Abstract
Microbial reductive dechlorination of commercial polychlorinated biphenyl (PCB) mixtures (e.g., Aroclors) in aquatic sediments is crucial to achieve detoxification. Despite extensive efforts over nearly two decades, the microorganisms responsible for Aroclor dechlorination remained elusive. Here we demonstrate that anaerobic bacteria of the Dehalococcoides group derived from sediment of the Housatonic River (Lenox, MA) simultaneously dechlorinate 64 PCB congeners carrying four to nine chlorines in Aroclor 1260 in the sediment-free JN cultures. Quantitative real-time PCR showed that the Dehalococcoides cell titer in JN cultures amended with acetate and hydrogen increased from 7.07 × 106 ± 0.42 × 106 to 1.67 × 108 ± 0.04 × 108 cells/ml, concomitant with a 64.2% decrease of the PCBs with six or more chlorines in Aroclor 1260. No Dehalococcoides growth occurred in parallel cultures without PCBs. Aroclor 1260 dechlorination supported the growth of 9.25 × 108 ± 0.04 × 108 Dehalococcoides cells per μmol of chlorine removed. 16S rRNA gene-targeted PCR analysis of known dechlorinators (i.e., Desulfitobacterium, Dehalobacter, Desulfuromonas, Sulfurospirillum, Anaeromyxobacter, Geobacter, and o-17/DF-1-type Chloroflexi organisms) ruled out any involvement of these bacterial groups in the dechlorination. Our results suggest that the Dehalococcoides population present in the JN cultures also catalyzes in situ dechlorination of Aroclor 1260 in the Housatonic River. The identification of Dehalococcoides organisms as catalysts of extensive Aroclor 1260 dechlorination and our ability to propagate the JN cultures under defined conditions offer opportunities to study the organisms' ecophysiology, elucidate nutritional requirements, identify reductive dehalogenase genes involved in PCB dechlorination, and design molecular tools required for bioremediation applications.
Polychlorinated biphenyls (PCBs), a family of 209 aromatic compounds (congeners) containing 1 to 10 chlorines, are toxic, recalcitrant, and ubiquitously distributed contaminants. Commercial PCBs were used worldwide from the 1920s through the 1970s. PCBs are included in the 12 worldwide priority persistent organic pollutants (or POPs) (57) and are ranked 5th (out of 275) on the U.S. Environmental Protection Agency Superfund Priority List of Hazardous Compounds (2) because of their propensity to bioaccumulate and biomagnify along the food chain and their suspected contributions to multiple health effects (3). Hundreds of thousands of metric tons of commercial PCBs (e.g., Aroclors) persist in aquatic sediments (49) and continue to pose a threat to human and ecosystem health. Aroclors, especially the highly chlorinated mixtures like Aroclors 1254 and 1260, present a particularly difficult biodegradation challenge because these complex mixtures consist of dozens of congeners.
Aerobic microbial degradation of PCBs by the biphenyl (or bph) pathway is well understood, but in situ degradation by this dioxygenase-based pathway is limited to PCBs with three or fewer chlorines (5). In contrast, anaerobic microbial reductive dechlorination, which replaces chlorine atoms with hydrogen atoms, preferentially attacks PCBs with two to nine chlorines. Reductive dechlorination is the key process for PCB detoxification in aquatic sediments (5, 9, 52). Significantly, reductive dechlorination acts on the congeners that bioaccumulate to the greatest extent and pose the greatest risks to human health (9, 12, 52, 62). The reduced products are less persistent and are more readily degraded by other organisms, in particular aerobic bacteria using the biphenyl pathway (5). In situ microbial reductive dechlorination of Aroclors was first reported in 1987 (15, 16) and has been documented in many contaminated river, harbor, and estuary sediments (5, 7, 9, 42), indicating that the organisms responsible for the dechlorination are widespread. Unfortunately, in situ PCB dechlorination at most sites has not achieved the full potential that laboratory experiments with microcosms have demonstrated (4, 12, 51). A major impediment to accelerating or enhancing reductive dechlorination processes that act on commercial PCBs has been the lack of knowledge of the organisms involved in Aroclor dechlorination.
Various bacterial groups contain members capable of respiring halogenated aromatic compounds in a process known as (de)halorespiration (32, 39). For example, Desulfitobacterium species are capable of dechlorinating a wide range of substituted (especially hydroxylated) chloroaromatics, including pentachlorophenol and hydroxylated PCBs (32). However, Dehalococcoides and the phylogenetically distinct (≤90% 16S rRNA gene sequence identity) Chloroflexi strains o-17 and DF-1 are the only organisms known that reductively dechlorinate chloroaromatics that exclusively carry chlorine substituents. For example, Dehalococcoides sp. strain CBDB1 respires chlorobenzenes with three to six chlorines and dechlorinates 1,2,3,7,8-pentachloro-p-dibenzodioxin (17, 34). Dehalococcoides ethenogenes strain 195 dechlorinates 2,3,4,5,6-chlorobiphenyl (23456-CB) to 2346-CB/2356-CB and 246-CB in the presence of growth-supporting tetrachloroethene (PCE) (25). Bacterial strains DF-1 and o-17 dechlorinate a limited range of individual PCB congeners and chlorobenzenes, but apparently cannot dechlorinate commercial PCB mixtures (45, 64).
Aroclor 1260 is a commercial PCB mixture of about 75 penta- to nonachlorobiphenyls with an average of 6.3 chlorines per biphenyl molecule (26). This mixture was widely used in electrical transformers, as a hydraulic fluid, as a plasticizer, and as a dedusting agent for more than 50 years (3). We previously reported the development of robust, stable, sediment-free mixed cultures (6), henceforth referred to as the JN cultures, from Aroclor 1260-contaminated Housatonic River sediment (Lenox, MA). These cultures extensively dechlorinate Aroclor 1260 at concentrations of 5 to 500 μg/ml (13.5 μM to 1.35 mM), including all PCB congeners containing 2,3,4-, 2,3,5-, 2,3,6-, 2,4,5-, 2,3,4,5-, 2,3,4,6-, and 2,3,5,6-chlorophenyl rings (6). Most of the chlorines removed are flanked meta-chlorines, but some para-dechlorination occurs as well. This predominantly meta-dechlorination activity is known as Process N (9, 51).
The objectives of the current study were to identify the organisms responsible for Process N dechlorination of Aroclor 1260 in the JN cultures and to determine if this dechlorination supports growth of the dechlorinators. Prior to this report, no microbes responsible for dechlorination of any commercial PCB mixture were known, and bacterial growth supported by dechlorination of commercial PCBs had never been shown. Here we demonstrate that a Dehalococcoides population is responsible for Process N dechlorination of Aroclor 1260 in the JN cultures, and most likely in the Housatonic River, and that these bacteria obtain energy for growth from this dechlorination.
MATERIALS AND METHODS
PCB nomenclature.
In this article, PCB congeners are named by listing the substituted positions on each ring separated by a hyphen and followed by CB (chlorobiphenyl). Thus, 24-24-CB is the congener substituted at positions 2, 2′, 4, and 4′.
Cultures.
