Abstract
The influence of the phytoplankton-specific organic compound glycolate on bacterial community structure was examined during the 2004 spring phytoplankton bloom (February to April) in Dabob Bay in Washington. The diversity of the bacteria able to utilize glycolate during the phytoplankton bloom was determined using previously developed PCR primers to amplify the gene for the D subunit of glycolate oxidase (glcD). Many of the glcD sequences obtained represented novel sequences that appeared to be specific to marine environments. Overall, the glcD sequence diversity decreased as the phytoplankton bloom progressed. Phylotype-specific glcD quantitative PCR primers were designed for the six most commonly detected glcD phylotypes that represented distinct phylogenetic groups of heterotrophic bacteria. Three patterns of phylotype abundance were detected: four phylotypes were most abundant during the onset of the bloom; the abundance of one phylotype increased as the bloom progressed; and one phylotype was abundant throughout the bloom. Quantitative reverse transcriptase PCR with the same phylotype-specific primers was used to determine the levels of day and night glcD RNA transcription over the course of the bloom. glcD transcripts, when detectable, were always more abundant in the day than at night for each phylotype, suggesting that the bacteria responded to the glycolate produced by phytoplankton during the day. The nearly constant low in situ glycolate concentrations suggested that bacteria rapidly utilized the available glycolate. This study provided evidence for direct phytoplankton-bacterium interactions and the resulting succession in a single functional group of marine bacteria.
About 50% of organic matter generated by phytoplankton is consumed and remineralized by bacteria (12, 39). Consequently, bacterial community structure in marine environments is affected by the composition of available organic matter (2). Shifts in bacterial community structure can be induced in microcosms by addition of high- or low-molecular-weight dissolved organic carbon (10) and protein (36). Such shifts likely arise because different bacterial lineages can specialize for different types of organic matter (8). Changes in bacterial community structure have been observed during diatom blooms (43), under diatom-versus-phytoflagellate regimens (37), and through enrichment with various alga-derived organic matter (22, 49), presumably due to the different types of dissolved organic matter associated with different phytoplankton species. Because much organic matter originates from phytoplankton, bacterial community structure may be related in large part to phytoplankton composition and the organic matter derived from different phytoplankton species under different environmental conditions.
Workers are starting to identify potential linkages between specific lineages of phytoplankton and bacteria using 16S rRNA gene diversity (22, 37, 43, 50). For example, the marine Roseobacter clade has been linked with utilization of dimethylsufoniopropionate (DMSP) (19, 29, 41). Subsequent studies using microautoradiography and fluorescence in situ hybridization confirmed that the Roseobacter clade is indeed responsible for metabolizing a significant proportion of DMSP (33, 50). In recent microcosm studies, DMSP was found to induce a change in the composition of the bacterial assemblage (38). Significant DMSP release in marine environments is facilitated by grazing and viral lysis rather than by active release by phytoplankton (30, 32, 44). Hence, the direct influence of phytoplankton and DMSP release on bacterial community dynamics can be confounded by these other processes. An ideal system for investigating direct phytoplankton influence on bacterial community structure would involve a phytoplankton-specific compound that is actively released by phytoplankton.
In our previous work (26), phytoplankton photorespiration and bacterial utilization of glycolate were identified as one such system for examining phytoplankton-bacterium interactions. Glycolate is a photorespiration-specific, labile compound that is rapidly utilized by marine heterotrophic bacteria, and it is apparently used primarily as an energy source (14, 53). Glycolate is present year-round and in many different nutrient regimens (9, 28), and at times it accounts for as much as 33% of the labile dissolved organic carbon pool (11, 28). Early culture studies using radioactive uptake showed that only a subset of marine bacteria could use glycolate (52, 53). Thus, phytoplankton-derived glycolate represents a reliable and potentially substantial source of energy available to only a subset of the marine bacterial community. Accordingly, its use may significantly influence bacterial community structure.
Through the development of PCR primers to amplify glcD (the gene encoding the D subunit of glycolate oxidase), we (26) recently discovered a genetic basis for the differential glycolate utilization observed in previous studies (52, 53). In both laboratory cultures and natural bacterial assemblages, only a subset of marine bacteria had the genetic potential for glycolate utilization (26). Nonetheless, the genetic potential for glycolate utilization was phylogenetically diverse and was present in marine bacterial communities from different nutrient regimens. We (26) also detected potential differences in glcD diversity both between and within the oligotrophic and eutrophic environments examined, suggesting that the glycolate-utilizing potential in bacterial communities may be structured by interactions with the phytoplankton community.
In this study, we examined shifts in diversity and succession in the glycolate-utilizing component of the bacterial community during a spring phytoplankton bloom in Dabob Bay in Washington. We determined diel glycolate concentrations and bacterial glcD diversity. To determine whether the glycolate utilizers might be responding to phytoplankton photorespiration, we also designed primers for the most common glcD phylotypes to use in quantitative PCR and determined the diel changes in glcD DNA and RNA copy numbers over the 7-week sampling period.
