Abstract
Saccharomyces boulardii, a yeast that was isolated from fruit in Indochina, has been used as a remedy for diarrhea since 1950 and is now a commercially available treatment throughout Europe, Africa, and South America. Though initially classified as a separate species of Saccharomyces, recent publications have shown that the genome of S. boulardii is so similar to Saccharomyces cerevisiae that the two should be classified as conspecific. This raises the question of the distinguishing molecular and phenotypic characteristics present in S. boulardii that make it perform more effectively as a probiotic organism compared to other strains of S. cerevisiae. This investigation reports some of these distinguishing characteristics including enhanced ability for pseudohyphal switching upon nitrogen limitation and increased resistance to acidic pH. However, these differences did not correlate with increased adherence to epithelial cells or transit through mouse gut. Pertinent characteristics of the S. boulardii genome such as trisomy of chromosome IX, altered copy number of a number of individual genes, and sporulation deficiency have been revealed by comparative genome hybridization using oligonucleotide-based microarrays coupled with a rigorous statistical analysis. The contributions of the different genomic and phenotypic features of S. boulardii to its probiotic nature are discussed.
Probiotics is traditionally used as a term to describe the use of live microorganisms as food supplements that benefit the host by improving the intestinal microbial balance (50). Probiotics are used as an alternative therapy for treatment of infectious gastroenteritis or in the prevention and cure of antibiotic-associated diarrhea (AAD), which is a frequent complication of antimicrobial treatment of hospitalized patients (7, 20, 37).
Lactic acid bacteria of the genera Lactobacillus and Bifidobacterium are the most common microorganisms used as probiotics. However, yeasts of the species Saccharomyces are used as well. Saccharomyces boulardii, in particular, has been very successful in prevention and treatment of AAD, being especially effective against Clostridium difficile, the cause of about a third of all AAD cases (7, 56). S. boulardii was isolated from fruit in Indochina, and because of its benefits in the treatment of diarrhea, it is now a commercially available alternative remedy in Europe, Africa, and South America (39, 45) for AAD, acute diarrhea in children, and traveler's diarrhea (22). Many in vitro and some in vivo studies have suggested that S. boulardii is able to prevent intestinal infections caused by the adherence or invasion of C. difficile, Escherichia coli, and Candida albicans to the epithelial layer of the gastrointestinal tract (4, 11, 12). The efficacy of S. boulardii reported in case studies and in vitro trials (11) has led to investigations of the mechanisms involved in the probiotic activity of this yeast. Several potential mechanisms of probiotic activity have been suggested, including secretion of proteases or inhibitory proteins, stimulation of immunoglobulin A, acquisition and elimination of secreted toxins, modification of the intestinal mucosa, and direct adherence to the epithelial layer causing competition for colonization sites (12, 20). However, the mechanisms identified so far do not reflect the full set of interactions of this yeast with the host. For a start, it is unclear whether the probiotic effect of S. boulardii is related to gut colonization. Administration of a single dose of lyophilized S. boulardii to gnotobiotic mice resulted in the shedding of yeast cells into the feces for more than 10 days after administration (49). However, common laboratory strains of Saccharomyces cerevisiae were rapidly excluded from the gut of normal mice in only a few days or hours, suggesting that the presence of a normal enteric microflora may prevent colonization (44).
While the probiotic properties of S. boulardii have been demonstrated over the past 50 years, there is also evidence that it may act as an opportunistic pathogen causing Saccharomyces fungemia (39). The cause of these cases has often been identified as infection by the yeast via inserted catheters (3, 8, 26, 38). However, these incidents have raised concerns regarding the advisability of administering S. boulardii to immunocompromised patients. As the potential mechanisms of probiotic action include interactions with the immune system as well as the direct stimulation of host tissue, the safety of such treatments for patients whose immune systems are incapacitated would appear in doubt. Therefore, to maximize the benefits of treatment with S. boulardii as well as to minimize the potential risks, the mechanisms of this yeast's probiotic action need to be identified, and any potential for virulence must be fully investigated.
S. boulardii was initially identified as a separate species of the hemiascomycete genus Saccharomyces (40). However, the rapid development of molecular phylogenetics in recent years has led to changes in the classification of many yeast species (17, 39, 57). Using comparative genomic hybridizations for whole-genome analysis, we concluded that S. cerevisiae and S. boulardii are members of the same species (17). However, we also demonstrated some distinctive features of the S. boulardii genome with respect to its Ty elements (yeast retrotransposons) and the copy number of genes in its subtelomeric regions (17). In this paper, we report a number of new findings concerning the genomic and phenotypic characteristics of S. boulardii that may have an indirect or, indeed, direct bearing on the probiotic effects of this yeast, and we identify potential benefits and risks that may be associated with them.
MATERIALS AND METHODS
Yeast strains, media, and plasmids.
The yeast strains used in this study are listed in Table 1. Standard nutrient media (yeast extract, peptone, and dextrose [YPD] medium; minimal synthetic dextrose [SD] or synthetic glycerol medium [SG]; presporulation or sporulation medium; or GAL indicator) were prepared as described previously (54). Synthetic low-ammonium dextrose (SLAD) medium was prepared as described by Gimeno et al. (21). Plasmids pAG26 (24) and pRS316-KanMX (N. Zhang, unpublished) were used in this investigation.
TABLE 1.
Yeast strains used in this investigation
| Strain | Genotype | Source or reference |
|---|---|---|
| S. cerevisiae BY3 | MATaura3-52 | 60 |
| S. cerevisiae FY23 | MATaura3-52 leu2Δ1 trp1Δ1 | 60 |
| S. cerevisiae FY73 | MATα ura3-52 his3Δ200 | 60 |
| S. cerevisiae FY1679 | MATa/α ura3-52/ura3-52 leu2Δ1/LEU2 trp1Δ1/TRP1 HIS3/his3Δ200 | 60 |
| S. cerevisiae Σ1278b | Wild type | S. G. Oliver |
| S. cerevisiae JO43 | MATα hisX | J. Hughes |
| S. cerevisiae BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ/leu2Δ lys2Δ/LYS2 met15Δ/MET15 ura3Δ/ura3Δ | Euroscarf |
| S. cerevisiae BY4743 ygr204w::kanMX4 | MATa/α his3Δ1/his3Δ1 leu2Δ/leu2Δ lys2Δ/LYS2 met15Δ/MET15 ura3Δ/ura3Δ ygr204w::kanMX4/ygr204w::kanMX4 | Euroscarf |
| S. cerevisiae BY4741 ygr204w::kanMX4 | MATahis3Δ1 leu2Δ met15Δ ura3Δ ygr204w::kanMX4 | Euroscarf |
| S. cerevisiae BY4742 ygr204w::kanMX4 | MATα his3Δ1 leu2Δ lys2Δ ura3Δ ygr204w::kanMX4 | Euroscarf |
| S. cerevisiae 243a | MATaura3Δ | L. Stateva |
| S. cerevisiae 243α | MATα ura3Δ | L. Stateva |
| S. boulardii UL | Wild type | P. Niederberger, Nestlé |
| S. boulardii UL#3509 | Wild type | P. Niederberger, Nestlé |
| S. boulardii UL[rho] | Petite mutant | This study |
| C. albicans CAI4 | Δura3::imm434/Δura3::imm434 | 19 |
Ethidium bromide mutagenesis to obtain mitochondrial mutants.