The sediment-free JN mixed cultures were established from microcosms of Aroclor 1260-contaminated sediment collected from the Woods Pond section of the Housatonic River (Lenox, MA) as described previously (6). We used a sulfide-free, bicarbonate-buffered mineral medium amended with selenite, tungstate, vitamins (including vitamin B12), a trace element solution (SL9), and 0.01% yeast extract (1, 6). The medium was reduced with titanium(III) chloride (0.1 M)-citrate (0.2 M [pH 7]) (1) and amended with 5 to 50 μg/ml Aroclor 1260 as described previously (6). Initially, the microcosms and cultures were also amended with 2,6-dibromobiphenyl (26-BB) (350 μM), which was completely dehalogenated to biphenyl, to promote PCB dechlorination (6). The addition of 26-BB was not required to achieve extensive PCB dechlorination in the sediment-free JN cultures and was discontinued unless indicated otherwise (6). Cultures (30 ml) were incubated in the dark at 22 to 24°C under a headspace of 20% CO2-80% N2 (vol/vol) in 50-ml serum bottles fitted with Teflon-lined butyl rubber septa and crimp seals. Cultures were initially incubated with acetate plus formate (5 mM each) and fed the same substrates every 3 to 4 weeks. Beginning with the fourth sediment-free transfer, hydrogen (7.5 mM nominal concentration) was added by injecting 5 ml with a sterile syringe. Hydrogen was replenished every 3 to 4 weeks. Beginning with the 6th transfer, formate was eliminated. All transfers were 10% (vol/vol). Samples for PCB extraction and for DNA analysis were taken by syringe after the syringe was rinsed with a sterile 5 mM sulfide solution to remove O2. Where indicated, vancomycin (Sigma) was added from a filter-sterilized, anoxic, aqueous stock solution (1 mg/ml).
Dilution-to-extinction experiments.
Tenfold serial dilutions of active cultures were carried out in 50-ml serum bottles containing 30 ml of medium amended with vancomycin (5 μg/ml), acetate (5 mM), hydrogen (7.5 mM nominal concentration), and 50 μg/ml Aroclor 1260 (6). Cultures were monitored periodically for PCB dechlorination and were fed acetate and hydrogen at intervals of 3 to 4 weeks.
Experiments to test growth with Aroclor 1260.
Triplicate JN cultures that had been transferred in sediment-free medium three times with 5 μg/ml (13.5 μM) Aroclor 1260 and 350 μM 26-BB were subcultured into three groups of triplicate cultures. Subsequent transfers did not receive 26-BB. The first group was transferred four additional times with Aroclor 1260 (5 μg/ml for the first transfer and 50 μg/ml for all subsequent transfers). The second group of replicates was transferred four times under identical conditions, but without PCBs. The third group of replicates was transferred three times without PCBs, followed by a final transfer to medium amended with Aroclor 1260 to determine if PCB dechlorinating activity could be restored after prolonged incubation without PCBs. PCB dechlorination was monitored and total genomic DNA was obtained at the time points indicated in Fig. 1. The extracted genomic DNA served as a template for Dehalococcoides cell number estimation by quantitative real-time PCR (qPCR) of Dehalococcoides 16S rRNA genes (53).
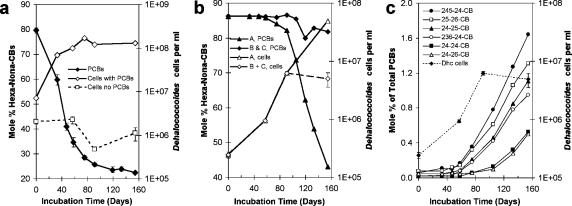
FIG. 1.
Relationship of Dehalococcoides cell titers to Aroclor 1260 dechlorination in JN subcultures. Dechlorination is presented as the percent decrease in PCB congeners with six to nine chlorines, which comprise ∼90% of the PCBs in Aroclor 1260. Data are means ± standard deviations. (a) PCB-dependent growth of Dehalococcoides in JN cultures incubated with Aroclor 1260 (50 μg/ml) and decrease of Dehalococcoides cell titers in cultures transferred four times without PCBs. (b) PCB-dependent growth in subcultures when PCBs were restored following three transfers without PCBs. (c) Correlation between PCB dechlorination and Dehalococcoides (Dhc) cell growth in replicate cultures B and C.
PCB extraction.
Aqueous samples (0.2 to 1 ml) from the cultures were transferred to 8-ml glass vials fitted with Teflon-lined screw caps, and PCBs were extracted with 4 to 5 ml of anhydrous diethyl ether by vigorous horizontal shaking on a platform shaker overnight at room temperature. Congener-specific PCB dechlorination was monitored by high-resolution capillary gas chromatography (GC) analysis with an Ni63 electron capture detector as previously described (12). Calibration standards were prepared from Aroclor 1260 supplemented with 43 PCB congeners previously identified as Process N dechlorination products (12). The concentrations of the individual components of Aroclor 1260 in the standards were calculated from previously determined weight percent distributions for the congeners in Aroclor 1260 (26). PCBs were quantified with a four-point external calibration for the customized PCB standard (542 to 8,668 ng/ml) with a quadratic fit forced through zero. We calculated the mole percent value for each individual peak; the distribution of ortho-, meta-, and para-chlorines per biphenyl; the total number of chlorines per biphenyl; and the PCB homolog distribution as reported previously (12).
DNA extraction, PCR, and qPCR.
Genomic DNA from the JN cultures was extracted as described previously (6). DNA for all time points was extracted from pooled samples of triplicate cultures (1 ml each for 3 ml total), except for the time point at 154 days for cultures A to C (Fig. 1b), for which DNA from each 1-ml sample was extracted separately. For the detection of 16S rRNA genes of known dechlorinating bacteria, a nested PCR approach was used. The initial amplification of 16S rRNA genes used the eubacterial primers 8F (5′-AGAGTTTGATCCTGGCTCAG) and 1541R (5′-AAGGAGGTGATCCAGCCGCA). A second round of PCR utilized 1:5 and 1:50 dilutions of the amplified 16S rRNA genes as template and dechlorinator-targeted primer pairs. Target organisms, primer sequences, and the references for the corresponding PCR conditions are summarized in Table 1. All PCRs were in a total volume of 25 μl and used 2.5 μl of DNA or diluted DNA as a template.
TABLE 1.
16S rRNA gene-targeted PCR screening of an Aroclor 1260-dechlorinating JN culture for known dechlorinating bacterial groups
| Dechlorinating group | PCR result at DNA dilution showna
|
Positive control | Primersb | |||||
|---|---|---|---|---|---|---|---|---|
| Direct
|
Nested
|
|||||||
| U | 1/10 | 1/100 | U | 1/5 | 1/50 | |||
| Dehalococcoides | + | + | + | D. ethenogenes 195 | DHC 1 (F) and DHC 1377 (R) (31) | |||
| Anaeromyxobacter | − | − | − | Anaeromyxobacter dehalogenans strain 2CP-C | A60-86F and A447-465R (21) | |||
| Desulfitobacterium | − | − | − | Desulfitobacterium sp. strain Viet 1 | Dd1 (F) and Dd2 (R) (23) | |||
| Desulfuromonas | − | − | − | Desulfuromonas michiganensis strain BB1 | BB1F and BB1R (41) | |||
| Dehalobacter | − | − | − | Dehalobacter sp. | deb 179f and deb 1007r (54) | |||
| Sulfurospirillum (formerly Dehalospirillum) | − | − | − | Sulfurospiriillum multivorans | JPDF 121 and JPDR 557 (50) | |||
| − | − | − | Sulfurospirillum halorespirans | DHSPM 576 and DHSPM 1210c | ||||
| Geobacter (SZ type) | − | − | − | Geobacter lovleyi strain SZ | 196F and 999R (58) | |||
| o-17/DF-1-type Chloroflexi | − | − | − | NDd | Clone of o-17 16S rRNA gene | 14F (22) and 1265R (61) | ||
U, undiluted.
