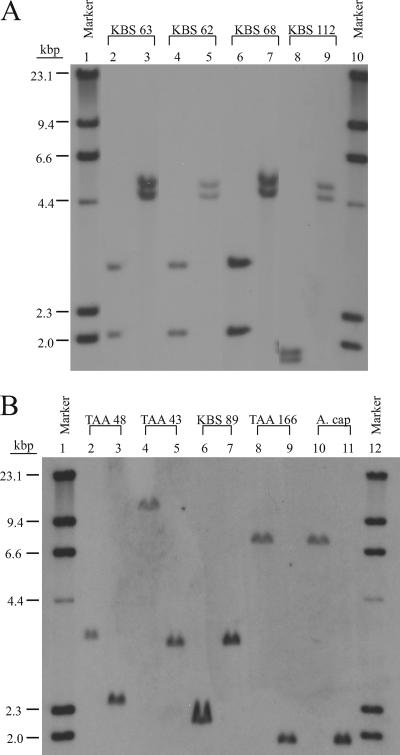
The relatively slow growth rate of the Acidobacteria strains is consistent with the presence of one (strains TAA 43, 48, 166, and KBS 89) or two (strains KBS 62, 63, 68, and 112) genes coding for the 16S rRNA (Fig. 5). Previous studies have indicated that the number of rRNA operons correlates with the rate at which bacteria respond to resource availability and with how efficiently those resources are utilized (35, 71).
FIG. 5.
Southern hybridization of digested genomic DNA from strains KBS 63, KBS 62, KBS 68, KBS 112, TAA 48, TAA 43, KBS 89, and TAA 166 and A. capsulatum A. cap). In panel A, isolates were cut with EcoRV (lanes 2, 4, and 6), SmaI (lane 3, 5, 7, and 9), and RsaI (lane 8). In panel B, isolates were cut with SmaI (lanes 2, 4, 6, 8, and 10) and ApaI (lanes 3, 5, 7, 9, and 11). The lambda DNA HindIII marker (lanes 1 and 10 in panel A and lanes 1 and 12 in panel B) provided size estimates.
The metabolism and growth of bacteria in soil environments are frequently limited by carbon availability but are subject to periodic fluctuations in resources resulting from events including rains or spring snow melt (74). Populations of Acidobacteria are more abundant in soil environments where resource availability is low (19); these bacteria can be described as oligotrophic bacteria because of this distribution in soil, their relatively slow growth rate, and the possession of either one or two copies of the 16S rRNA-encoding gene (a genomic marker of oligotrophic bacteria) (35). Although the acidobacteria characterized in this study do not fit the definition of an obligate oligotroph, because they are capable of growth on rich medium, they do fit into the broader notion of oligotrophs as organisms adapted to low-nutrient environments (37).
All strains were able to grow in a 2% oxygen atmosphere as well as atmospheric concentrations of oxygen, with or without elevated levels of carbon dioxide, but they could not grow anaerobically with nitrate, iron [Fe(III)], or AQDS as an external electron acceptor, nor could they grow by fermentation of glucose, lysine, or ornithine (Table 1). Growth under low levels of oxygen would be advantageous in soil, since oxygen is depleted in the soil environment, particularly in soil aggregates, as soil moisture increases (5, 55, 74).
There were nominal differences in profiles of carbon utilization: all strains were able to oxidize d-glucose, d-fructose, d-galactose, d-mannose, d-xylose, d-arabinose, sucrose, d-maltose, d-raffinose, d-cellobiose, sodium succinate, d-glucuronate, and d-gluconic acid; however, they were not able to oxidize d-mannitol, methyl cellulose, carboxymethyl cellulose, sodium acetate, sodium pyruvate, sodium formate, maleic acid, the organic acid mix, d-galacturonate, monomeric constituents of lignins, or humic acids. Only two differences in the carbon utilization pattern were noticed; strain KBS 89 was able to oxidize ribose and sorbitol while strains TAA 43 and TAA 48 were the only strains capable of oxidizing the mixture of purines and pyrimidines (Table 1).