Cells from an overnight YPD culture were incubated in minimal SD medium with ethidium bromide at 10 μg/ml overnight and subsequently spread onto minimal agar until single colonies developed, usually after 2 to 3 days. Colonies were replica plated on minimal SD and synthetic glycerol agar, and those able to utilize only glucose but not glycerol were retained as respiratory-deficient petite ([rho]) mutants.
Mating and sporulation analysis.
Parental strains grown overnight on YPD agar were crossed by replica plating onto the same medium and incubated for another 24 h. Hybrids were isolated on appropriate selective minimal SD medium. To induce sporulation, the hybrids were first grown in presporulation medium for 2 days at 30°C, and then an aliquot of the biomass was transferred to sporulation agar and incubated at room temperature (∼25°C) for up to 2 weeks. For tetrad dissection, a small amount of the sporulated biomass was suspended in 1 M sorbitol containing 1 mg/ml zymolyase and incubated at 30°C for 20 min to digest the ascus walls. Four-spored asci were dissected with a Micromanipulator (Singer Instruments, United Kingdom), and the individual spores were incubated at 30°C until growth was observed.
Induction of pseudohyphal growth.
Overnight cultures, grown in minimal SD medium at 30°C, were harvested, and the cells were plated on SLAD agar. Plates were incubated at 30°C and photographed after 24 h, 48 h, and 1 week.
CGH.
Genomic DNA from S. boulardii UL and S. cerevisiae BY4743 was prepared and used for comparative genomic hybridization (CGH) against the Affymetrix GeneChips as described previously (61). The data were normalized using RMAExpress (see below) and then analyzed with Genespring as described previously (17).
Statistical analysis of Affymetrix GeneChip hybridization data.
Spatial biases due to arrangement of probe sets on Affymetrix GeneChips are known to affect intensities (47). The probe set spatial arrangement is correlated with chromosomal organization, and so any spatial bias present may be confounded with the potential changes we are attempting to detect. Affymetrix chip probes are either perfect match (PM) or mismatch (MM), with the spatial position of a particular probe being given by x and y coordinates. Probes corresponding to neighboring open reading frames (ORFs) on a chromosome are predominantly neighbors on the chip with respect to their x coordinates. We noted, in agreement with Qian et al. (47), that this tends to introduce periodic short-ranged intensity correlations along each chromosome. This was considered to be an artifact, and so for each hybridization the median logarithm of the PM intensity value for each column (defined by a common x coordinate) was calculated. The median of these column medians was evaluated to obtain a chip-wide median. Columns of logarithms of PM values were then centered to this chip-wide median. A similar column-based median centering was performed on the logarithms of the MM intensity values. The exponential of the median-centered logarithms of the PM and MM values were calculated to construct a within-chip normalized CEL file. Between-chip normalization was performed using RMAExpress (6) on the modified CEL files. The subset of probe sets which could be mapped to an identified annotated ORF was selected for further analysis.
For each ORF, average RMA (robust multiarray average)-normalized logarithms of intensities were calculated for S. boulardii and S. cerevisiae BY4743 strains by taking an average of the three corresponding replicate normalized hybridizations. Residuals about the average values for S. boulardii and S. cerevisiae BY4743 were then calculated. The logarithm of the copy number ratio was estimated by taking the difference in the average logarithms of the intensities for S. boulardii and S. cerevisiae BY4743. The median value, over all ORFs analyzed, of the logarithm of the copy number ratio was centered to zero. Copy number for S. boulardii was calculated by evaluating the exponential of the logarithm of the copy number ratio and taking a copy number of 2 for each S. cerevisiae BY4743 ORF. Bootstrap confidence intervals (CIs) of 95% were calculated, under a homoscedastic assumption, by resampling suitably scaled residuals (31) to produce 100,000 bootstrap data sets. Marginal CIs are reported rather than multiply adjusted ones, since the 17 genes listed in Table 2 were not selected solely on the basis of estimated copy number. To aid detection of substantial regions of copy number gain or loss, a local weighted average copy number was also obtained for each ORF, thereby providing a smoother picture of copy number variation across each chromosome. The weighted average was calculated using a tri-cube window, taking the sequential ORF number within the chromosome as the ordinal coordinate and a window width of 10 ORFs. To test chromosomes for statistically significant enrichment of high-copy-number ORFs, type I error rates (P values) for median copy number within each chromosome were calculated by random permutation of ORF labels to produce 100,000 permuted data sets. We have not corrected for multiple testing, but even with stringent Bonferroni corrections, unadjusted P values that are less than 10−5 will be significant.
TABLE 2.
Copy number values of individual genes predicted by CGH using the Affymetrix GeneChip oligonucleotide microarrays
| Gene name | Chra | Copy number (95% CI)
|
||
|---|---|---|---|---|
| Average | Lower limit | Upper limit | ||
| ADE1 (YAR015W) | I | 1.83 | 1.56 | 2.14 |
| ADE8 (YDR408C) | IV | 2.02 | 1.72 | 2.36 |
| HIS5 (YIL116W) | IX | 2.91 | 2.48 | 3.40 |
| CUP1 (YHR053C) | VIII | 0.95 | 0.81 | 1.11 |
| CDC16 (YKL022C) | XI | 1.21 | 1.04 | 1.42 |
| DMC1 (YER179W) | V | 1.47 | 1.31 | 1.63 |
| STE11 (YLR362W) | XII | 1.51 | 1.29 | 1.77 |
| SKM1 (YOL113W) | XV | 1.06 | 0.90 | 1.23 |
| RAS1 (YOR101W) | XV | 1.01 | 0.87 | 1.18 |
| CDC24 (YAL041W) | I | 2.44 | 2.09 | 2.85 |
| CDC42 (YLR229C) | XII | 2.43 | 2.07 | 2.84 |
| CDC25 (YLR310C) | XII | 2.40 | 2.05 | 2.80 |
| DFG16 (YOR030W) | XV | 2.50 | 2.14 | 2.93 |
| RGS2 (YOR107W) | XV | 2.38 | 2.03 | 2.78 |
| HSP26 (YBR072W) | II | 2.47 | 2.11 | 2.90 |
| RMD6 (YEL072W) | V | 3.10 | 2.65 | 3.62 |
| CYR1 (YJL005W) | X | 2.62 | 2.24 | 3.06 |
Chr, chromosome.
Yeast-to-epithelia adherence analysis.
Assessment of yeast's adherence to human buccal epithelial cells was performed as described by Douglas et al. (16). Briefly, yeast cells grown in YPD medium at 30°C were washed in Dulbecco's phosphate-buffered saline (PBS) (Gibco, United Kingdom), counted with a hemocytometer, and resuspended at a concentration of 1 × 108 cells/ml. Buccal epithelial cells obtained by swabs of the inner cheek were also washed with PBS and resuspended at a concentration of 1 × 106 cells/ml. Mixtures of 100 μl each of yeast and epithelial cell suspensions were incubated for 1 h at 37°C and then vacuum filtered onto a polycarbonate filter with 12-μm pores (Whatman) and washed with PBS. The filters were air dried, fixed with methanol, and stained with Gram stain.
Alternatively, yeast suspensions, prepared as described above, were mixed with a suspension of Caco2 cells. The Caco2 cells grown in Dulbecco's modified Eagle medium (DMEM; Gibco, United Kingdom) were removed from the flasks by trypsinization at 37°C with 0.25% (vol/vol) trypsin and 0.2% EDTA (wt/vol) in Hank's balanced salt solution, washed twice with DMEM, and resuspended in DMEM at a concentration of 1 × 106 cells/ml. The mixture of yeast and Caco2 cells was treated as described above.
Animal work.