References for some primers are shown in parentheses.
R. C. Ebersole and E. R. Hendrickson, U.S. patent application 20030077601.
ND, not determined. Nested PCR was not performed because the 14F primer shares the same recognition site as the bacterial universal primer 27F used in the initial round of PCR amplification.
qPCR targeting total Dehalococcoides 16S rRNA genes was performed with primers and conditions described previously in a 20-μl reaction volume using an ABI 7000 thermocycler and ABI TaqMan Universal PCR master mix (53). Each reaction was run in triplicate. PCR reagents were purchased from Applied Biosystems (ABI, Carlsbad, CA), and primers and probes were from Integrated DNA Technologies (IDT, Coralville, IA). Standard curves were prepared using 10-fold serial dilutions of genomic DNA from Dehalococcoides sp. strain GT (60) ranging from 3.8 × 10−5 to 3.8 ng/μl (20 to 2 × 106 copies per μl of DNA) as template, as well as plasmid DNA carrying a cloned 16S rRNA gene fragment of Dehalococcoides sp. strain FL2 (29), ranging from 1.1 × 10−7 to 1.1 ng/μl (20 to 2 × 108 copies per μl of plasmid DNA) as a template. qPCR for total Geobacteraceae 16S rRNA genes was performed in a 20-μl reaction volume using the QuantiTect SYBR green PCR kit master mix (ABI) and primers Geo564F (5′-AAGCGTTGTTCGGAWTTAT) and Geo840R (5′-GGCACTGCAGGGGTCAATA) as described previously (58). These primers were originally used in a TaqMan assay (18) and were adapted for use with the SYBR green-based approach by Sung (58). The thermocycler conditions included 50°C for 2 min, 95°C for 15 s, and 40 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s. We observed that these primers also amplified Anaeromyxobacter 16S rRNA genes and used dissociation curve analysis to distinguish the amplicons. A dissociation curve was run at 95°C for 15 s, 64°C for 1 min, and 99°C for 15 s, and a peak melting temperature of 82.2 to 82.9°C was indicative of Geobacter spp. while a melting temperature of 81.5 to 81.9°C was diagnostic for Anaeromyxobacter spp. The standard curve was generated from a 10-fold dilution series of 1 ng/μl plasmid DNA (ranging from 18 to 1.8 × 108 gene copies) containing the cloned 16S rRNA gene from Geobacter lovleyi strain SZ and had a slope of −3.38, a y intercept of 33.57, and an r2 of 0.9988. Cell numbers for Dehalococcoides and Geobacter were calculated assuming that these bacteria contain 1 and 2 16S rRNA gene copies, respectively, as the fully sequenced genomes have demonstrated (37, 47, 55; see TIGR database at http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi.).
DNA cloning and community analysis.
Genomic DNA of a sediment-free JN culture that had been transferred eight times in the presence of vancomycin (5 μg/ml) was used as a PCR template to generate a 16S rRNA gene clone library, which was screened by restriction fragment length polymorphism analysis (RFLP) as previously described (6).
Nucleotide sequence accession numbers.
Cloned 16S rRNA gene fragments representative of each distinct RFLP pattern were sequenced and classified (17a, 27) as previously described (6) and submitted to GenBank. The accession numbers are listed in Table 3.
TABLE 3.
Community analysis of an Aroclor 1260-dechlorinating JN culture transferred repeatedly in the presence of vancomycin
| Clone name (accession no.)e | No. of clones | Norm %a | Bergey's classification (RDP II)b
|
No. of 16S rRNA genesc | Closest match in GenBank as determined by RDP Sequence Match and BLAST tools | % Identity | |
|---|---|---|---|---|---|---|---|
| JN18Phylogenetic group | Closest classified relative (% certainty) | ||||||
| JN18 V4 B (EF059527) | 7 | 25.8-27 | Chloroflexi | Dehalococcoides (100) | 1 | Dehalococcoides sp. strains CBDB1, FL2, GT (AF230641, AF357918 | >99.9 |
| JN18 V12 B (EF059528) | AY914178) | ||||||
| JN18 V35 B (EF059529) | |||||||
| JN18 V108 B (EF059530) | |||||||
| JN18 V2 A* (DQ168642) | 54 | 49.7-50 | Betaproteobacteria | Thauera (94) | 4d | Azoarcus sp. strain LU1 (AJ007007) | 98-99 |
| JN18 V17 A3 (DQ168643) | |||||||
| JN18 V62 (DQ168641) | |||||||
| JN18 V65 E (EF059531) | 14 | 1.7-2.8 | Gammaproteobacteria | Acinetobacter (100) | 7 | Acinetobacter sp. strain LY1 (AJ007008) | 99 |
| JN18 V5 C (EF059532) | |||||||
| JN18 A13 Q (DQ168644) | 3 | 3.7-3.8 | Gammaproteobacteria | Pseudomonas (100) | 4-7 | Uncultured bacterial clone 69-7G (AY955095) | 99 |
| JN18 A17 R (DQ168645) | |||||||
| JN18 A62 G* (DQ168649) | 9 | 7.4-7.7 | Bacteroidales | Porphyromonadaceae (83, 100) | 4 | Uncultured bacterial clone PL-7B7 (AY570639); Bacteroides sp. strain Z4 (AY949860) | 99 |
| JN18 V15 G (EF059535) | |||||||
| JN18 V95 J (EF059536) | 1 | 1.8-1.9 | Deltaproteobacteria | Geobacteraceae (98) | 2 | Uncultured bacterial clone W31 (AY770971) | 96 |
| JN18 V27 I (EF059533) | 2 | 0.6-1.5 | Clostridiales | Sedimentibacter (100) | 5-12 | Sedimentibacter sp. strain C7 (AY766466) | 96 |
| JN18 V41 S (DQ168656) | 3 | 1-2.2 | Clostridiales | Lachnospiraceae (94) | 5-12 | Anaerotruncus colihominis HKU19 (DQ082932) | 94 |
| JN18 V56 P (EF059534) | 2 | 0.6-1.5 | Clostridiales | Clostridiaceae (95) | 5-12 | Clostridium sp. strain FCB90-3 (AJ229251) | 98 |
| JN18 A7 F* (DQ168648) | 1 | 3.7-3.8 | Eubacteria | Proteobacteria (38) | ? | Uncultured bacterial strain SHA-53 (AJ249111) | 99 |
Percentage of clones normalized for estimated rRNA copy number.
Each distinct operational taxonomic unit identified by RFLP analysis was phylogenetically classified according to Bergey's Manual (27) using the naïve Bayesian rRNA classifier hosted on the RDP II site (17a). This classifier is trained on all known type strain 16S rRNA gene sequences. The query sequence is assigned a taxonomic hierarchy and the calculation is repeated for 100 trials to assign an estimate of the certainty of that taxonomic assignment.