MATERIALS AND METHODS
Study site and sample collection.
Samples were collected on 24 and 25 February, 3 and 4 March, 10 and 11 March, 22 to 25 March, and 7 and 8 April 2004 from one site in Dabob Bay in Washington (47°46.14′N, 122°50.10′W) by researchers on board R/V Clifford A. Barnes. Dabob Bay was chosen to monitor bacterial populations during daily and seasonal cycles because it has minimal tidal exchange and estuarine circulation and low freshwater input from nearby rivers (3, 13).
Water samples were collected from the surface (1.5 to 3.5 m) at 2- to 8-h intervals over the course of 24 to 72 h. The sampling time was normalized to sunrise and sunset times based on calculated sunrise and sunset times using the NOAA Surface Radiation Research Branch Sunrise/Sunset Calculator (www.srrb.noaa.gov/highlights/sunrise/sunrise.html). In most cases chlorophyll a (Chl a) fluorescence was measured using Seabird instruments mounted on a Niskin bottle rosette; the only exception was the sample collected at noon on 25 February, when water was collected with a vacuum pump and no reading was obtained. Chl a fluorescence values were calibrated with Chl a concentrations (J. Pierson, personal communication). Seawater samples were prefiltered with 10-μm Nitex mesh into a dark bottle before subsampling. Seawater samples used for bacterial counting were fixed with formaldehyde (final concentration, 3.7%) and stored at 4°C prior to analysis. Seawater samples used for glycolate measurement were filtered through 0.2-μm HT Tuffryn membrane Acrodisc syringe filters (Pall Corp.) with rubber-free syringes (Norm-Ject) into muffled glass bottles (I-Chem) and stored at 4°C prior to analysis. For molecular analyses, seawater was filtered through 0.2-μm Isopore membrane filters (Millipore) for 20 min in 50-ml increments. The filters were immediately placed in liquid nitrogen and transferred to −80°C in the laboratory prior to extraction of DNA and RNA.
Total bacterial counts.
Bacterial abundance was determined by epifluorescence microscopy of samples stained with 4′,6′-diamidino-2-phenylindole (DAPI) and acridine orange (AO) (6). Samples were vortexed vigorously with 50 μl of 0.5% Triton X-100 prior to incubation with filtered (0.2 μm) AO (final concentration in artificial seawater [ASW], 0.01%) for 3 min. A sample volume that yielded 20 to 50 cells per field was filtered onto black polycarbonate 0.8-μm filters (Poretics) for counting. These filters were used rather than 0.2-μm filters due to incorrect labeling by the vendor that did not become apparent until well after all samples were filtered and counted. For the purposes of this study, the counts obtained were used as estimates of the total bacterial cell counts because they likely underestimated the total bacterial abundance. One milliliter of filtered (0.2 μm) DAPI (20 μg/ml in ASW) was added to each filter and incubated for 7 min, and then the filter was rinsed with filtered (0.2 μm) ASW. Twenty random fields and at least 200 cells were counted for each filter using the DAPI filter set, and staining ambiguity was resolved using the AO filter set.
DNA extraction, clone libraries, and DNA sequencing.
DNA was extracted for a time 2 to 3 h after sunset for each cruise using the gram-positive protocol of an DNeasy tissue kit (QIAGEN) and was quantified using a PicoGreen double-stranded DNA quantitation kit (Molecular Probes, Inc.). For clone library construction and sequencing of glcD we used the protocol for field samples described previously (26), with the following modifications. The PCR mixtures contained 2.0 mM MgCl2, and the PCRs consisted of 26 to 28 cycles of amplification. In addition, PCR products from 10 20-μl reaction mixtures were pooled and then purified and concentrated (19:3) with Quantum Prep PCR Kleen spin columns (Bio-Rad) and a QIAquick PCR purification kit (QIAGEN). The DNA template used for cycle sequencing of clones was generated with a Templiphi amplification kit (Amersham Biosciences Corp). Contigs were formed using Sequencher (GeneCodes). The glcD DNA sequences, amplified from both ends, were 895 to 918 bp long.
Phylogenetic analysis.