CF-1 male mice, approximately 10 weeks old, were individually housed under specific-pathogen-free conditions. Food was withdrawn 18 h prior to administration of the yeast, although mice continued to have free access to water throughout. The experimental animals were administered approximately 108 CFU of the appropriate yeast cell suspension by gastric gavage in a total volume of 200 μl before being returned to food. Feces collected from the bottom of the cage at regular intervals were suspended in PBS (100 mg/ml) for 1 h to soften before being suspended in PBS, and after appropriate dilution the suspensions were spread onto YPD plates containing strain-relevant antibiotic supplements plus 200 μg/ml ampicillin to inhibit the growth of natural gut flora. To check for colonization in the gut, the animals were euthanized by cervical dislocation, and the gastrointestinal tract was removed and divided into stomach, jejunum, ileum, cecum, and colon. PBS was flushed through each section, followed by scraping with a glass slide to isolate the mucosa. Mucosal isolates were briefly homogenized, and aliquots of both the flushings and the mucosal isolates were spread onto YPD agar plus appropriate antibiotic to determine CFU of any associated yeast cells. All animal procedures were undertaken under Home Office approval and supervision.
For the in vivo studies, S. boulardii and S. cerevisiae BY3 were transformed with plasmid pAG26 conferring resistance to hygromycin B, while S. cerevisiae Σ1278b was transformed with pRS316-KanMX conferring resistance to G418. Strains were maintained on YPD agar containing either 400 μg/ml hygromycin B or 600 μg/ml G418, as appropriate. For oral administration to mice, the three plasmid-containing strains were grown for 24 h in YPD medium containing the appropriate antibiotic. The cells were washed twice in sterile water and once in PBS before being suspended in PBS to a density of 108 CFU/200 μl and stored on ice until required.
Acid sensitivity.
Yeast grown overnight in YPD medium were suspended in PBS adjusted to different pH values (pH 6, pH 3, pH 2, pH 1, or pH 7), and following 1 h incubation at 37°C, appropriate dilutions of the cell suspensions were spread on YPD agar to measure survival.
Microarray data accession number.
The S. boulardii UL and S. cerevisiae BY4743 microarray data have been deposited in the Array Express database (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MAXD-15.
RESULTS
CGH analysis reveals significant genomic differences between S. boulardii and the sequenced strain of S. cerevisiae.
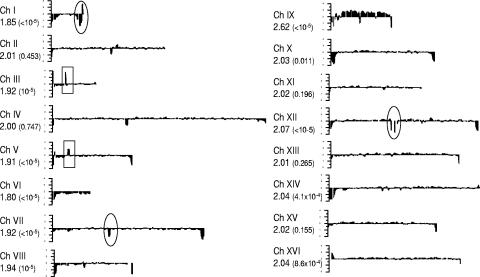
Recently, we reported that S. boulardii is a strain of S. cerevisiae that has lost most of its Ty1/2 elements (17). In the current study, we carried out a more detailed investigation of the S. boulardii genome employing CGH analysis using oligonucleotide microarrays (Affymetrix GeneChips) rather than the glass slide arrays with full-length PCR products for each ORF that we had employed previously. While the PCR product arrays are most suitable for making between-species comparisons (17), the higher-resolution short-oligonucleotide arrays are more useful for within-species (interstrain) studies (61). When the S. boulardii data were visualized using the Genespring software (data not shown), it was observed that there were significantly higher levels of hybridization to the sets of oligonucleotides corresponding to chromosome IX, compared to those representing the other 15 chromosomes. This preliminary observation was further supported by a rigorous statistical analysis, illustrated in Fig. 1, which plots the local average gene copy number for each ORF, following normalization and smoothing as described in Materials and Methods. As chromosome IX appeared to be present in a higher copy number in S. boulardii, the median log-ratio for each chromosome was determined relative to S. cerevisiae BY4743, a balanced diploid strain with two copies of each chromosome. As seen in Fig. 1, the copy number of each chromosome except chromosome IX was approximately 2, supporting the idea that S. boulardii is an aneuploid, containing three copies of chromosome IX. This interpretation is strengthened further by the permutation test which gave a P value of less than 10−5 for chromosome IX.
FIG. 1.
Chromosome plot of copy number distribution over the S. boulardii genome. Using the window-averaged copy number for each ORF, the results have been plotted using the starting nucleotide position of the ORF. The x axis represents the length of each chromosome (Ch) and the y axis shows copy number. The circled areas indicate the three most prominent internal regions of low copy number on chromosomes I, VII, and XII. The rectangles indicate the ORFs deleted in S. cerevisiae BY4743 causing the appearance of overrepresented genes. The value for each chromosome is the copy number as revealed by CGH. The P values shown in brackets are estimates of the probability of obtaining a median chromosomal copy number of the magnitude observed, by chance, if ORF copy number values were uniformly distributed across the genome.
Further sources of genome differences were revealed from the internal regions of lower copy number in the respective chromosomes of S. boulardii shown by the circles in Fig. 1. The three regions contain, respectively, YAR031W, YAR033W, YAR047C, YAR050W, YAR053W, YAR060W, and YAR061W on chromosome I; YGL052W and YGL051W on chromosome VII; and YLR155C and YLR156W on chromosome XII. Interestingly, YGL051W (chromosome VII), YAR031W, and YAR033W (chromosome I) are members of a recently described gene family (46), which encodes nonessential membrane-specific proteins specific to Saccharomyces sensu stricto species. These proteins have been shown to act as part of complexes: Mst27p (encoded by YGL051W) acts with Mst28p (encoded by YAR033W), and Prm9p (encoded by YAR031W) acts with a protein encoded by YGL053W (51). Furthermore, YAR050W (chromosome I) encodes a lectin-like protein involved in flocculation (34). YLR155C (ASP3) encodes a nitrogen catabolite-regulated cell wall l-asparaginase II (32). There are four copies of ASP3 in the S288C genome, but as our study suggests, the number could be lower in the genome of S. boulardii. The rest of the genes in this group, with the exception of YAR061W (a pseudogene), encode unknown, hypothetical proteins. In addition, these results support our previous findings (17) that there is more variation within the subtelomeric region of each chromosome (Fig. 1).
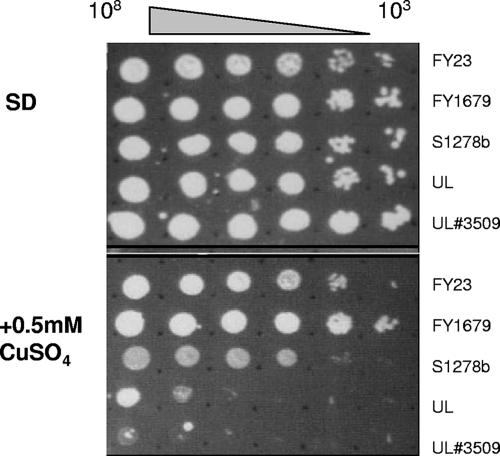
Another intriguing example of gene copy number differences was revealed in the case of CUP1—a gene known to be variably amplified in strains of S. cerevisiae from different origins (59) and present in two copies in the S288C strain. In S. boulardii, a lower copy number of CUP1 (YHR053C) was revealed by the present analysis, which predicted an average copy number of 0.95, a figure which is two times lower than the average for chromosome VIII (Fig. 1 and Table 2). This difference manifested itself in the form of the higher sensitivity of S. boulardii to copper ions at 0.5 mM than the standard laboratory S. cerevisiae strains (FY23, FY1679, and Σ1278b) that we tested (Fig. 2).