Azoarcus, a close relative, possesses four 16S rRNA gene copies.
RESULTS AND DISCUSSION
Targeted PCR of known reductively dechlorinating bacteria.
Analysis of a 16S rRNA gene clone library generated from genomic DNA of an Aroclor 1260-dechlorinating JN culture indicated that Dehalococcoides and Geobacter organisms were present (6). The known Dehalococcoides strains are strict halorespirers and require a halogenated compound as an electron acceptor for growth (39, 40). Members of the Geobacter genus are well-known iron reducers that are distributed in anoxic sediments but are not recognized for their ability to perform reductive dechlorination. Only recently, a “dechlorinating clade” comprised of Geobacter (formerly Trichlorobacter) thiogenes and Geobacter lovleyi strain SZ was described (20, 56, 59). To more fully investigate the presence of dechlorinators in the JN cultures, genomic DNA was used as template in PCR with primers specific for the 16S rRNA genes of eight known groups of dechlorinating bacteria. Dehalococcoides organisms were readily detected by direct PCR, but no other known dechlorinating bacteria were detected even when the more sensitive nested PCR approach was used (Table 1). These results rule out any significant role of Dehalobacter, Desulfitobacterium, Desulfuromonas, Sulfurospirillum, Anaeromyxobacter, SZ-type Geobacter, and o-17/DF-1-type Chloroflexi organisms in the Aroclor 1260 dechlorination in the JN culture. Our results differ from findings reported with PCB-impacted estuarine sediments where 16S rRNA genes of o-17/DF-1-type Chloroflexi were found but Dehalococcoides organisms were apparently absent (61).
The Geobacter-targeted primers used for the qualitative PCR analysis specifically targeted the tetrachloroethene-dechlorinator Geobacter lovleyi strain SZ. Because other dechlorinating Geobacteraceae such as Geobacter thiogenes (20, 56) would not have been detected, these results do not rule out the presence of other dechlorinating Geobacter species. Therefore, additional experiments were performed with primers that target all Geobacteraceae.
Growth experiments with Aroclor 1260 as electron acceptor.
To conclusively determine whether Geobacter spp. were involved in PCB dechlorination and grew at the expense of Aroclor 1260 dechlorination, we used qPCR targeting all Geobacteraceae to monitor total Geobacter cell titers in JN cultures incubated with and without Aroclor 1260. Geobacter cell titers were similar (1.1 × 106 ± 0.18 × 106 to 1.8 × 106 ± 0.25 × 106 cells/ml) (average ± standard deviation; n = 3) in all cultures regardless of whether or not Aroclor 1260 was present. Notably, the Geobacter cell numbers were 200-fold lower than the Dehalococcoides cell numbers in cultures amended with PCBs (see below). The absence of any correlation of an increase in Geobacter cell titers with PCB dechlorination indicates that the Geobacter population was not involved in Aroclor 1260 reductive dechlorination.
All known Dehalococcoides organisms have a highly restricted metabolism and can derive energy only from reductive dechlorination reactions (37, 39, 40, 55). Hence, the presence of Dehalococcoides in the JN cultures suggested that these bacteria derive energy for growth from the reductive dechlorination of Aroclor 1260. To conclusively determine whether Dehalococcoides organisms were the catalysts of PCB dechlorination and grew at the expense of Aroclor 1260 dechlorination, their cell titers over the course of Aroclor 1260 dechlorination were monitored with qPCR (53). Dehalococcoides cell titers increased from 7.07 × 106 ± 0.42 × 106 cells/ml at day 0 to 1.67 × 108 ± 0.04 × 108 cells/ml at day 75, concomitant with a 64.2% decrease in the PCBs with six or more chlorines in Aroclor 1260 (Fig. 1a).
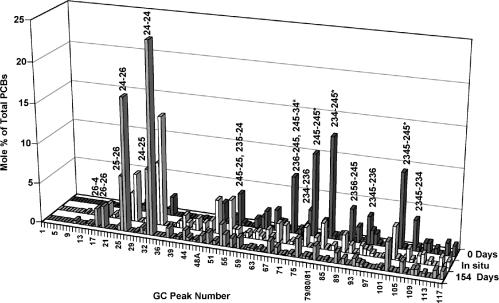
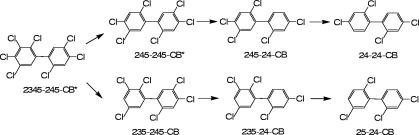
Table 2 depicts all PCB substrates and products that were identified by GC analysis. By day 154, nearly all of the PCBs with six or more chlorines were dechlorinated to four main products: 24-24-CB, 24-26-CB, 24-25-CB, and 25-26-CB (Fig. 2). The observed dechlorination pathways for a major heptachlorobiphenyl in Aroclor 1260 are shown in Fig. 3. Significantly, the four PCB congeners that accumulate to the highest levels in humans, PCBs 118, 138, 153, and 180 (i.e., 245-34-CB, 245-245-CB, 234-245-CB, and 2345-245-CB) (62), were all extensively dechlorinated to 24-4-CB, 24-24-CB, 24-24-CB, and both 24-24-CB and 24-25-CB, respectively (Fig. 2 and 3 and Table 2). These dechlorination products are far less persistent in vertebrates (14). Hence, these results suggest that Process N dechlorination observed in the JN cultures contributes to Aroclor 1260 detoxification. In addition, many of the Process N dechlorination products can be further dechlorinated by Process LP, another PCB dechlorination process which occurs elsewhere in the Housatonic River (10) and removes unflanked para-chlorines (8). Process LP converts 24-24-CB to 2-2-CB, 24-25-CB to 25-2-CB, and 24-26-CB to 26-2-CB (8), all of which are degradable by aerobic bacteria (11, 13). Hence, as a result of sequential anaerobic dechlorination by Process N and then Process LP, highly chlorinated PCB congeners can be dechlorinated to PCB congeners with a low degree of chlorination that are susceptible to aerobic degradation and mineralization (i.e., complete detoxification).
TABLE 2.
Confirmed PCB substrates and terminal products of Process N dechlorination of Aroclor 1260 by Dehalococcoides in JN cultures
| Hepta-, octa, or nona-CB substrate | Hexa-CB substrate | Penta- or tetra-CB substrate | Terminal product |
|---|---|---|---|
| 23456-2345 | 2345-24 | 234-24 | 24-24 |
| 23456-234 | 2345-25 | 234-25 | 24-25 |
| 23456-245 | 2345-26 | 234-34 | 25-25 |
| 2345-2345 | 2345-34 | 235-24 | 24-26 |
| 2345-2346 | 2346-23 | 235-25 | 25-26 |
| 2345-2356 | 2346-24 | 235-26 | 26-26 |
| 23456-25 | 234-234 | 236-23 | 246-24 |
| 23456-34 | 234-235 | 236-24 | 246-25 |
| 2345-234 | 234-236 | 236-25 | 246-26 |
| 2345-235 | 234-245 | 236-34 | 24-2 |
| 2345-236 | 235-235 | 245-24 | 24-4 |
| 2345-245 | 235-236 | 245-25 | 25-2 |
| 2345-345 | 235-245 | 245-26 | 26-2 |
| 2346-234 | 2346-25 | 245-34 | 26-3 |
| 2346-236 | 2346-34 | 25-34 | 26-4 |
| 2346-245 | 2356-23 | 26-34 | |
| 2356-234 | 2356-24 | 245-4 | |
| 2356-235 | 2356-25 | ||
| 2356-236 | 2356-34 | ||
| 2356-245 | 236-236 | ||
| 2356-345 | 236-245 | ||
| 236-246 | |||
| 236-345 | |||
| 245-245 | |||
| 245-246 | |||
| 245-345 |
FIG. 2.