An operational taxonomic unit (OTU) was defined by DNA sequences that were ≥97% identical to one another. An amino acid sequence was inferred from a representative DNA sequence for each OTU. The two most closely related amino acid sequences identified for each OTU with the BLASTX program (http://www.ncbi.nih.gov/BLAST/) were retrieved from the GenBank database. GlcD sequences representative of a broad range of phylogenetic groups were also retrieved from the GenBank database and the Integrated Microbial Genome (IMG) database (http://img.jgi.doe.gov/pub/main.cgi) for phylogenetic tree construction. The only GlcD sequence that has been verified experimentally to date is the sequence of Escherichia coli; therefore, all database sequences (or GlcD sequences from the same species but different genomes) were verified to be “GlcD” sequences by checking for E. coli GlcD among the clustered bidirectional best hits in the IMG database (http://img.jgi.doe.gov/pub/main.cgi). GlcD sequences from the GenBank and IMG databases that were not bidirectional best hits to E. coli GlcD were excluded. As a result, 18 (3%) environmental glcD sequences were also excluded from further analyses based on phylogenetic grouping (data not shown) with these excluded database sequences. The amino acid sequences used for tree construction were aligned using ClustalW (47). Columns with gaps after alignment were removed when gaps were present in all but one sequence. This resulted in 296 and 289 amino acid positions that were analyzed for the trees shown in Fig. 2A and B, respectively (see Results). All trees were constructed using the maximum-likelihood method with PROML (Phylip, version 3.65) (15) with 100 bootstrap replicates. The parameters used for tree construction were the Jones-Taylor-Thornton probability model, the constant rate of change, randomized sequence addition, and the Swofford-Rogers search technique. For final tree construction, sequences were split into two groups based on a bootstrap value of 65 from an initial tree containing all sequences. The consensus tree was constructed with the majority rule algorithm.
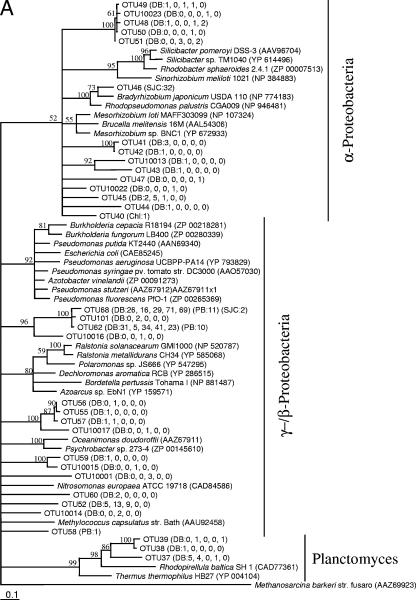
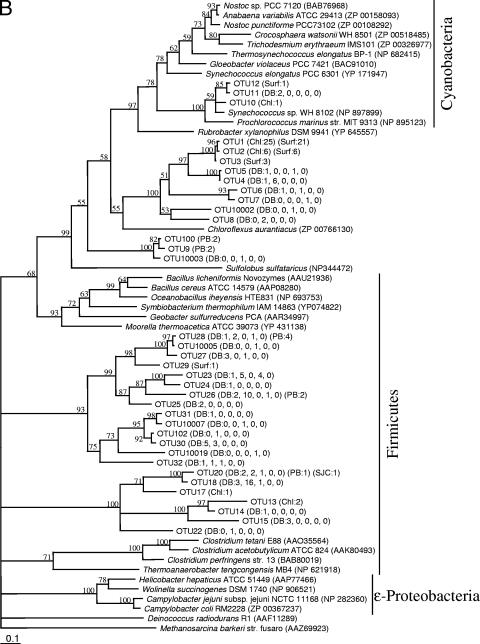
FIG. 2.
Maximum-likelihood phylogenetic trees for the GlcD amino acid sequences from Dabob Bay (DB) and from previously described samples collected in 2000 from Parks Bay in Washington (PB), San Juan Channel in Washington (SJC), and Chl a maximum (Chl) and surface (Surf) from an Atlantic Gulf Stream Ring. An initial tree containing all sequences supported (bootstrap value, 65) (not shown) the separation of clades for construction of two separate trees (A and B). The numbers in parentheses are the numbers of clones observed for the OTUs. For Dabob Bay, the five numbers in parentheses are in order for the five sequential clone libraries. Scale bars represent 1 substitution per 10 bases. OTUs for which phylotype-specific primers were designed for qPCR analyses are indicated by bold type. Methanosarcina barkeri strain fusaro was used as the outgroup for both trees. Unsupported branches (bootstrap values, <50) have been collapsed.
Diversity analyses.
glcD diversity was analyzed with EstimateS with 100 randomization runs (version 6.0b1) (http://purl.oclc.org/estimates). Sample-based rarefaction curves were constructed using the Mao Tau estimator (7). The means of 100 runs were determined for the Shannon and Simpson diversity indices (20).
Quantitative PCR (qPCR).
In addition to the DNA extracted from filters for clone library construction, DNA was also extracted, as described above, from filters collected at the time closest to noon following the after-sunset collection the previous evening. Thus, two times, about 2 to 3 h after sunset and around noon (1130 to 1300) the next day, were analyzed for each of five cruises.