FIG. 2.
Copper sensitivity. Cells were plated out at the concentrations indicated onto minimal SD medium plates containing all required amino acids with or without CuSO4. The plates were then incubated at 30°C for 3 days prior to examination. The experiment was performed in duplicate, and the images are a representation of the complete results.
The two higher-copy-number regions on chromosomes III and V, highlighted by the rectangles in Fig. 1, correspond to LEU2 and URA3, respectively. As both copies of these genes are deleted in the S. cerevisiae diploid reference strain BY4743, their values appear about twofold higher in S. boulardii, a result which validates the reliability of this method for copy number determination.
S. boulardii produces fertile hybrids when crossed with other haploid and diploid strains of S. cerevisiae.
Previously we reported that S. boulardii contains copies of both mating type loci, MATa and MATα (17). As shown in the current study, it is also trisomic for chromosome IX and contains different copy numbers for a set of genes (Table 2) that could have phenotypic consequences, as demonstrated by the enhanced sensitivity to copper. Taken together, these results could help explain previous reports that have suggested that S. boulardii is unable to sporulate (39, 40). While our own experiments confirmed the sporulation deficiency of S. boulardii, some changes in cell morphology were observed (data not shown) after prolonged incubation on sporulation medium. Cells appeared enlarged and highly granular, suggestive of initiation of the sporulation process coupled with an inability to complete meiosis.
Possible genomic reasons for the sporulation deficiency of S. boulardii were revealed among the set of genes with values significantly lower than the average of the respective chromosome. Most noteworthy was CDC16 (YKL022C), identified as having a significantly lower value than the average of chromosome XI (Fig. 1 and Table 2). It encodes a subunit of the anaphase-promoting complex, which is required for sporulation (62), and has been shown to interact with Cdc23p and Cdc27p, both subunits of the same complex. Moreover, DMC1 (YER179W), encoding a meiosis-specific protein required for repair of double-strand breaks and pairing between homologous chromosomes, was also characterized by a value lower than that of chromosome V.
The observations described above suggested that the sporulation deficiency of S. boulardii, if caused by a lower copy number or, indeed, loss-of-function mutations (we cannot at present distinguish between these two possibilities) in one or more of the potential candidates, should be complemented in the presence of the corresponding functional wild-type allele(s). In order to test this hypothesis, we took advantage of the rare mating that occurs at low but measurable frequency between S. cerevisiae heterozygous MATa/MATα diploids and haploids of either mating type. As there were no auxotrophic strains of S. boulardii to use in the selection of hybrids, a mitochondrial petite mutant derivative was generated by ethidium-bromide mutagenesis. Such mutants were readily obtained in the current study although other workers have reported that their attempts to isolate petite mutants had failed (39). The S. boulardii mitochondrial petite mutant (UL[rho]) was crossed with known mating tester strains of S. cerevisiae. Crosses were also performed with wild-type S. boulardii strains, taking advantage of their elevated sensitivity to geneticin (data not shown) for selection purposes. Putative hybrids were selected on minimal medium containing geneticin, in the latter case, or on minimal medium containing glycerol, in the former. In contrast to the sporulation-deficient S. boulardii strains, successful sporulation occurred with all hybrids, and tetrad dissection showed that spore viability was significantly higher than 5% (Table 3), a figure that has been suggested to represent the borderline value that distinguishes inter- and intraspecific hybrids (13). Taken together, these data suggest that the sporulation deficiency of S. boulardii strains is most likely a result of mutant and/or lower-copy-number sporulation gene(s) since it could be successfully complemented by hybridization to several sporulation-competent S. cerevisiae strains. The experiment also provides further independent confirmation that S. boulardii is indeed a strain of S. cerevisiae, regardless of a number of genotypic differences.
TABLE 3.
Viability of spores isolated from hybrids between S. boulardii and S. cerevisiae strains
| Mating strains | No. of spores plated | No. of viable spores | % Viability |
|---|---|---|---|
| S. cerevisiae strain pairs | |||
| FY23 × JO43α | 144 | 5 | 3.5 |
| FY23 × BY4742 ygr204wα | 28 | 5 | 17.9 |
| FY23 × BY4743 ygr204wa/α | 52 | 9 | 17.3 |
| FY1679 × BY4743 ygr204wa/α | 28 | 26 | 92.9 |
| S. boulardii strain × S. cerevisiae strain pairs | |||
| UL[rho] × JO43α | 192 | 140 | 72.9 |
| UL[rho] × BY4741 ygr204wa | 28 | 2 | 7.1 |
| UL[rho] × BY4742 ygr204wα | 52 | 7 | 13.5 |
| UL[rho] × 243a | 48 | 4 | 8.3 |
| UL[rho] × 243α | 56 | 4 | 7.1 |
| UL[rho] × FY73α | 56 | 4 | 7.1 |
| UL × BY4741 ygr204wa | 40 | 5 | 12.5 |
| UL × BY4742 ygr204wα | 52 | 5 | 9.6 |
| UL × BY4743 ygr204wa/α | 64 | 56 | 87.5 |
S. boulardii has an enhanced ability for pseudohyphal switching in response to nitrogen limitation.
The sets of ORFs with predicted copy number values that were significantly different than the average of their respective chromosomes revealed several that affect pseudohyphal growth (Table 2 and Fig. 1). They include CDC24 (encoding the guanine nucleotide exchange factor for Cdc42), CDC42 (encoding small rho-like GTPase), DFG16 (encoding a membrane protein involved in pseudohyphal growth), RGS2 (negative regulator of cyclic AMP [cAMP] signaling), CYR1 (encoding adenyate cyclase), and CDC25 (encoding the guanine nucleotide exchange factor for Ras2p and Ras1p), whose values were higher. In contrast, STE11 (encoding the MEK kinase involved in pheromone and pseudohyphal/invasive growth mitogen-activated protein kinase signal transduction pathways), SKM1 (encoding a member of the PAK family of Ser/Thr protein kinases), and RAS1 (GTPase involved in the cAMP pathway) had lower values than those of their respective chromosomes.
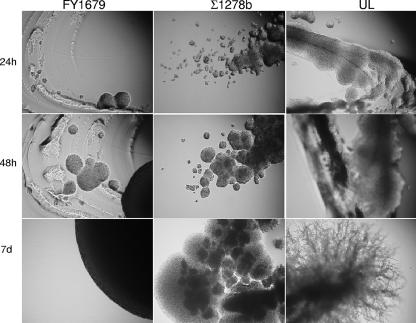
The predicted genomic differences in S. boulardii suggested most likely changes of the cAMP pathway. Previously, this pathway has been shown to be hyperactivated in the genetic background of S. cerevisiae Σ1278b (55), resulting in enhanced pseudohyphal development. In view of this, it was decided to test the ability of S. boulardii UL to form pseudohyphae in comparison to that of S. cerevisiae strains FY1679 (filamentation negative) and Σ1278b (filamentation positive) on SLAD agar at 30°C. The results (Fig. 3) demonstrated not only that S. boulardii has the ability to make this morphological switch but also that its response was both more rapid and more extensive than that of S. cerevisiae Σ1278b.
FIG. 3.
Pseudohyphal switching upon nitrogen limitation. Shown are the photographs of the SLAD agar plates taken at a magnification of ×10 following inoculation by spreading with a loop and incubation at 30°C for up to 1 week. After 7 days (d), the edges of the colonies were compared.