Change in Aroclor 1260 congener distribution (Process N dechlorination) in the JN cultures. Data for days 0 and 154 are from the JN cultures depicted in Fig. 1a and represent the averages of triplicate determinations. Parallel cultures transferred nine times in the presence of vancomycin showed the same congener profile in only 54 days. The center profile shows Aroclor 1260 dechlorination in situ in a sediment sample collected from the Housatonic River. A complete list of the congener assignments for all GC peaks is given by Frame et al. (26). Congeners marked with an asterisk are those reported to accumulate to the highest levels in humans (62).
FIG. 3.
Observed Process N dechlorination pathways of an environmentally relevant heptachlorobiphenyl by Dehalococcoides bacteria in the JN cultures. Asterisks denote two of the four PCB congeners that accumulate to the highest levels in humans (62).
From the data shown in Fig. 1a, we calculated that the release of 1 μmol of chloride from Aroclor 1260 by Process N supports the growth of 9.25 × 108 ± 0.04 × 108 Dehalococcoides cells. This compares favorably with the cell yield reported for Dehalococcoides strain GT, which was 2.5 × 108 ± 0.13 × 108 cells per μmol Cl− released when grown with vinyl chloride as an electron acceptor (60).
In contrast to the results for cultures incubated with Aroclor 1260, the Dehalococcoides cell numbers decreased nearly 50% (from 2.05 × 106 ± 0.24 × 106 cells/ml to 1.11 × 106 ± 0.37 × 106 cells/ml) in cultures transferred four times without PCBs (Fig. 1a). The Dehalococcoides cell titer after four consecutive transfers without PCBs was higher than expected. Assuming that neither growth nor decay of Dehalococcoides cells occurred in cultures without PCBs, four consecutive transfers should have yielded a cell titer of about 103 cells/ml; however, we measured cell titers of about 106 cells/ml. A likely explanation for the higher-than-expected Dehalococcoides cell numbers is the carryover of Aroclor 1260 and especially 26-BB, which was present in a higher concentration, from the original culture. Published most-probable-number experiments with microcosms of Housatonic River sediment indicate that the populations catalyzing Aroclor 1260 dechlorination by Process N also dehalogenate 26-BB (63). We calculated that the carryover of halogenated biphenyls was sufficient to support some growth and sustain a cell titer of at least 2.8 × 105 cells/ml after four transfers. Nevertheless, our data conclusively demonstrate that the Dehalococcoides cell numbers decrease in the absence of PCBs and therefore indicate that Dehalococcoides growth in the JN cultures hinges on the presence of PCBs as electron acceptors.
A third set of subcultures was included to determine if PCB dechlorination could be restored after a prolonged incubation period without PCBs, and, if so, if the restoration of PCB dechlorination would be accompanied by an increase in Dehalococcoides cell titers. These cultures were transferred three times without PCBs for a total of 196 days before Aroclor 1260 was restored. After PCBs were restored, there was a 60-day lag before PCB dechlorination was detected (Fig. 1b), and then all three replicates began to dechlorinate PCBs as evidenced by the appearance of dechlorination intermediates and products (Fig. 1c). The Dehalococcoides cell titers increased concomitantly with the onset of PCB dechlorination. PCB dechlorination in replicate A coincided with a 200-fold increase in Dehalococcoides cell numbers from 2.44 × 105 ± 0.30 × 105 cells/ml at day 0 to 4.94 × 107 ± 0.35 × 107 at day 154 (Fig. 1b). PCB dechlorination in replicates B and C did not progress to the same extent seen in replicate A, possibly due to oxygen contamination (i.e., introduction of air) during sampling events. Dehalococcoides cells are very oxygen sensitive and are inactivated by even brief exposure to oxygen. Nevertheless, the Dehalococcoides cell numbers in replicates B and C did increase nearly 30-fold to 5 × 106 ± 1.4 × 106 cells/ml between days 60 and 90 (Fig. 1b and c). Based on our calculations, even the small amount of dechlorination that occurred in replicates B and C (release of about 1.25 nmol/ml Cl−) was sufficient to support the growth of at least 1.25 × 106 cells/ml, which is consistent with our cell number observations for these replicates. Collectively, the data establish that reductive dechlorination of Aroclor 1260 supports the growth of Dehalococcoides organisms present in the JN cultures, i.e., the Dehalococcoides cells halorespire Aroclor 1260.
Aroclor 1260 (50 μg/ml) dechlorination in the JN cultures was not detectable by GC analysis until the Dehalococcoides population reached a minimum near 106 cells/ml (Fig. 1b and c), indicating that substantial Process N dechlorination of Aroclor 1260 (i.e., at least 30% decrease of hexa- through nonachlorobiphenyls) requires a minimum Dehalococcoides cell titer. The extrapolation of these laboratory results to the field suggests that substantial in situ dechlorination of Aroclors at similar concentrations requires a threshold titer of active Dehalococcoides cells, which may explain why substantial Aroclor dechlorination is rarely seen in situ.
PCB dechlorination in the JN cultures is resistant to vancomycin, an antibiotic that targets primarily gram-positive bacteria but does not affect Dehalococcoides (46). JN cultures parallel to those reported in Fig. 1a were subcultured with Aroclor 1260 eight times in the presence of 5 μg/ml of vancomycin. Comparison of the community structure following enrichment in the presence (Table 3) and absence of vancomycin (see Table 5 in reference 6) shows that the antibiotic decreased the proportions of Bacteroidales and Clostridiales, but doubled the proportion of Dehalococcoides to about 25% of the total bacteria. Vancomycin also accelerated the rate of PCB dechlorination, and the cultures maintained the same PCB dechlorination specificity on Aroclor 1260 (i.e., Process N dechlorination), thus providing further evidence that Dehalococcoides populations are responsible for Process N dechlorination. Cultures transferred repeatedly in the presence of vancomycin reached a dechlorination performance corresponding to that shown in Fig. 2 about three times faster (i.e., in 54 days versus 154 days) than that of cultures incubated in the absence of the antibiotic. The higher dechlorination rates observed in the presence of vancomycin suggest that the antibiotic alleviated competitive interactions with other populations (e.g., Bacteroidales and Clostridiales), which may compete for electron donors (i.e., hydrogen) or nutrients with the dechlorinating Dehalococcoides strains.
We also recovered extensive Process N Aroclor 1260 dechlorination activity after 131 days of incubation from vancomycin-treated 108 dilution tubes established from active JN cultures in dilution-to-extinction experiments. In the 106 and 107 dilutions, dechlorination commenced after 76 and 91 days of incubation, respectively. These findings also demonstrate Aroclor 1260-dependent growth of Dehalococcoides and corroborate the qPCR data that Dehalococcoides cell titers reached at least 108 cells/ml in the JN cultures (Fig. 1). Furthermore, the recovery of active cultures from 108 dilution tubes suggests that the dilution-to-extinction principle may be productive for further enriching the Aroclor 1260-dechlorinating Dehalococcoides organisms.