The whole-cell fatty acid compositions of the newly isolated strains and A. capsulatum are listed in Table 2. The dominant fatty acids for the new strains are 15:0 iso (36.33 to 47.09%) and 16:1 ω7c/15 iso 2-OH (summed feature 3) (26.18 to 30.24%). These results were compared to the Microbial ID database but did not correspond to any known species. A. capsulatum was submitted as a source of comparison, and its dominant fatty acids were 15:0 iso (40.28%) and 18:1 ω9c (21.21%).
TABLE 2.
Whole-cell fatty acid composition of Terriglobus and Acidobacteriaceae strains and A. capsulatuma
| Fatty acid type | Whole-cell fatty acid composition (%)
|
||||
|---|---|---|---|---|---|
|
Terriglobus strain
|
Acidobacteriaceae strain
|
A. capsulatum | |||
| KBS 63 | TAA 43 | KBS 89 | TAA 166 | ||
| Saturated | |||||
| 14:0 | 3.42 | 4.25 | 1.16 | 3.68 | 1.58 |
| 15:0 | 0.64 | 0.74 | 0.00 | 0.60 | 0.57 |
| 16:0 | 7.77 | 9.14 | 7.33 | 8.71 | 4.29 |
| 16:0 N alcohol | 0.52 | 3.54 | 0.50 | 5.01 | 4.61 |
| 17:0 | 0.70 | 0.00 | 0.93 | 0.00 | 1.85 |
| 18:0 | 0.38 | 0.68 | 0.75 | 0.59 | 2.34 |
| 20:0 | 0.76 | 0.00 | 4.07 | 0.00 | 0.00 |
| Unsaturated | |||||
| 14:1 ω5c | 1.07 | 0.67 | 1.00 | 0.57 | 0.00 |
| 15:1 ω6c | 0.83 | 0.92 | 0.43 | 0.71 | 0.00 |
| 16:1 ω5c | 0.43 | 0.00 | 0.00 | 0.56 | 0.00 |
| 17:1 ω8c | 0.00 | 0.00 | 0.00 | 0.00 | 3.97 |
| 18:1 ω9c | 0.00 | 0.00 | 0.00 | 0.00 | 21.21 |
| 18:1 ω7c | 0.00 | 0.84 | 0.00 | 1.27 | 0.45 |
| 18:1 ω5c | 1.05 | 0.00 | 0.00 | 0.00 | 2.24 |
| 19:0 cyclo ω8c | 0.00 | 0.00 | 0.00 | 1.10 | 0.00 |
| 20:2 ω6,9c | 0.67 | 0.00 | 0.00 | 0.00 | 0.00 |
| Methyl-branched | |||||
| 13:0 iso | 4.02 | 0.00 | 4.12 | 0.00 | 0.00 |
| 15:0 iso | 43.88 | 36.33 | 47.09 | 37.06 | 40.28 |
| 15:0 anteiso | 0.48 | 0.70 | 0.00 | 1.03 | 0.00 |
| 16:0 iso | 0.00 | 0.00 | 0.00 | 0.58 | 0.00 |
| iso 17:1 ω9c | 0.00 | 0.00 | 1.25 | 0.00 | 0.90 |
| iso 17:1 ω5c | 4.24 | 7.27 | 2.33 | 0.00 | 7.40 |
| 17:0 iso | 0.53 | 0.79 | 1.60 | 0.78 | 2.15 |
| 17:0 anteiso | 0.27 | 0.60 | 0.00 | 0.74 | 0.49 |
| 19:0 anteiso | 0.00 | 1.82 | 0.00 | 0.00 | 0.00 |
| Hydroxy | |||||
| 15:0 iso 3-OH | 0.00 | 0.00 | 0.58 | 0.00 | 0.00 |
| 17:0 iso 3-OH | 0.00 | 0.00 | 0.00 | 0.00 | 0.77 |
| Summed feature | |||||
| 15:1 iso H/13:0 3-OH | 0.59 | 0.00 | 0.36 | 0.00 | 0.32 |
| 16:1 ω7c/15 iso 2-OH | 27.08 | 30.24 | 26.18 | 26.99 | 4.30 |
| 17:1 iso i/anteiso-b | 0.00 | 0.00 | 0.00 | 8.07 | 0.00 |
| 18:2 ω6,9c/18:0 anteiso | 0.64 | 1.47 | 0.31 | 1.96 | 0.00 |
| Unknown 11.799 | 0.00 | 0.00 | 0.00 | 0.00 | 0.28 |
Strains were grown in glucose-yeast extract medium. The most abundant fatty acids that distinguish the Terriglobus strains from A. capsulatum are shown in boldface.