S. boulardii is unable to adhere to human epithelial cells in vitro and does not colonize the mouse gut in vivo.
S. boulardii has been reported to be rapidly eliminated from the gastrointestinal (GI) tract (33) in the presence of a normal enteric flora. This would suggest that it is unlikely to adhere to the gut epithelial cells, but to our knowledge this has not been determined directly. We therefore tested this using first the method described by Douglas et al. (16). The strain of C. albicans used as a positive control adhered successfully to the epithelial cells; however, none of the S. cerevisiae or S. boulardii strains displayed this ability (data not shown). Similar studies were performed in the polarized colonic cell line which has been widely used as a model of the intestinal epithelium, particularly with regard to its interaction with commensal and pathogenic microorganisms (9). However, although there appeared to be some binding of the yeast cells to Caco2 cells, this was not directly to the epithelia but to some extracellular factor, possibly mucus secreted by the cultures (data not shown).
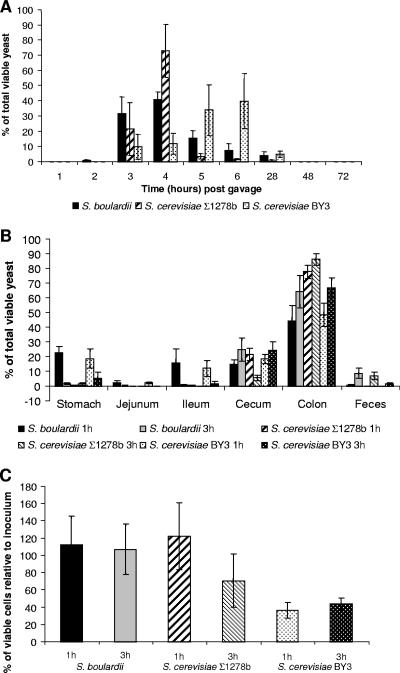
Differences in the intestinal handling of the yeast strains were assessed by analyzing their transit time through the mouse gut. In order to differentiate the test yeast strains from the resident gut microflora, S. boulardii and S. cerevisiae strains were transformed with appropriate plasmids, the presence of which conferred resistance to antibiotics (see Materials and Methods). As shown in Fig. 4A, the three yeast strains were all excreted within the first 28 h, with the bulk of the cells excreted between 3 and 6 h after gavage. Qualitative differences in excretion between the strains were observed with some evidence that S. boulardii and S. cerevisiae Σ1278b were excreted more rapidly than S. cerevisiae BY3 although this was not statistically significant. No viable yeast cells were found in the feces 48 h postgavage for any of the strains. There was also no evidence that these strains were able to colonize the gut after a single administration as judged by the low levels of viable yeast cells found in gut flushings (<25,000 CFU/flushing) or mucosal scrapings (<200 CFU/scraping) 72 h after administration.
FIG. 4.
Survival of yeast strains in the mouse gut. The different S. cerevisiae strains were distinguished from each other by plasmids conferring distinct antibiotic resistance phenotypes. CF-1 male mice were administered 108 CFU of the appropriate yeast suspension by gastric gavage in 200 μl. (A) The percentage of yeast recovered in the feces at different times (1 to 72 h) after administration; n = 6 mice for each time point. (B) Percentage of yeast recovered from different gut compartments, 1 h or 3 h after administration of different yeast strains. Animals were euthanized by cervical dislocation, and the GI tract was removed and divided into the gut components as indicated. Aliquots of the mucosal homogenates and the flushing and fecal homogenates were spread onto YPD medium with appropriate antibiotics to determine the number of CFU of the yeast strains. For each strain and time point, n = 6 mice. (C) Percentage of viable cells recovered from the mice following inoculation and incubation for 1 or 3 h. An increase indicated continued cell growth and division following inoculation.
To investigate the transit of the yeast cells through the gut in more detail, mice were sacrificed at 1 h and 3 h postgavage (n = 6 for each strain at each time point), and the contents from five sequential sections of the gut (stomach, jejunum, ileum, secum, and colon) were cultured to build a profile of passage of each strain. Feces collected at the same time intervals after yeast administration were also analyzed. Typically, the majority of viable yeast cells were present in the cecum and colon after only 1 h postgavage (Fig. 4B). By 3 h, almost no yeast cells were present in the stomach or small intestine, and the first cells were being excreted in the feces. For both experiments, there was a considerable variation between individual mice in both the percentages of cells surviving and growing from the original inoculum (Fig. 4C) and in the apparent rates at which the cells traversed the gut. Similar variation has been also observed in analogous experiments in humans (33). This is likely to be due to a number of factors, ranging from experimental technique (variation in the volumes successfully introduced by gavage and proportion of fecal pellets successfully retrieved) to individual variation in the mice's physiology and feeding habits after the gavage procedure. This meant that no statistically significant difference was found between the survival of the different strains in either the feces or the gut flushings (Student's t test, P < 0.05 for all pair-wise comparisons of strains in each condition) or the rate at which the different strains traversed the gastric tract.
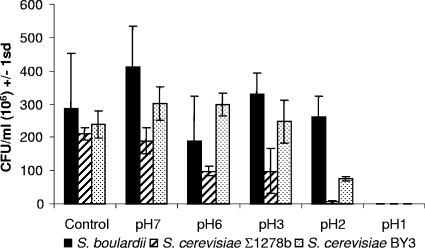
S. boulardii survives significantly better at low pH than other strains of S. cerevisiae.
Our study demonstrated that in terms of adherence and survival in the gut, S. boulardii strain did not differ significantly from the commonly used laboratory strains. However, we observed differences in the extent of survival of Saccharomyces yeast strains to the acidic conditions of the stomach when an acid sensitivity assay was performed. In comparison to the controls, both S. boulardii and S. cerevisiae BY3 displayed an increased number of CFU/ml after 1 h at pH 7 at 37°C, indicating that cells currently undergoing budding were able to complete their cycle and form 2 viable CFU (Fig. 5). As the pH decreased, the viability of S. boulardii remained similar to that of the control, down to pH 2. In contrast, the number of viable CFU for S. cerevisiae Σ1278b, decreased to less than half of the control at pH 6 and pH 3, and less than 4% of the control at pH 2. S. cerevisiae BY3 maintained similar or greater numbers of viable CFU relative to the control strain but less than one-third survived at pH 2. None of the yeast strains showed any survival at pH 1 (<300,000 CFU/ml plated).
FIG. 5.
Acid tolerance of yeast strains. Appropriate dilutions of the cell suspensions incubated under different pH conditions at 37°C were spread on YPD agar, and the plates were incubated until colonies appeared. Survival is expressed as the number of CFU/ml observed following incubation.
DISCUSSION
S. boulardii is a strain of S. cerevisiae with some specific genome features including chromosome IX trisomy and different copy numbers of some genes.