Evidence that Dehalococcoides cells carry out Process N dechlorination of Aroclor 1260 in situ.
Two decades ago, Brown and colleagues first reported the in situ dechlorination of Aroclors in anaerobic sediments (15, 16). They proposed that this dechlorination was due to anaerobic microorganisms that obtained energy for growth by using the PCBs as electron acceptors (15, 16). The analysis of Aroclor dechlorination in various sediment microcosms has distinguished eight different microbial dechlorination processes on the basis of congener selectivity, chlorines targeted, and dechlorination products (5, 9). Various investigators have proposed that each of these distinct PCB dechlorination processes is associated with a different microbial population, but none of these populations has ever been identified. We have now, for the first time, identified the bacteria responsible for dechlorination of a commercial PCB mixture. We have identified Dehalococcoides as the catalyst of Process N dechlorination of Aroclor 1260 and have demonstrated that this dechlorination supports the growth of Dehalococcoides by halorespiration.
Significantly, Process N is the primary PCB dechlorination process observed in Housatonic River sediments in situ (10; D. L. Bedard and L. A. DeRose, unpublished data). Figure 2 compares an example of the in situ dechlorination of Aroclor 1260 observed in Housatonic River sediment with the dechlorination pattern observed in the JN cultures, which were derived from Housatonic River sediment. This comparison demonstrates that the predominant dechlorination pattern in situ is very similar to that observed in the JN cultures (i.e., Process N). Unfortunately, in situ dechlorination has not progressed this far throughout most of the sediment in the Housatonic River and Process N dechlorination is incomplete (10). However, we consistently observed extensive Process N dechlorination of Aroclor 1260 in anaerobic microcosms of Housatonic River sediment when a halogenated “primer” such as 26-BB was used to stimulate PCB dechlorination (12). Direct PCR with Dehalococcoides 16S rRNA gene-targeted primers consistently detected Dehalococcoides in these sediment microcosms, even with 100-fold template dilutions (G. V. S. Jerzak and D. L. Bedard, unpublished data). Our results do not exclude the possibility that other microbial populations contribute to PCB dechlorination in situ. However, our findings indicate that Dehalococcoides cells most likely catalyze the dominant dechlorination process acting on Aroclor 1260 (Process N) under in situ conditions in the Housatonic River as they do in the sediment-free JN enrichments derived from the same site.
Dehalococcoides bacteria belong to the Chloroflexi, a deeply branching group on the bacterial phylogenetic tree (39). Although a large number of environmental clone sequences associated with the Chloroflexi group are deposited in GenBank, few dechlorinating isolates have been obtained, all of which are Dehalococcoides spp. Most of these dechlorinate chlorinated aliphatic compounds (e.g., chlorinated ethenes), although Dehalococcoides ethenogenes 195 and Dehalococcoides strain CBDB1 dechlorinate several chlorinated aromatic compounds (17, 25, 34). Two other Chloroflexi strains, o-17 and DF-1, which are phylogenetically distinct from Dehalococcoides (≤90% 16S rRNA gene sequence identity) and dechlorinate some PCB congeners, have also been obtained in culture (19, 64). Bacteria of the o-17/DF-1 group have been detected in PCB-impacted sediments from several estuarine sites, and it was suggested that they may play a role in the dechlorination of commercial PCB mixtures at those sites (61). Bacterium DF-1 removes doubly flanked chlorines, and bacterium o-17 removes doubly flanked meta-chlorines and flanked ortho-chlorines, but neither strain has been shown to dechlorinate commercial PCB mixtures (45, 64). The o-17 culture dechlorinated 8 of 26 PCB congeners that were tested. Seven of these were chlorinated on only one ring. Furthermore, the o-17 culture could not dechlorinate Aroclor 1260 and the presence of Aroclor 1260 in o-17 cultures inhibited the dechlorination of its known PCB substrates (45). In contrast, the Dehalococcoides strains in the JN cultures dechlorinated 64 PCB congeners present in Aroclor 1260, all of which are chlorinated on both rings and 47 of which carry six or more chlorines.
Presence of multiple Dehalococcoides strains in the JN cultures.
The 16S rRNA gene-based analysis suggests that the JN cultures may contain several distinct Dehalococcoides strains (Table 3) (6). The presence of multiple Dehalococcoides strains in dechlorinating enrichments is not uncommon (60). Each of several different Dehalococcoides 16S rRNA gene sequences obtained from the JN cultures was ≥99.8% identical over a stretch of 1,420 bp to those of the chlorobenzene-dechlorinating Dehalococcoides sp. strain CBDB1 and the chloroethene-dechlorinating Dehalococcoides strains FL2, GT, and BAV1 (29, 30, 37, 53, 60). Dehalococcoides isolates that share identical or nearly identical (i.e., 99.9%) 16S rRNA genes differ in the range of halogenated substrates that they use as growth-supporting electron acceptors (30, 33, 53, 60). Genomic studies and the analysis of entire genomes have revealed that individual Dehalococcoides strains contain multiple nonidentical reductive dehalogenase (RDase) genes (33, 37, 55). For example, Dehalococcoides strain CBDB1 contains 32 putative RDase genes. To date, function has been assigned to only three RDase genes, all involved in chlorinated ethene reductive dechlorination (36, 43, 48), and no RDase genes implicated in PCB reductive dechlorination have been identified. The Dehalococcoides strains active in the JN cultures provide an excellent opportunity to identify the first RDase gene(s) involved in PCB dechlorination and to determine the specificities and substrate ranges of the enzymes that they encode.
Conclusions.
Dehalococcoides strains are relevant for both the natural attenuation and bioremediation of chlorinated ethenes (40), and their presence has been documented at many sites contaminated with chlorinated ethenes (31). Our findings suggest that Dehalococcoides bacteria may also play a major role in the in situ dechlorination of commercial PCB mixtures. The sediment-free JN cultures with robust PCB dechlorination activity will allow in-depth studies of the pathways and requirements of Dehalococcoides strains that dechlorinate environmentally relevant commercial PCB mixtures (e.g., Aroclors) and will facilitate the isolation of these strains. With increasing Dehalococcoides genome information and the application of high-throughput microarray technologies, the discovery of novel RDase genes responsible for commercial PCB dechlorination is in reach.
The work reported here is also a major step toward the development of cost-effective methods for promoting in situ PCB remediation and toward the design of molecular biological tools for the detection, quantification, and monitoring of PCB-dechlorinating Dehalococcoides strains and their activity in contaminated environments. It will be interesting to learn whether Dehalococcoides populations are widespread in PCB-contaminated sediments and are also responsible for catalyzing some of the other major PCB dechlorination processes that have been documented and described (5, 9, 51) as well as Process N. Our findings suggest that increasing the cell titer of the PCB-dechlorinating Dehalococcoides population to >106 cells/ml of sediment in the Housatonic River and possibly other sites contaminated with Aroclor 1260 or Aroclor 1254 may promote extensive Process N PCB dechlorination in situ. The increased understanding of the microbiology contributing to chlorinated ethene detoxification has directed successful aquifer bioremediation efforts (24, 38, 44). As our understanding of the microbiology involved in Aroclor dechlorination increases, the engineering challenge will be the implementation of enhanced bioremediation approaches (i.e., biostimulation and bioaugmentation) in freshwater and estuarine sediments.