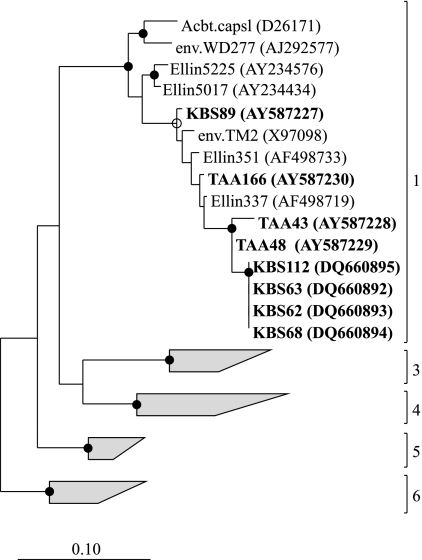
Six of the strains (KBS 62, KBS 63, KBS 68, KBS 112, TAA 43, and TAA 48) are sufficiently similar to one another but distinct from previously isolated acidobacteria (Fig. 6) to warrant creation of a new genus. The similarity of the 16S rRNA gene sequences within these six strains is 96% and, on average, only 92% similar to A. capsulatum. Moreover, unlike A. capsulatum, these new strains do not require extremely low pH conditions (pH 3.0 to 6.0), they cannot grow at 37°C, and they contain 16:1 ω7c/15:0 iso 2-OH as a dominant fatty acid and not 18:1 ω9c, which is dominant in A capsulatum (Table 2). Hence, we propose that these six strains be regarded as members of a new genus, Terriglobus, with KBS 63 as the type strain of the new species, T. roseus, which also includes the other pink-pigmented strains (KBS 62, KBS 68, and KBS 112). The nomenclature and description of the genus and species are given below. We are postponing application of specific epithets to strains TAA 43 and TAA 48 until further characterization is done for species identification. Although the characteristics of strains KBS 89 and TAA 166 are consistent with the description of Terriglobus, there is insufficient metabolic information currently available about the previously isolated and closely related strains Ellin 351 and Ellin 337 (61) to include all four strains in the genus Terriglobus at this time.
FIG. 6.
Maximum-likelihood tree of the Acidobacteria subdivisions 1, 3, 4, 5, and 6 (indicated to the right of the group) based on the 16S rRNA gene using sequences obtained from cultivated representatives and environmental clones. G. fermentans and H. foetida of subdivision 8 were used as outgroups (not shown). Strains from this study are in bold font. Each gray trapezoid represents a different subdivision with approximately eight representative sequences. Internal nodes supported by a bootstrap value of >95% are indicated with a filled circle, and those supported by a bootstrap value of >75% are indicated with an open circle. The scale bar indicates 0.10 changes per nucleotide.
Description of Terriglobus gen. nov.
Terriglobus gen. nov. (Ter·ri·glo′ bus. L. fem n. terra, earth; L. masc. n. globus ball, clump; N.L. masc. n. Terriglobus clump of earth) literally translated as “clump of earth” in the phylum Acidobacteria. Gram-negative, short, plump, rods (ca. 1 μm × 0.6 μm) when grown in R2B medium. Moderate acidophiles (pH range for growth is 5.0 to 7.0, optimal for KBS 63 is 6.0 and for TAA 43 is 5.0). Produce an extracellular matrix which apparently facilitates cell floculation in liquid medium incubated under an atmosphere containing either 2% or 20% molecular oxygen. G+C content ca. 59 mol% (Table 1); 15:0 iso and 16:1 ω7c/15:0 iso 2-OH as dominant fatty acids (Table 2). Members of the phylum Acidobacteria, subdivision 1. The type species is Terriglobus roseus.