RNA on another set of filters for the same 10 times was extracted using an RNAqueous-4PCR kit (Ambion). The extracted RNA was treated with 3 U of RNase-free DNase (Ambion) for 1 h at 37°C. DNA contamination of the extracted RNA was examined by performing a qPCR with the OTU 68 primers (see below) prior to quantification and reverse transcription. Incubation with DNase was repeated as needed. DNase was inactivated using the procedure recommended by the manufacturer. Extracted RNA was quantified using a RiboGreen RNA-specific quantitation kit (Molecular Probes, Inc.). RNA was reverse transcribed into cDNA using an Advantage RT-for-PCR kit (BD BioSciences-Clontech) using random hexamers and 12.5 μl of extracted RNA (see below for calculation of transcript abundance).
Standards for quantification were created with plasmids extracted from one clone of each OTU using a QiaPrep spin miniprep kit (QIAGEN). Extracted plasmids were linearized with 5 U of SpeI (New England BioLabs, Inc.) by incubation for 2 h at 37°C, followed by a 20-min enzyme inactivation step at 65°C. Linearized plasmids were quantified using a PicoGreen double-stranded DNA quantitation kit (Molecular Probes, Inc.).
Specific primers for the six most abundant OTUs identified in the clone libraries were designed using ARB (31) (Table 1). The primers were designed to recognize (no mismatches in either the forward or reverse primer sequence) at least 95% of the sequences in each OTU and not to recognize any of the other OTUs in the five clone libraries. A single set of OTU 68-specific primers could not be designed to amplify 95% of the sequences without sacrificing specificity; therefore, two sets of OTU 68 primers were designed to differentially recognize the sequence encoding two specific amino acids that subdivide OTU 68 into OTU 68a and OTU 68b. Only minor cross-amplification occurred with these two primers. The cross-amplification value for the OTU 68b plasmid standard with the OTU 68a primers was 8.39 × 10−6 copies detected per copy present, and for the OTU 68a plasmid standard with the OTU 68b primers it was 8.17 × 10−4 copies detected per copy present. Extracted DNA and cDNA of RNA were quantified using an iCycler iQ real-time PCR detection system (Bio-Rad Laboratories, Inc.). For four of the OTUs (OTU 18, OTU 26, OTU 37, and OTU 52), iQ SybrGreen Supermix (Bio-Rad) was used with 0.4 mM of each primer. The amplification protocol consisted of an initial 3-min enzyme activation step at 95°C, followed by 50 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 20 s. For the other three OTUs (OTU 62, OTU 68a, and OTU 68b), the PCR amplification mixture contained 0.3 mM deoxynucleoside triphosphates, 0.2 mM of each primer, 1.5 mM MgCl2, 0.05 U/ml DNA polymerase (Takara Ex Taq R-PCR Ver. 2.1; Takara Bio Inc.), 10 nM fluorescein, and 0.4× Sybr Green (Molecular Probes), and the amplification protocol consisted of a 30-s initial enzyme activation step at 95°C, followed by 50 cycles of 95°C for 50 s, 60°C for 15 s, and 72°C for 15 s. At least five standard amounts ranging from 2 × 100 to 2 × 106 copies were run in duplicate or triplicate for each set of analyses. Regression of all standard curves yielded an r2 value of 0.999. All samples were run in triplicate.
TABLE 1.
Bacterial clades analyzed by qPCR, primers used, and amplicon sizes
| OTUa | Forward primer (5′→3′) | Reverse primer(5′→3′) | Amplicon size (bp) |
|---|---|---|---|
| 18 | TACAGCCTCCGAGCAACTG | ATCGACTCGCTCGCGATTC | 209 |
| 26 | GTGCTCAACCTCGAAGTGG | TTGGGCCACGGCAGCAGT | 163 |
| 37 | GAGGTGGTGCAGTTTGGC | GCGACCGATAGCCAACAAG | 162 |
| 52 | ATGTAGCCAAGGCGGGCG | GCCACTTCATCTGCAGTGC | 180 |
| 62 | GCAACTGAAGCTCGTCTCG | AACGGTACCTGCTGCTTCAT | 213 |
| 68a | CTAGCTGCCGTGCTAGCGA | ACCCGCTTTATTTGCGTCA | 126 |
| 68b | TTTTCCGAGCAGCACGGC | ACCAACTTCGATGCAGAGGT | 155 |
OTUs were based on a level of DNA sequence similarity of 97%.
The amplification efficiencies for plasmid standards, DNA, and cDNA were calculated with LinReg PCR (40). Differences in amplification efficiencies between standards and samples were examined using the appropriate t test (54) after the variance between the standard and sample efficiencies was assessed using an F test (54). Sample reactions with efficiencies significantly different from the efficiencies of the standards were removed from analyses and redone to obtain triplicate measurements for each sample. The starting copy numbers of glcD in DNA and cDNA were calculated based on regression parameters of standard curves, and concentrations were calculated based on the volume of seawater filtered, the volume of extracted DNA or RNA, and the volume of DNA or cDNA used per reaction.