The probiotic yeast S. boulardii, classified originally as a separate species, has recently been shown to be a strain of S. cerevisiae (17, 39, 41, 42, 43), regardless of its strain-specific differences (12, 17, 22, 40). The present study extended the earlier findings, showing that S. boulardii appears to be trisomic for chromosome IX. S. cerevisiae now encompasses a wide range of strains, with various unique characteristics. These traits have generally developed in response to the environmental conditions to which each strain has adapted. The common laboratory strains have been extensively modified for ease of use and manipulation, but strains taken from their original sources and used for specific purposes vary greatly in ploidy as well as other traits. Wine and beer brewing strains, for example, are identified as strains of S. cerevisiae (10), even though their molecular karyotype, Ty distribution, and ploidy vary greatly. Aneuploidy, in particular, is rife among brewing and wine-making strains which traditionally have been selected for their desirable properties (2). A good example is the extra copy of chromosome XIII (25) found in strains proficient in “flor” formation used in the sherry-type wine industry. Variations in S. cerevisiae strains can also be traced to yeasts that have a different genetic background. The S. cerevisiae strain Σ1278b, whose genetic background is apparently characterized by an overactive Ras pathway (55), is used commonly as the model organism for pseudohyphal development in yeast, a trait not observed in the strain with the background of the standard (sequenced) strain, S288C, due to a mutation in the FLO8 gene (35). Recent work indicates that S. cerevisiae can control and maintain aneuploidy at the individual chromosome level (58). One potential reason for the maintenance of aneuploidy may be the selective advantage to the cell from the presence of extra copies of gene(s) contained within the duplicated chromosome (1, 52). In the pathogenic yeast C. albicans, aneuploidy has also been proposed to provide a selective advantage under certain environmental conditions: cells adapt to utilization of the secondary carbon source l-sorbose by reversible loss of chromosome V (29, 30). However, it remains to be seen if chromosome IX trisomy is characteristic of other strains of S. boulardii.
In addition to the overrepresentation of the entire chromosome IX, there is also a noticeable presence of genes related to protein synthesis (RPL31A, RPL41A, RPS24B, RPL2B, and RSA3) and stress responses (HSP26, SSA3, SED1, HSP42, HSP78, and PBS2) among the genes with higher values than the average copy number for the respective chromosomes. These genes' potentially elevated copy number could be contributing to the increased growth rate (data not shown), enhanced pseudohyphal switching (Fig. 3), and better survival at acid pH (Fig. 5) observed with S. boulardii. At the same time, our investigation suggests a lower copy number for some genes, such as CUP1, which was further supported by altered sensitivity to copper (Fig. 2) and members of the DUP240 multigene family (Fig. 1) and confirms the previously reported lower copy number of Ty 1/2 elements (data not shown). Copy number is not only a major source of variation between fungal species but it also, as shown recently, represents a significant mechanism of variation in humans (28, 53).
The success of S. boulardii as a probiotic might be due to its enhanced ability for pseudohyphal switching and better survival at acid pH.
The enhanced ability of S. boulardii to form pseudohyphae could be potentially linked to the mechanisms of its probiotic activity. The morphological response of S. boulardii appeared to be more extensive than that of Σ1278b upon exposure to low nitrogen, and this could in part be explained by genomic changes as revealed by CGH (Table 2). High survival at low pH (Fig. 5) is another physiologically important feature which would be expected to allow more organisms to survive passage through the stomach and therefore increase probiotic potential by increasing the number of yeast cells reaching the intestine. However, the data from the gut transit studies do not support this, with the different strains showing similar levels in the intestine.
S. boulardii has been found in the fecal matter of healthy human subjects several weeks after administration, suggesting that it was able to establish itself within the GI tract after treatment with antibiotics (33). The other yeasts that are known to be able to achieve this are opportunistic pathogens of the genus Candida. During colonization, many Candida species undergo a morphological transition to filamentous forms (true hyphae and pseudohyphae), which are better able to penetrate the lining of the GI tract (and, indeed, other tissues), and this ability to switch forms may be a significant virulence factor. S. cerevisiae is unable to form true hyphae. However, while strains related to the sequenced isolate of S. cerevisiae FY1679 (23), a derivative of S288c, are unable to form even pseudohyphae, strains of different genetic background (e.g., Σ1278b) are able to adopt a filamentous mode of growth upon exposure to either a poor nitrogen source (21, 55) or fusel alcohols (15). Our study demonstrates that strains of the S. boulardii genetic background also display enhanced pseudohyphal growth ability, most likely as a result of higher activity of the cAMP pathway.
The ability to switch readily between yeast and filamentous forms of growth combined with adherence to host tissues has long been considered a virulence trait in fungal pathogens such as C. albicans. However, S. boulardii has generally been shown to be beneficial to most hosts. A key issue in relation to its probiotic properties is the way in which S. boulardii is handled by the mammalian gut, particularly in regard to the rate at which it is excreted and also its ability to colonize. Although it was thought that S. boulardii would have the ability to adhere to epithelial cells and subsequently to sustain longer term colonization of the gut, the current study found no evidence that this occurs. The transit time of all the yeast strains tested in mouse were very short, with total clearance of the yeast burden within 28 h (6 to 28 h), an observation which supports previous reports (18, 33, 44, 48). In contrast, Rodrigues and coworkers showed that S. boulardii colonized the gut of gnotobiotic mice after a single oral dose of lyophilized S. boulardii cells, shedding yeast in the feces for more than 10 days after administration (49). This suggests that S. boulardii may be sensitive to competition from the resident microflora in the gut. The finding that repeated reinoculation of S. boulardii was necessary to maintain high intestinal populations over several days in conventional mice supports this view (18, 48). A similar effect was observed with human volunteers (33); single doses of S. boulardii were eliminated rapidly in the stools while repeated daily doses allowed the establishment of a steady-state concentration after about 3 days. In this study, ampicillin was also administered to the volunteers to disrupt the commensal flora of the gut, which significantly increased mean fecal concentrations of S. boulardii. Our own study demonstrated rapid elimination kinetics of single doses of different Saccharomyces strains, including S. boulardii, in the mouse. This is an important observation which should be considered if this animal model is to be used to evaluate probiotic and other applications of S. boulardii, including the recently suggested use of yeast as a novel delivery system for different pharmaceuticals (5).
The current study suggests genomic reasons for the sporulation deficiency phenotype of S. boulardii.
It had previously been reported that S. boulardii was unable to sporulate (39, 40). Our study confirmed this, and on the basis of comparative genomic hybridization analysis predicted several reasons for this phenotype. CDC16, DMC1 (Table 2), and MND2 (encoding a subunit of the anaphase-promoting complex) as well were characterized by values lower than those of their respective chromosome's average predicted copy number. This suggests either copy number difference or sequence divergence, and we cannot at present distinguish between the two possibilities. However, the successful complementation of the S. boulardii sporulation deficiency by hybridizing to sporulation-competent strains of S. cerevisiae (Table 3) suggests that the most likely reason for the lower values of CDC16, DMC1, and MND2 is sequence divergence. Cdc16p has been shown to interact with Cdc23p and Cdc27p to form a macromolecular complex, most probably via association through their tetratricopeptide repeats (36). It could be that the divergence affects the sequence of this repeat; however, further investigation is required to test this hypothesis. Interestingly down-regulation of the cAMP pathway by PDE2 overexpression has been shown to rescue the temperature sensitivity of the cdc16-1 mutant (27). As our study suggests that the cAMP pathway is activated in S. boulardii, this could be an additional contributing factor to the inability of strains of this genetic background to sporulate.
Genomic reasons for the sporulation deficiency of S. boulardii were also revealed among the set of genes with values significantly higher than the average of the respective chromosome. Genes in this category, which according to the current Gene Ontology nomenclature are involved in sporulation, included YSW1 (encoding a protein expressed specifically in spores), IME2 (encoding a Ser/Thr kinase involved in activation of meiosis), ADY4 (encoding a structural component of the meiotic outer plaque), SLK19 (encoding a kinetochore-associated protein required for normal chromosome segregation in meiosis), and SRC1 (encoding a protein with a putative role in sister chromatid segregation). All of them were characterized by copy number values significantly higher than the average for their respective chromosomes. These changes together with the observed chromosome IX aneuploidy could be another contributing factor for the sporulation deficiency of S. boulardii. In a recent study (14), three quantitative trait loci which control and determine the efficiency of sporulation in yeast were mapped. It is noteworthy that one of these, TAO3, is located on chromosome IX.