Description of Terriglobus roseus sp. nov.
Terriglobus roseus sp. nov. (ro′ se·us. L. masc. adj. roseus rose-colored, pink), “a pink Terriglobus” (type strain KBS 63). Pink-pigmented colonies due to the presence of carotenoids; catalase positive; oxidase negative; and two copies of the rrs. Isolated from soils in a never-tilled, successional plant community at the KBS LTER. The type strain KBS 63 was deposited into the USDA Agricultural Research Service Culture Collection (NRRL B-41598T) and Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSM 18391).
Acknowledgments
This work was funded by the USDA (2001-35107-09939), the NSF (MCB-0135880), and the NSF Long-Term Ecological Research Program at the Kellogg Biological Station and the Michigan Agricultural Experiment Station.
We thank Kwi S. Kim for her assistance with determining the rrs copy number, Les Dethlefsen for his help with the statistical analysis, Frank B. Dazzo for his assistance with the CMEIAS software, Kristin M. Huizinga and Jorge L. M. Rodrigues for their help with soil collection, Joseph R. Graber with his help with determination of G+C mole percent, Zachary Blount for his helpful suggestions on the manuscript, and John P. Eichorst and Hans G. Trüper for their help with the nomenclature and Latinized specific epithets.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Albrecht, M., S. Takaichi, S. Steiger, Z.-Y. Wang, and G. Sandmann. 2000. Novel hydroxycarotenoids with improved antioxidative properties produced by gene combination in Escherichia coli. Nat. Biotechnol. 18:843-846. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, G. A. 1997. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu. Rev. Microbiol. 51:629-659. [DOI] [PubMed] [Google Scholar]
- 3.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloem, J. 1995. Fluorescent staining of microbes for total direct counts, section 4.1.8, p. 1-12. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Brady, N. C., and R. R. Weil. 1999. Elements of the nature and property of soils, 1st ed. Prentice Hall, Upper Saddle River, NJ.
- 6.Britton, G. 1983. The biochemistry of natural pigments. Cambridge University Press, Cambridge, United Kingdom.
- 7.Britton, G., S. Liaaen-Jensen, and H. Pfander. 1995. Carotenoids, vol. 1B: spectroscopy. Birkhauser Verlag, Boston, MA.
- 8.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 9.Coates, J., D. J. Ellis, C. V. Gaw, and D. R. Lovley. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Evol. Microbiol. 49:1615-1622. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conn, P. F., C. Lambert, E. J. Land, W. Schalch, and T. G. Truscott. 1992. Carotene-oxygen radical interactions. Free Radic. Res. Commun. 16:401-408. [DOI] [PubMed] [Google Scholar]
- 12.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crocetti, G. R., J. F. Banfield, J. Keller, P. L. Bond, and L. L. Blackall. 2002. Glycogen-accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology 148:3353-3364. [DOI] [PubMed] [Google Scholar]
- 14.Davis, K. E. R., S. J. Joseph, and P. H. Janssen. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dedysh, S. N., T. A. Pankratov, S. E. Belova, I. S. Kulichevskaya, and W. Liesack. 2006. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic sphagnum peat bog. Appl. Environ. Microbiol. 72:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans, W. R., D. E. Fleischman, H. E. Calvert, P. V. Pyati, G. M. Alter, and N. S. Subba Rao. 1990. Bacteriochlorophyll and photosynthetic reaction centers in Rhizobium strain BTAi 1. Appl. Environ. Microbiol. 56:3445-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fierer, N., M. A. Bradford, and R. B. Jackson. Towards an ecological classification of soil bacteria. Ecology, in press. [DOI] [PubMed]
- 20.Franzluebbers, A. J., R. L. Haney, C. W. Honeycutt, H. H. Schomberg, and F. M. Hons. 2000. Flush of carbon dioxide following rewetting of dried soil relates to active organic pools. Soil Sci. Soc. Am. J. 64:613-623. [Google Scholar]
- 21.Gans, J., M. Wolinsky, and J. Dunbar. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387-1390. [DOI] [PubMed] [Google Scholar]
- 22.Garrity, G. M., J. A. Bell, and T. G. Lilburn. 2004. Taxonomic outline of the prokaryotes. Bergey's manual of systematic bacteriology, 2nd ed., release 5.0. Springer-Verlag, NY. doi: 10.007/bergeysoutline200405. [DOI]
- 23.Gremion, F., A. Chatzinotas, and H. Harms. 2003. Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ. Microbiol. 5:896-907. [DOI] [PubMed] [Google Scholar]
- 24.Haynes, R. J., and M. H. Beare. 1996. Aggregation and organic matter storage in meso-thermal, humid soil, p. 213-262. In M. R. Carter, B. A. Stewart (ed.), Structure and organic matter storage in agricultural soils. CRC Press, Boca Raton, FL.