Glycolate measurement.
Glycolate concentrations were determined by high-performance liquid chromatography, using minor modifications of previously described protocols (1, 34, 35). Briefly, filtered seawater samples were bubbled with nitrogen gas without pyridine buffer for 15 min. The seawater samples were then derivatized with 2-nitrophenylhydrazine (Sigma) and 1-[3-(dimethylamino)propyl]-3-ethylcarbodiimide hydrochloride (Aldrich) for 1.5 h at room temperature. The derivatized samples were loaded and concentrated onto a 1.5-cm Brownlee C18 column (Perkin-Elmer) in the sample loop of a Gilson model 231 autosampler (Gilson Inc., Middleton, WI). After the concentrating column was rinsed with Nanopure water, the samples were run through a 25-cm Beckman Ultrasphere C18 column (Beckman Coulter Inc.) with the ion-pairing buffers described by Albert and Martens (1) and detected with an SPD-10Avvp UV-visible detector (Shimadzu Scientific Instruments) at 400 nm. Glycolate concentrations were determined by using glycolate (Sigma-Aldrich) standards diluted in Nanopure water. Experiments with standards and samples were performed in triplicate.
Statistical analyses.
Statistical analyses were performed with SPSS for Windows (version 13.0; Apache Software Foundation). Pairwise comparisons of parameters (Chl a concentration, glycolate concentration, OTU abundance, and total bacterial abundance) were done with two-tailed Pearson correlations. The Dunn-Sidak method was used to adjust the P values from the multiple comparisons (45). Night and day values for OTU abundance, RNA transcript levels, and total bacterial abundance were compared with the Wilcoxon signed-rank test.
Nucleotide sequence accession numbers.
All glcD sequences determined in this study have been deposited in the GenBank database under accession numbers DQ871607 to DQ872155.
RESULTS
Bloom dynamics.
The 7-week sampling period spanned phytoplankton prebloom to mid- to late-bloom conditions (Table 2). The Chl a concentrations were relatively low during the first week of sampling, with no evidence of a subsurface Chl a maximum. In the following week, the water column began to stratify, and the surface Chl a concentrations increased about 10-fold with the onset of the spring phytoplankton bloom. By the third week, a distinct subsurface Chl a maximum was detected. Over the course of the sampling period, a succession of different species of dominant phytoplankton was observed, including the diatoms Chaetoceros debilis, Thalassiosira spp., and Skeletonema costatum and the prymnesiophyte Phaeocystis cf. pouchetii (Table 2).
TABLE 2.
Phytoplankton bloom conditions during the sampling period
| Cruise dates | Week of sampling | Surface Chl a concn (mg/m3) | Subsurface Chl a
|
Dominant phytoplankton speciesa | Average bacterial abundance (105 cells ml−1)b | |
|---|---|---|---|---|---|---|
| Depth (m) | Concn (mg/m3) | |||||
| 24 and 25 February | 1 | 2.00-3.23 | NAc | NPd | Thalassiosira pacifica, Thalassiosira aestivalis | 4.82 (0.479) |
| 3 and 4 March | 2 | 15.32-30.39 | 6-8 | 7.12-19.11 | Chaetoceros debilis, Phaeocystis spp., Thalassiosira spp. | 6.06 (0.482) |
| 10 and 11 March | 3 | 4.00-13.55 | 4-8 | 9.75-21.30 | Phaeocystis spp., Thalassiosira rotula | 1.95 (0.145) |
| 22 to 26 March | 5 | 6.83-17.79 | 4-8 | 10.25-16.51 | Phaeocystis spp. | 3.41 (0.139) |
| 7 and 8 April | 7 | 3.67-6.07 | 8-11 | 16.41-25.44 | Skeletonema costatum | 7.72 (0.602) |
See reference 21.
Based on the 0.8- to 10-μm size fraction. The values in parentheses are standard deviations.
NA, not applicable.
NP, not present.
Bacterial abundance (0.8- to 10-μm fraction) varied over the course of the phytoplankton bloom, although no clear diel (data not shown) or weekly trends in abundance were apparent (Table 2). There was no significant correlation between bacterial abundance and Chl a concentration (P > 0.05). In weeks 3 and 5 the bacterial abundance was lower than the bacterial abundance in the other weeks, coinciding with a greater abundance of Phaeocystis spp.
Bacterial glcD diversity.