Our present investigation did not cover the whole genome as both the previously used glass slide microarrays and the currently used Affymetrix chips contain probes representative of only the annotated ORFs but not the intergenic regions. A more detailed investigation employing the recently developed tiling microarrays may reveal additional genome differences between yeast strains, which the current study has been unable to identify.
In conclusion, we have shown that S. boulardii strains differ significantly from laboratory strains of S. cerevisiae at both the genomic and physiological levels with regard to sporulation, individual chromosome and gene copy numbers, ability for pseudohyphal switching, and survival at low acid pH, the latter two features having a direct bearing on the probiotic nature of S. boulardii. On the other hand, our study found no significant differences in terms of the ability of S. boulardii strains to adhere to human epithelial cells and of transit time through the mouse gut. It remains to be seen how these differences affect the relationship of this yeast strain with the host.
Acknowledgments
We are grateful to Peter Warn for help and advice with the reverse transcription quantitative PCR experiments, A. Hayes for help with the microarray hybridizations, D. Delneri for help with the tetrad analysis, N. Zhang for plasmids, P. Niederberger (Nestlé) for S. boulardii strains, and the anonymous reviewers for helpful comments.
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC) (to L.S., S.G.O., and G.W.). L.E.-I. and I.C. are grateful recipients of BBRSC studentships.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Adams, J., S. Puskas-Rozsa, J. Simlar, and C. M. Wilke. 1992. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr. Genet. 22:13-19. [DOI] [PubMed] [Google Scholar]
- 2.Bakalinsky, A. T., and R. Snow. 1990. The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast 6:367-382. [DOI] [PubMed] [Google Scholar]
- 3.Bassetti, S., R. Frei, and W. Zimmerli. 1998. Fungemia with Saccharomyces cerevisiae after treatment with Saccharomyces boulardii. Am. J. Med. 105:71-72. [DOI] [PubMed] [Google Scholar]
- 4.Berg, R., P. Bernasconi, D. Fowler, and M. Gautreaux. 1993. Inhibition of Candida albicans translocation from the gastrointestinal tract of mice by oral administration of Saccharomyces boulardii. J. Infect. Dis. 168:1314-1318. [DOI] [PubMed] [Google Scholar]
- 5.Blanquet, S., S. Marol-Bonnin, E. Beyssac, D. Pompon, M. Renaud, and M. Alric. 2001. The “biodrug” concept: an innovative approach to therapy. Trends Biotechnol. 19:393-400. [DOI] [PubMed] [Google Scholar]
- 6.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 7.Brassart, D., and E. J. Schiffrin. 1997. The use of probiotics to reinforce mucosal defence mechanisms. Trends Food Sci. Technol. 8:321-326. [Google Scholar]
- 8.Cassone, M., P. Serra, F. Mondello, A. Girolamo, S. Scafetti, E. Pistella, and M. Venditti. 2003. Outbreak of Saccharomyces cerevisiae subtype boulardii fungemia in patients neighboring those treated with a probiotic preparation of the organism. J. Clin. Microbiol. 41:5340-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, E., C. Hoare, J. Tanianis-Hughes, G. L. Carlson, and G. Warhurst. 2005. Interferon gamma induces translocation of commensal Escherichia coli across gut epithelial cells via a lipid raft-mediated process. Gastroenterology 128:1258-1267. [DOI] [PubMed] [Google Scholar]
- 10.Codon, A. C., T. Benitez, and M. Korhola. 1998. Chromosomal polymorphism and adaptation to specific industrial environments of Saccharomyces strains. Appl. Microbiol. Biotechnol. 49:154-163. [DOI] [PubMed] [Google Scholar]
- 11.Czerucka, D., S. Dahan, B. Mograbi, B. Rossi, and P. Rampal. 2000. Saccharomyces boulardii preserves the barrier function and modulates the signal transduction pathway induced in enteropathogenic Escherichia coli-infected T84 cells. Infect. Immun. 68:5998-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czerucka, D., and P. Rampal. 2002. Experimental effects of Saccharomyces boulardii on diarrheal pathogens. Microbes Infect. 4:733-739. [DOI] [PubMed] [Google Scholar]
- 13.Delneri, D., I. Colson, S. Grammenoudi, I. N. Roberts, E. J. Louis, and S. G. Oliver. 2003. Engineering evolution to study speciation in yeasts. Nature 422:68-72. [DOI] [PubMed] [Google Scholar]
- 14.Deutschbauer, A. M., and R. W. Davis. 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37:1333-1340. [DOI] [PubMed] [Google Scholar]
- 15.Dickinson, J. R. 1996. “Fusel” alcohols induce hyphal-like extensions and pseudohyphal formation in yeasts. Microbiology 142:1391-1397. [DOI] [PubMed] [Google Scholar]
- 16.Douglas, L. J., J. G. Houston, and J. McCourtie. 1981. Adherence of Candida albicans to human buccal epithelial cells after growth on different carbon sources. FEMS Microbiol. Lett. 12:241-243. [Google Scholar]
- 17.Edwards-Ingram, L. C., M. E. Gent, D. C. Hoyle, A. Hayes, L. I. Stateva, and S. G. Oliver. 2004. Comparative genomic hybridization provides new insights into the molecular taxonomy of the Saccharomyces sensu stricto complex. Genome Res. 14:1043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filho-Lima, J. V., E. C. Vieira, and J. R. Nicoli. 2000. Antagonistic effect of Lactobacillus acidophilus, Saccharomyces boulardii and Escherichia coli combinations against experimental infections with Shigella flexneri and Salmonella enteritidis subsp. typhimurium in gnotobiotic mice. J. Appl. Microbiol. 88:365-370. [DOI] [PubMed] [Google Scholar]
- 19.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fooks, L. J., and G. R. Gibson. 2002. Probiotics as modulators of the gut flora. Br. J. Nutr. 88(Suppl. 1):S39-S49. [DOI] [PubMed] [Google Scholar]
- 21.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 22.Gismondo, M. R., L. Drago, and A. Lombardi. 1999. Review of probiotics available to modify gastrointestinal flora. Int. J. Antimicrob. Agents 12:287-292. [DOI] [PubMed] [Google Scholar]
- 23.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546, 563-567. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 25.Guijo, S., J. C. Mauricio, J. M. Salmon, and J. M. Ortega. 1997. Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and “flor” film ageing of dry sherry-type wines. Yeast 13:101-117. [DOI] [PubMed] [Google Scholar]
- 26.Hennequin, C., A. Thierry, G. F. Richard, G. Lecointre, H. V. Nguyen, C. Gaillardin, and B. Dujon. 2001. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heo, S. J., K. Tatebayashi, and H. Ikeda. 1999. The budding yeast cohesin gene SCC1/MCD1/RHC21 genetically interacts with PKA, CDK and APC. Curr. Genet. 36:329-338. [DOI] [PubMed] [Google Scholar]
- 28.Iafrate, A. J., L. Feuk, M. N. Rivera, M. L. Listewnik, P. K. Donahoe, Y. Qi, S. W. Scherer, and C. Lee. 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36:949-951. [DOI] [PubMed] [Google Scholar]