Acknowledgments
We thank Ben Amos, Sarah Thomas, and Youlboong Sung for assistance with targeted PCR analyses and Steve Zinder and Kevin Sowers for providing cloned 16S rRNA genes of D. ethenogenes strain 195 and bacterium o-17, respectively. We thank Richard Bopp and Harry Roy for helpful comments on the manuscript.
D.L.B. acknowledges support from National Science Foundation grant 0077837 and a grant from GE Corporate Environmental Programs. The National Science Foundation provided additional support under grant no. 0090496 (Career Award to F.E.L.). F.E.L. and K.M.R. acknowledge support from Regenesis.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Adrian, L., U. Szewzyk, and H. Görisch. 2000. Bacterial growth based on reductive dechlorination of trichlorobenzenes. Biodegradation 11:73-81. [DOI] [PubMed] [Google Scholar]
- 2.Agency for Toxic Substances and Disease Registry. 2006. CERCLA priority list of hazardous compounds. Agency for Toxic Substances and Disease Registry, Atlanta, GA. http://www.atsdr.cdc.gov/cercla/05list.html.
- 3.Agency for Toxic Substances and Disease Registry. 2000. Toxicological profile for polychlorinated biphenyls (update). Agency for Toxic Substances and Disease Registry, Atlanta, GA. http://www.atsdr.cdc.gov/toxprofiles/tp17.html. [PubMed]
- 4.Alder, A. C., M. M. Häggblom, S. R. Oppenheimer, and L. Y. Young. 1993. Reductive dechlorination of polychlorinated biphenyls in anaerobic sediments. Environ. Sci. Technol. 27:530-538. [Google Scholar]
- 5.Bedard, D. L. 2003. Polychlorinated biphenyls in aquatic sediments: environmental fate and outlook for biological treatment, p. 443-465. In M. M. Häggblom and I. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Press, Dordrecht, The Netherlands.
- 6.Bedard, D. L., J. J. Bailey, B. L. Reiss, and G. V. S. Jerzak. 2006. Development and characterization of stable sediment-free anaerobic bacterial enrichment cultures that dechlorinate Aroclor 1260. Appl. Environ. Microbiol. 72:2460-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedard, D. L., and R. J. May. 1996. Characterization of the polychlorinated biphenyls in the sediments of Woods Pond: evidence for microbial dechlorination of Aroclor 1260 in situ. Environ. Sci. Technol. 30:237-245. [Google Scholar]
- 8.Bedard, D. L., E. A. Pohl, J. J. Bailey, and A. Murphy. 2005. Characterization of the PCB substrate range of microbial dechlorination Pocess LP. Environ. Sci. Technol. 39:6831-6839. [DOI] [PubMed] [Google Scholar]
- 9.Bedard, D. L., and J. F. Quensen III. 1995. Microbial reductive dechlorination of polychlorinated biphenyls, p. 127-216. In L. Y. Young and C. E. Cerniglia (ed.), Microbial transformation and degradation of toxic organic chemicals. Wiley-Liss, New York, NY.
- 10.Bedard, D. L., L. A. Smullen, and R. J. May. 1996. Microbial dechlorination of highly chlorinated PCBs in the Housatonic River, p. 117. In Proceedings of the 1996 International Symposium on Subsurface Microbiology. Swiss Society of Microbiology and Institute of Plant Biology, University of Zurich, Davos, Switzerland.
- 11.Bedard, D. L., R. Unterman, L. H. Bopp, M. J. Brennan, M. L. Haberl, and C. Johnson. 1986. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol. 51:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedard, D. L., H. Van Dort, and K. A. DeWeerd. 1998. Brominated biphenyls prime extensive microbial reductive dehalogenation of Aroclor 1260 in Housatonic River sediment. Appl. Environ. Microbiol. 64:1786-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bopp, L. H. 1986. Degradation of highly chlorinated biphenyls by Pseudomonas strain LB400. J. Ind. Microbiol. 1:23-29. [Google Scholar]
- 14.Brown, J. F., Jr. 1994. Determination of PCB metabolic, excretion, and accumulation rates for use as indicators of biological response and relative risk. Environ. Sci. Technol. 28:2295-2305. [DOI] [PubMed] [Google Scholar]
- 15.Brown, J. F., Jr. D. L. Bedard, M. J. Brennan, J. C. Carnahan, H. Feng, and R. E. Wagner. 1987. Polychlorinated biphenyl dechlorination in aquatic sediments. Science 236:709-711. [DOI] [PubMed] [Google Scholar]
- 16.Brown, J. F., Jr., R. E. Wagner, H. Feng, D. L. Bedard, M. J. Brennan, J. C. Carnahan, and R. J. May. 1987. Environmental dechlorination of PCBs. Environ. Toxicol. Chem. 6:579-593. [Google Scholar]
- 17.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 17a.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummings, D. E., O. L. Snoeyenbos-West, D. T. Newby, A. M. Niggemyer, D. R. Lovley, L. A. Achenbach, and R. F. Rosenzweig. 2003. Diversity of Geobacteraceae species inhabiting metal-polluted freshwater lake sediments ascertained by 16S rDNA analyses. Microb. Ecol. 46:257-269. [DOI] [PubMed] [Google Scholar]
- 19.Cutter, L. A., J. E. M. Watts, K. R. Sowers, and H. D. May. 2001. Identification of a microorganism that links its growth to the reductive dechlorination of 2,3,5,6-chlorobiphenyl. Environ. Microbiol. 3:699-709. [DOI] [PubMed] [Google Scholar]
- 20.De Wever, H., J. R. Cole, M. R. Fettig, D. A. Hogan, and J. M. Tiedje. 2000. Reductive dehalogenation of trichloroacetic acid by Trichlorobacter thiogenes gen. nov., sp. nov. Appl. Environ. Microbiol. 66:2297-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dollhopf, S. L., J.-H. Hyun, A. C. Smith, H. J. Adams, S. O'Brien, and J. E. Kostka. 2005. Quantification of ammonia-oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Appl. Environ. Microbiol. 71:240-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Fantroussi, S., J. Mahillon, H. Naveau, and S. N. Agathos. 1997. Introduction of anaerobic dechlorinating bacteria into soil slurry microcosms and nested-PCR monitoring. Appl. Environ. Microbiol. 63:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis, D. E., E. J. Lutz, J. M. Odom, R. J. Buchanan, C. L. Bartlett, M. D. Lee, M. R. Harkness, and K. A. DeWeerd. 2000. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ. Sci. Technol. 34:2254-2260. [Google Scholar]
- 25.Fennell, D. E., I. Nijenhuis, S. F. Wilson, S. H. Zinder, and M. M. Häggblom. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38:2075-2081. [DOI] [PubMed] [Google Scholar]
- 26.Frame, G. M., R. E. Wagner, J. C. Carnahan, J. F. Brown, Jr., R. J. May, L. A. Smullen, and D. L. Bedard. 1996. Comprehensive, quantitative, congener-specific analyses of eight Aroclors and complete PCB congener assignments on DB-1 capillary GC columns. Chemosphere 33:603-623. [Google Scholar]
- 27.Garrity, G. M. 2001. Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, NY.