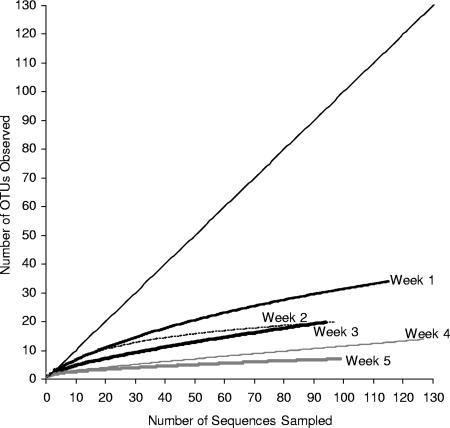
Approximately 100 clones from each of the five clone libraries were sampled to obtain a total of 532 sequences (Table 3). These sequences grouped into 55 OTUs. The rarefaction curve for each library started to plateau (although all slopes were greater than 0), indicating that the diversity in each clone library was well represented (Fig. 1). Over the bloom period, the glcD diversity appeared to decrease. The slopes of the rarefaction curves showed that more unsampled diversity remained in the early stages of the bloom (higher slopes) than in the later stages of the bloom (lower slopes). The Shannon and Simpson diversity indices also decreased over the course of the bloom, as did the number of OTUs observed (Table 3).
TABLE 3.
Number of glcD clones sequenced, number of OTUs observed, and diversity indices for each clone library
| Clone library | Week of sampling | No. of clones | No. of OTUs observed | Shannon diversity index | Simpson diversity index |
|---|---|---|---|---|---|
| DB1 | 1 | 115 | 35 | 2.69 | 7.82 |
| DB44 | 2 | 97 | 20 | 2.57 | 11.06 |
| DB106 | 3 | 94 | 20 | 1.93 | 4.33 |
| DB162 | 5 | 127 | 14 | 1.22 | 2.42 |
| DB288 | 7 | 99 | 7 | 0.89 | 1.87 |
FIG. 1.
Rarefaction curves for the five glcD clone libraries constructed over the course of the bloom. An OTU was defined by using a level of DNA sequence similarity of 97%.
Several OTUs dominated the five clone libraries, and 27 OTUs were detected only once. OTUs 68 and 62 comprised 40 and 25% of the clones sequenced, respectively. OTUs 52, 18, 26, and 37 were the four next most abundant OTUs sampled (5.1, 3.8, 2.4, and 1.9%, respectively). Detection of these six OTUs in the clone libraries was not uniform during the sampling period. OTUs 52, 18, 26, and 37 were detected relatively more frequently during the early stages of the bloom; OTU 68 was detected relatively more frequently during the later stages of the bloom; and OTU 62 was detected relatively frequently throughout the sampling period except during week 2 (Fig. 2).
GlcD phylogeny has been shown to agree well with 16S rRNA phylogeny (26). The GlcD sequences in this study belonged to the heterotrophic α-, β-, δ-, and γ-Proteobacteria, Firmicutes, and Planctomyces and to the photosynthetic cyanobacteria; some clades showed unresolved phylogenetic affiliations (Fig. 2). All six of the most abundant OTUs grouped with heterotrophic bacteria, although a few OTUs grouped with cyanobacteria. Many environmental sequences regardless of the bacterial division grouped together, forming clades that did not have close relatives among GlcD sequences of known bacteria (Fig. 2). OTUs 68 and 62 together with OTU 101 formed a clade that contained the most sequences sampled and belonged to either the γ- or β-Proteobacteria. For sequences in these three OTUs the maximum difference in glcD DNA sequences was 6%, which was found to represent species- to genus-level DNA sequence similarity (26). The other four most abundant OTUs sampled belonged to other phylogenetic groups (Fig. 2), indicating that the most abundant OTUs were phylogenetically diverse.
GlcD sequences from Dabob Bay have been observed in other marine environments. Five phylogenetically diverse OTUs were found in Parks Bay in Washington and in the San Juan Channel in Washington in July 2000 (Fig. 2) (26). Three of these common OTUs coincided with the three most abundant OTUs found in the five clone libraries from Dabob Bay (OTUs 68, 62, and 26). The other resampled OTUs were OTU 28 and OTU 20. In addition, two more distantly related clades contained sequences from both Dabob Bay and an Atlantic Gulf Stream Ring sampled in 2000; one clade comprised OTUs 1 to 5, grouping most closely with Chloroflexus aurantiacus, and the other comprised OTUs 10 to 12 and grouped with marine cyanobacteria.
glcD OTU abundance and expression.