- 25.Hiraishi, A., N. Kishimoto, Y. Kosako, N. Wakao, and T. Tano. 1995. Phylogenetic position of the menaquinone-containing acidophilic chemo-organotroph Acidobacterium capsulatum. FEMS Microbiol. Lett. 132:91-94. [DOI] [PubMed] [Google Scholar]
- 26.Hobel, C. F. V., V. T. Marteinsson, G. O. Hreggvidsson, and J. K. Kristjansson. 2005. Investigation of the microbial ecology of intertidal hot springs by using diversity analysis of 16S rRNA and chitinase genes. Appl. Environ. Microbiol. 71:2771-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoejberg, O., N. P. Revsbach, and J. M. Tiedje. 1994. Denitrificatioin in soil aggregates analyzed with microsensors for nitrous oxide and oxygen. Soil Sci. Soc. Am. J. 58:1691-1698. [Google Scholar]
- 28.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, K. S., T. G. Lilburn, M. J. Renner, and J. A. Breznak. 1998. arfI and arfII, two genes encoding alpha-l-arabinofuranosidases in Cytophaga xylanolytica. Appl. Environ. Microbiol. 64:1919-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishimoto, N., Y. Kosako, and T. Tano. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 34.Kishimoto, N., and T. Tano. 1987. Acidophilic heterotrophic bacteria isolated from acidic mine drainage, sewage, and soils. J. Gen. Appl. Microbiol. 33:11-25. [Google Scholar]
- 35.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch, A. L. 2001. Oligotrophs versus copiotrophs. Bioessays 23:657-661. [DOI] [PubMed] [Google Scholar]
- 38.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 119-175. In E. S. M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., New York, NY.
- 40.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaPara, T. M., C. H. Nakatsu, L. Pantea, and J. E. Alleman. 2000. Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl. Environ. Microbiol. 66:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenski, R. E., M. R. Rose, S. C. Simpson, and S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Natural. 138:1315-1341. [Google Scholar]
- 43.Liesack, W., F. Bak, J.-U. Kreft, and E. Stackebrandt. 1994. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85-90. [DOI] [PubMed] [Google Scholar]
- 44.Lipson, D. A., and S. K. Schmidt. 2004. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl. Environ. Microbiol. 70:2867-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, J., F. B. Dazzo, O. Glagoleva, B. Yu, and A. K. Jain. 2001. CMEIAS: a computer-aided system for the image analysis of bacterial morphotypes in microbial communities. Microb. Ecol. 41:173-194. [DOI] [PubMed] [Google Scholar]
- 46.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K.-H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 47.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madigan, M. T., J. M. Martinko, and J. Parker. 2006. Brock biology of microorganisms, 11th ed. Prentice Hall, Upper Saddle River, NJ.