- 29.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines l-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95:5150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janbon, G., F. Sherman, and E. Rustchenko. 1999. Appearance and properties of L-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics 153:653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol 7:819-837. [DOI] [PubMed] [Google Scholar]
- 32.Kim, K. W., J. Q. Kamerud, D. M. Livingston, and R. J. Roon. 1988. Asparaginase II of Saccharomyces cerevisiae. Characterization of the ASP3 gene. J. Biol. Chem. 263:11948-11953. [PubMed] [Google Scholar]
- 33.Klein, S. M., G. W. Elmer, L. V. McFarland, C. M. Surawicz, and R. H. Levy. 1993. Recovery and elimination of the biotherapeutic agent, Saccharomyces boulardii, in healthy human volunteers. Pharm. Res. 10:1615-1619. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi, O., N. Hayashi, R. Kuroki, and H. Sone. 1998. Region of FLO1 proteins responsible for sugar recognition. J. Bacteriol. 180:6503-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kron, S. J. 1997. Filamentous growth in budding yeast. Trends Microbiol. 5:450-454. [DOI] [PubMed] [Google Scholar]
- 36.Lamb, J. R., W. A. Michaud, R. S. Sikorski, and P. A. Hieter. 1994. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 13:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, Y. K., and S. Salminen. 1995. The coming age of probiotics. Trends Food Sci. Technol. 6:241-245. [Google Scholar]
- 38.Lherm, T., C. Monet, B. Nougiere, M. Soulier, D. Larbi, C. Le Gall, D. Caen, and C. Malbrunot. 2002. Seven cases of fungemia with Saccharomyces boulardii in critically ill patients. Intensive Care Med. 28:797-801. [DOI] [PubMed] [Google Scholar]
- 39.McCullough, M. J., K. V. Clemons, J. H. McCusker, and D. A. Stevens. 1998. Species identification and virulence attributes of Saccharomyces boulardii (nom. inval.). J. Clin. Microbiol. 36:2613-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFarland, L. V. 1996. Saccharomyces boulardii is not Saccharomyces cerevisiae. Clin. Infect. Dis. 22:200-201. [DOI] [PubMed] [Google Scholar]
- 41.Mitterdorfer, G., W. Kneifel, and H. Viernstein. 2001. Utilization of prebiotic carbohydrates by yeasts of therapeutic relevance. Lett. Appl. Microbiol. 33:251-255. [DOI] [PubMed] [Google Scholar]
- 42.Mitterdorfer, G., H. K. Mayer, W. Kneifel, and H. Viernstein. 2002. Clustering of Saccharomyces boulardii strains within the species S. cerevisiae using molecular typing techniques. J. Appl. Microbiol. 93:521-530. [DOI] [PubMed] [Google Scholar]
- 43.Mitterdorfer, G., H. K. Mayer, W. Kneifel, and H. Viernstein. 2002. Protein fingerprinting of Saccharomyces isolates with therapeutic relevance using one- and two-dimensional electrophoresis. Proteomics 2:1532-1538. [DOI] [PubMed] [Google Scholar]
- 44.Pecquet, S., D. Guillaumin, C. Tancrede, and A. Andremont. 1991. Kinetics of Saccharomyces cerevisiae elimination from the intestines of human volunteers and effect of this yeast on resistance to microbial colonization in gnotobiotic mice. Appl. Environ Microbiol. 57:3049-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penna, F. J., L. A. Filho, A. C. Calcado, H. R. Junior, and J. R. Nicolli. 2000. Up-to-date clinical and experimental basis for the use of probiotics. J. Pediatr. (Rio J.) 76(Suppl. 1):S209—S217. [In Portuguese.] [DOI] [PubMed] [Google Scholar]
- 46.Poirey, R., L. Despons, V. Leh, M. J. Lafuente, S. Potier, J. L. Souciet, and J. C. Jauniaux. 2002. Functional analysis of the Saccharomyces cerevisiae DUP240 multigene family reveals membrane-associated proteins that are not essential for cell viability. Microbiology 148:2111-2123. [DOI] [PubMed] [Google Scholar]
- 47.Qian, J., Y. Kluger, H. Yu, and M. Gerstein. 2003. Identification and correction of spurious spatial correlations in microarray data. BioTechniques 35:42-44, 46, 48. [DOI] [PubMed] [Google Scholar]
- 48.Rodrigues, A. C., R. M. Nardi, E. A. Bambirra, E. C. Vieira, and J. R. Nicoli. 1996. Effect of Saccharomyces boulardii against experimental oral infection with Salmonella typhimurium and Shigella flexneri in conventional and gnotobiotic mice. J. Appl. Bacteriol. 81:251-256. [DOI] [PubMed] [Google Scholar]
- 49.Rodrigues, A. C., D. C. Cara, S. H. Fretez, F. Q. Cunha, E. C. Vieira, J. R. Nicoli, and L. Q. Vieira. 2000. Saccharomyces boulardii stimulates sIgA production and the phagocytic system of gnotobiotic mice. J. Appl. Microbiol. 89:404-414. [DOI] [PubMed] [Google Scholar]
- 50.Salminen, S., A. Ouwehand, Y. Benno, and Y. K. Lee. 1999. Probiotics: how should they be defined? Trends Food Sci. Technol. 10:107-110. [Google Scholar]
- 51.Sandmann, T., J. M. Herrmann, J. Dengjel, H. Schwarz, and A. Spang. 2003. Suppression of coatomer mutants by a new protein family with COPI and COPII binding motifs in Saccharomyces cerevisiae. Mol. Biol. Cell 14:3097-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schatz, P. J., F. Solomon, and D. Botstein. 1986. Genetically essential and nonessential alpha-tubulin genes specify functionally interchangeable proteins. Mol. Cell. Biol. 6:3722-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sebat, J., B. Lakshmi, J. Troge, J. Alexander, J. Young, P. Lundin, S. Maner, H. Massa, M. Walker, M. Chi, N. Navin, R. Lucito, J. Healy, J. Hicks, K. Ye, A. Reiner, T. C. Gilliam, B. Trask, N. Patterson, A. Zetterberg, and M. Wigler. 2004. Large-scale copy number polymorphism in the human genome. Science 305:525-528. [DOI] [PubMed] [Google Scholar]
- 54.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Stanhill, A., N. Schick, and D. Engelberg. 1999. The yeast Ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol. Cell. Biol. 19:7529-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Surawicz, C. M., L. V. McFarland, R. N. Greenberg, M. Rubin, R. Fekety, M. E. Mulligan, R. J. Garcia, S. Brandmarker, K. Bowen, D. Borjal, and G. W. Elmer. 2000. The search for a better treatment for recurrent Clostridium difficile disease: use of high-dose vancomycin combined with Saccharomyces boulardii. Clin. Infect. Dis. 31:1012-1017. [DOI] [PubMed] [Google Scholar]
- 57.Vaughan-Martini, A. 2003. Reflections on the classification of yeasts for different end-users in biotechnology, ecology, and medicine. Int. Microbiol. 6:175-182. [DOI] [PubMed] [Google Scholar]
- 58.Waghmare, S. K., and C. V. Bruschi. 2005. Differential chromosome control of ploidy in the yeast Saccharomyces cerevisiae. Yeast 22:625-639. [DOI] [PubMed] [Google Scholar]
- 59.Winge, D. R., K. B. Nielson, W. R. Gray, and D. H. Hamer. 1985. Yeast metallothionein. Sequence and metal-binding properties. J. Biol. Chem. 260:14464-14470. [PubMed] [Google Scholar]
- 60.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 61.Winzeler, E. A., C. I. Castillo-Davis, G. Oshiro, D. Liang, D. R. Richards, Y. Zhou, and D. L. Hartl. 2003. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163:79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039-2058. [DOI] [PubMed] [Google Scholar]