The relative abundance values for the six glcD OTUs determined by qPCR were consistent with the relative frequencies observed in the clone libraries, although the absolute percentages differed (Table 4). For example, OTU 62 was consistently undersampled and OTU 68 was consistently oversampled in the clone libraries. Nonetheless, the three patterns observed for these six OTUs in the clone libraries were confirmed by the qPCR data. OTUs 18, 26, 37, and 52 were always less abundant than OTUs 62 and 68 (Table 5). The abundance of OTU 62 was generally the highest abundance of the six OTUs, and the abundance of OTU 68 was higher only during the day in week 7 (Table 5). None of the OTU abundance values correlated significantly with total bacterial abundance (P > 0.05; n = 10). OTU 26, 37, and 52 abundance values correlated significantly with Chl a concentrations (P < 0.05; n = 9). The abundance values for OTUs 18, 26, 37, and 52 all correlated with each other (P < 0.05, n = 10).
TABLE 4.
Comparisons of the frequencies of observation for six OTUs based on clone library sequencing and qPCR quantificationa
| Week | % OTU 18
|
% OTU 26
|
% OTU 37
|
% OTU 52
|
% OTU 62
|
% OTU 68
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone library | qPCR | Clone library | qPCR | Clone library | qPCR | Clone library | qPCR | Clone library | qPCR | Clone library | qPCR | |
| 1 | 4.3 | 0.31 | 2.9 | 2.7 | 7.2 | 3.6 | 7.2 | 1.3 | 45 | 80 | 33 | 13 |
| 2 | 25 | 1.5 | 16 | 11 | 6.2 | 13 | 20 | 5.0 | 7.8 | 55 | 25 | 15 |
| 3 | 1.4 | 0.12 | 0 | 0.87 | 0 | 3.2 | 13 | 2.4 | 47 | 80 | 39 | 13 |
| 5 | 0 | 0.03 | 0.88 | 0.63 | 0.88 | 0.40 | 0 | 1.0 | 36 | 78 | 62 | 20 |
| 7 | 0 | 0.02 | 0 | 0.004 | 0 | 0.33 | 0 | 0.12 | 25 | 67 | 75 | 33 |
Clone library percentages are based only on observations of the six OTUs analyzed; therefore, the sum of the clone library percentages for each week is 100%. The larger of the two percentages for each pairwise comparison (clone library and qPCR) is indicated by bold type.
TABLE 5.
glcD copy numbers per milliliter of seawater for six OTUs determined by qPCR and total numbers of bacterial cells per milliliter of seawater determined by DAPI countinga
| Week | No. of copies of glcD/ml
|
||||||
|---|---|---|---|---|---|---|---|
| OTU 18 (100) | OTU 26 (100) | OTU 37 (101) | OTU 5 (101) | OTU 62 (103) | OTU 68 (103) | Total bacteria (105)b | |
| Night | |||||||
| 1 | 46.5 (13.7) | 408 (40.8) | 54.2 (4.29) | 19.1 (1.18) | 12.0 (2.55) | 1.88 (0.474) | 2.73 (0.609) |
| 2 | 256 (15.7) | 1,900 (313) | 223 (38.2) | 84.5 (15.8) | 9.32 (1.50) | 2.48 (0.352) | 3.65 (0.568) |
| 3 | 10.2 (5.20) | 77.0 (13.7) | 28.3 (5.56) | 20.8 (2.76) | 7.09 (0.570) | 1.14 (0.0768) | 2.44 (0.364) |
| 5 | 9.04 (1.86) | 212 (46.2) | 13.3 (1.80) | 32.9 (9.39) | 26.3 (5.13) | 6.57 (0.371) | 4.61 (0.891) |
| 7 | 9.74 (2.61) | 2.05 (0.189) | 17.4 (1.81) | 6.36 (1.00) | 35.2 (10.1) | 17.2 (2.06) | 8.70 (1.66) |
| Day | |||||||
| 1 | 26.2 (7.24) | 177 (17.2) | 42.4 (3.74) | 6.93 (0.663) | 1.39 (0.199) | 0.266 (0.0224) | 6.26 (1.90) |
| 2 | 124 (19.3) | 1,780 (232) | 181 (24.3) | 140 (47.6) | 37.0 (2.64) | 2.40 (0.326) | 7.98 (1.49) |
| 3 | 4.19 (1.00) | 31.1 (12.2) | 4.08 (0.742) | 21.7 (2.44) | 1.94 (0.327) | 0.620 (0.0463) | 2.47 (0.292) |
| 5 | 13.1 (4.61) | 205 (13.2) | 24.8 (2.57) | 5.20 (0.687) | 10.5 (3.11) | 5.20 (0.769) | 2.53 (0.634) |
| 7 | 21.8 (1.89) | BDc | 75.4 (13.9) | 9.23 (1.84) | 13.7 (4.05) | 22.3 (3.06) | 9.03 (1.40) |
Night indicates that samples were collected 2 to 3 h after sunset, and day indicates that samples were collected at “noon” the following day. The values in parentheses are standard deviations.
Based on the 0.8- to 10-μm size fraction.
BD, below the limit of detection.