- 49.Martiny, A. C., H.-J. Albrechtsen, E. Arvin, and S. Molin. 2005. Identification of bacteria in biofilm and bulk water samples from a nonchlorinated model drinking water distribution system: detection of a large nitrite-oxidizing population associated with Nitrospira spp. Appl. Environ. Microbiol. 71:8611-8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McVeigh, H. P., J. Munro, and T. M. Embley. 1996. Molecular evidence for the presence of novel actinomycete lineages in a temperate forest soil. J. Ind. Microbiol. 17:197-204. [Google Scholar]
- 51.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid of high-performance liquid chromatography. Int. J. Syst. Evol. Microbiol. 39:159-167. [Google Scholar]
- 52.Mikell, A. T., B. C. Parker, and E. M. Gregory. 1986. Factors affecting high-oxygen survival in heterotrophic microorganisms from an antarctic lake. Appl. Environ. Microbiol. 52:1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norris, T. B., J. M. Wraith, R. W. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paul, E. A., and F. E. Clark. 1996. Soil microbiology and biochemistry. Academic Press, Inc., New York, NY.
- 56.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 58.Robertson, G. P., D. C. Coleman, C. S. Bledsoe, and P. Sollins. 1999. Standard soil methods for long-term ecological research. Oxford University Press, New York, NY.
- 59.Rosch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sait, M., K. E. R. Davis, and P. H. Janssen. 2006. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 72:1852-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 62.Schabereiter-Gurtner, C., C. Saiz-Jimenez, G. Pinar, W. Lubitz, and S. Rolleke. 2002. Altamira cave Paleolithic paintings harbor partly unknown bacterial communities. FEMS Microbiol. Lett. 211:7-11. [DOI] [PubMed] [Google Scholar]
- 63.Schabereiter-Gurtner, C., C. Saiz-Jimenez, G. Pinar, W. Lubitz, and S. Rolleke. 2002. Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its Palaeolithic paintings. Environ. Microbiol. 4:392-400. [DOI] [PubMed] [Google Scholar]
- 64.Schabereiter-Gurtner, C., C. Saiz-Jimenez, G. Pinar, W. Lubitz, and S. Rolleke. 2004. Phylogenetic diversity of bacteria associated with Paleolithic paintings and surrounding rock walls in two Spanish caves (Llonin and La Garma). FEMS Micrbiol. Ecol. 47:235-247. [DOI] [PubMed] [Google Scholar]
- 65.Seeley, H. W., P. J. Vandemark, and J. J. Lee. 1995. Microbes in action, a laboratory manual of microbiology, 4th ed. W.H. Freeman and Company, New York, NY.
- 66.Sexstone, A. J., N. P. Revsbech, T. B. Parkin, and J. M. Tiedje. 1985. Direct measurement of oxygen profiles and denitrification rates in soil aggregates. Soil Sci. Soc. Am. J. 49:645-651. [Google Scholar]
- 67.Sievert, S. M., J. Kuever, and G. Muyzer. 2000. Identification of 16S ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 66:3102-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 69.Smith, S. M., R. M. M. Abed, and F. Garcia-Pichel. 2004. Biological soil crusts of sand dunes in Cape Cod National Seashore, Massachusetts, USA. Microb. Ecol. 48:200-208. [DOI] [PubMed] [Google Scholar]
- 70.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stevenson, B. S., and T. M. Schmidt. 2004. Life history implications of rRNA gene copy number in Escherichia coli. Appl. Environ. Microbiol. 70:6670-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4.0b10. Sinauer Associates, Sunderland, MA.
- 73.van Elsas, J. D., and K. Smalla. 1997. Methods for sampling soil microbes, p. 383-390. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, DC.
- 74.van Elsas, J. D., J. T. Trevors, and E. M. H. Wellington. 1997. Modern soil microbiology. Marcel Dekker Inc., New York, NY.
- 74a.Wertz, J. T., B. S. Stevenson, and J. A. Breznak. 2003. Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. N-223.
- 75.Zimmermann, J., J. M. Gonzalez, and C. Saiz-Jimenez. 2006. Epilithic biofilms in Saint Callixtus Catacombs (Rome) harbour a broad spectrum of Acidobacteria. Antonie Leeuwenhoek 89:203-208. [DOI] [PubMed] [Google Scholar]
- 76.Zimmermann, J., J. M. Gonzalez, C. Saiz-Jimenez, and W. Ludwig. 2005. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in Altamira Cave using 23S rRNA sequence analyses. Geomicrobiol. J. 22:379-388. [Google Scholar]