- 28.Hallin, P. F., and D. Ussery. 2004. CBS Genome Atlas Database: a dynamic storage for bioinformatic results and sequence data. Bioinformatics 20:3682-3686. [DOI] [PubMed] [Google Scholar]
- 29.He, J., Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti, and F. E. Löffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 30.He, J. Z., K. M. Ritalahti, K. L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 31.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holliger, C., C. Regeard, and G. Diekert. 2003. Dehalogenation by anaerobic bacteria, p. 115-157. In M. M. Häggblom and I. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Press, Dordrecht, The Netherlands.
- 33.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. von Wintzingerode, H. Görisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jayachandran, G., H. Görisch, and L. Adrian. 2003. Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 180:411-416. [DOI] [PubMed] [Google Scholar]
- 35.Klappenbach, J. A., P. R. Saxman, J. T. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krajmalnik-Brown, R., T. Hölscher, I. N. Thomson, F. M. Saunders, K. M. Ritalahti, and F. E. Löffler. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kube, M., A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian. 2005. Genome sequence of the chlorinated compound respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269-1273. [DOI] [PubMed] [Google Scholar]
- 38.Lendvay, J. M., F. E. Löffler, M. Dollhopf, M. R. Aiello, G. Daniels, B. Z. Fathepure, M. Gebhard, R. Heine, R. Helton, J. Shi, R. Krajmalnik-Brown, C. L. Major, M. J. Barcelona, E. Petrovskis, R. Hickey, J. M. Tiedje, and P. Adriaens. 2003. Bioreactive barriers: a comparison of bioaugmentation and biostimulation for chlorinated solvent remediation. Environ. Sci. Technol. 37:1422-1431. [Google Scholar]
- 39.Löffler, F. E., J. R. Cole, K. M. Ritalahti, and J. M. Tiedje. 2003. Diversity of dechlorinating bacteria, p. 53-87. In M. M. Häggblom and I. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Press, Dordrecht, The Netherlands.
- 40.Löffler, F. E., and E. A. Edwards. 2006. Harnessing microbial activities for environmental cleanup. Curr. Opin. Biotechnol. 17:274-284. [DOI] [PubMed] [Google Scholar]
- 41.Löffler, F. E., Q. Sun, J. Li, and J. M. Tiedje. 2000. 16S rRNA gene-based detection of tetrachloroethene-dechlorinating Desulfuromonas and Dehalococcoides species, Appl. Environ. Microbiol. 66:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magar, V. S., R. C. Brenner, G. W. Johnson, and J. F. Quensen III. 2005. Long-term recovery of PCB-contaminated sediments at the Lake Hartwell superfund site: PCB dechlorination. 2. Rates and extent. Environ. Sci. Technol. 39:3548-3554. [DOI] [PubMed] [Google Scholar]
- 43.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Major, D. W., M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Henrickson, M. G. Starr, J. A. Payne, and L. W. Buonamici. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106-5116. [DOI] [PubMed] [Google Scholar]
- 45.May, H. D., L. A. Cutter, G. S. Miller, C. E. Milliken, J. E. Watts, and K. R. Sowers. 2006. Stimulatory and inhibitory effects of organohalides on the dehalogenating activities of PCB-dechlorinating bacterium o-17. Environ. Sci. Technol. 40:5704-5709. [DOI] [PubMed] [Google Scholar]
- 46.Maymó-Gatell, X., Y. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 47.Methé, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. Van Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 48.Müller, J. A., B. M. Rosner, G. von Abendroth, G. Meshulam-Simon, P. L. McCarty, and A. M. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.National Research Council. 1979. Polychlorinated biphenyls. National Academy of Sciences, Washington, DC.
- 50.Pietari, J. M. H. 2002. Characterization of tetrachloroethene dechlorinating cultures and isolation of a novel tetrachloroethene to cis-1,2-dichloroethene halorespiring bacterium. Ph.D. thesis. University of Washington, Seattle.
- 51.Quensen, J. F., III, S. A. Boyd, and J. M. Tiedje. 1990. Dechlorination of four commercial polychlorinated biphenyl mixtures (Aroclors) by anaerobic microorganisms from sediments. Appl. Environ. Microbiol. 56:2360-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quensen, J. F., III, M. A. Mousa, S. A. Boyd, J. T. Sanderson, K. L. Froese, and J. P. Giesy. 1998. Reduction of aryl hydrocarbon receptor-mediated activity of polychlorinated biphenyl mixtures due to anaerobic microbial dechlorination. Environ. Toxicol. Chem. 17:806-813. [Google Scholar]
- 53.Ritalahti, K. M., B. K. Amos, Y. Sung, Q. Wu, S. S. Koenigsberg, and F. E. Löffler. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlötelburg, C., C. von Wintzingerode, R. Hauck, F. von Wintzingerode, W. Hegemann, and U. B. Göbel. 2002. Microbial structure of an anaerobic bioreactor population that continuously dechlorinates 1,2-dichloropropane. FEMS Microbiol. Ecol. 39:229-237. [DOI] [PubMed] [Google Scholar]
- 55.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methé, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. T. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 56.Snoeyenbos-West, O., C. G. Van Praagh, and D. R. Lovley. 2001. Trichlorobacter thiogenes should be renamed as a Geobacter species. Appl. Environ. Microbiol. 67:1020-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stockholm Convention on Persistent Organic Pollutants (POPs). 1971. http://www.pops.int/. (Accessed 10 December 2006.)
- 58.Sung, Y. 2005. Isolation and ecology of bacterial populations involved in reductive dechlorination of chlorinated solvents, p. 149, appendix G. Ph.D. thesis. Georgia Institute of Technology, Atlanta.
- 59.Sung, Y., K. E. Fletcher, K. M. Ritalahti, R. P. Apkarian, N. Ramos- Hernández, R. A. Sanford, N. M. Mesbah, and F. E. Löffler. 2006. Geobacter lovleyi sp. nov. strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl. Environ. Microbiol. 72:2775-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sung, Y., K. M. Ritalahti, R. P. Apkarian, and F. E. Löffler. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watts, J. E. M., S. K. Fagervold, H. D. May, and K. R. Sowers. 2005. A PCR-based specific assay reveals a population of bacteria within the Chloroflexi associated with the reductive dehalogenation of polychlorinated biphenyls. Microbiology 151:2039-2046. [DOI] [PubMed] [Google Scholar]
- 62.Wolff, M. S., J. Thornton, A. Fischbein, R. Lilis, and I. J. Selikoff. 1982. Disposition of polychlorinated biphenyl congeners in occupationally exposed persons. Toxicol. Appl. Pharmacol. 62:294-306. [DOI] [PubMed] [Google Scholar]
- 63.Wu, Q., D. L. Bedard, and J. Wiegel. 1999. 2,6-Dibromobiphenyl primes extensive dechlorination of Aroclor 1260 in contaminated sediment at 8-30°C by stimulating growth of PCB-dehalogenating microorganisms. Environ. Sci. Technol. 33:595-602. [Google Scholar]
- 64.Wu, Q., J. E. M. Watts, K. R. Sowers, and H. D. May. 2002. Identification of a bacterium that specifically catalyzes the reductive dechlorination of polychlorinated biphenyls with doubly flanked chlorines. Appl. Environ. Microbiol. 68:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]