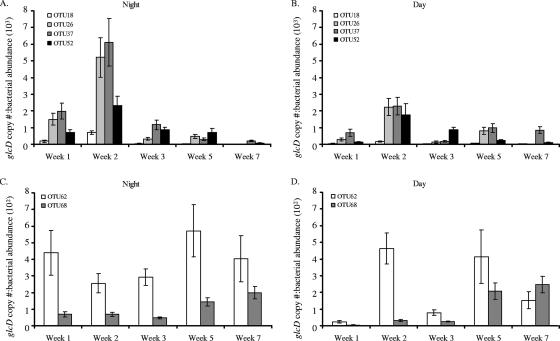
Three different trends were observed during the spring phytoplankton bloom when glcD abundance was normalized to bacterial cell counts. For the four lower-abundance OTUs (OTUs 18, 26, 37, 52), there was a relative abundance peak during week 2 (Fig. 3A and B). The relative abundance of OTU 68 generally increased over the course of the phytoplankton bloom, and the abundance of OTU 62 remained relatively high throughout the sampling period (Fig. 3C and D). In general, the relative abundance for all six OTUs represented a higher proportion of the bacterial community during the night than during the day (P < 0.05). However, the abundance of OTU 62 was proportionately lower during the day in weeks 1 and 3 (Fig. 3D).
FIG. 3.
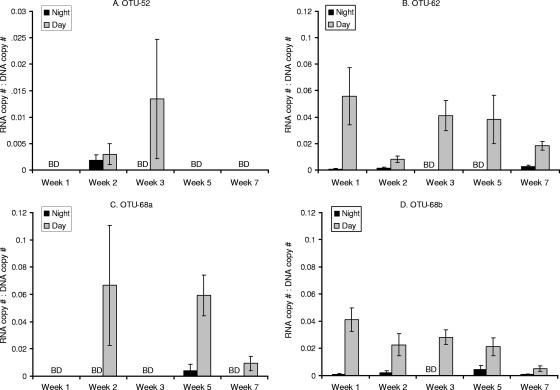
glcD copy number normalized to bacterial abundance (0.8- to 10-μm fraction) for one night and the following “noon” sample for each week of sampling. (A and B) Percentages of lower-abundance OTUs detected at night (A) and around noon (B) the next day. (C and D) Percentages of higher-abundance OTUs detected at night (C) and around noon (D) the next day. The OTU 68 values are the sums of the values for the two subgroups (OTU 68a and OTU 68b). The error bars indicate standard deviations for triplicate qPCR measurements of one sample per time. BD, below the limit of detection. Note that the y axis scales are different for different graphs.
glcD RNA transcripts were detected for OTUs 52, 63, 68a, and 68b, but the levels for OTUs 18, 26, and 37 were below the detection limit (2 × 103 copies per liter of seawater) at all times. glcD transcript abundance values were normalized to the corresponding DNA copy abundance values to determine whether changes in transcript abundance resulted from an increase in the number of cells or from higher levels of glcD mRNA per cell. The normalized transcript amounts were always larger during the day than at night for all four OTUs, indicating that there was a diel pattern for glcD mRNA levels (P < 0.005) (Fig. 4). The normalized transcript amounts were always less than 0.1 regardless of the phylotype. No trends in transcript amounts over the course of the phytoplankton bloom were detected for any of the OTUs or for all the OTUs. The normalized transcript amounts (when detectable) seemed to differ even within the same OTU (OTU 68a and OTU 68b) (Fig. 4C and D).
FIG. 4.
glcD RNA transcript abundance normalized to the corresponding DNA abundance for OTU 52 (A), OTU 62 (B), OTU 68a (C), and OTU 68b (D). Data for OTU 68 are presented separately for the two subgroups amplified with different primer sets. The error bars indicate standard deviations for triplicate qPCR measurements of one sample per time for both DNA and RNA. BD, below the limit of detection. Note that the y axis scale for panel A differs from the y axis scale for panels B to D.
Glycolate concentrations.
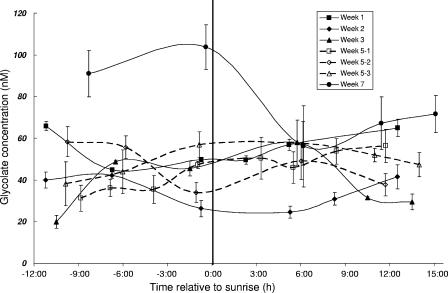
In contrast to the glcD transcription patterns, the glycolate concentrations did not exhibit a diel or seasonal pattern and did not appear to correlate with any species of phytoplankton. With the exception of the two times before sunrise in week 7, the glycolate concentrations were relatively constant throughout the phytoplankton bloom, ranging from 20 to 60 nM (Fig. 5). The glycolate concentration correlated negatively with the Chl a concentration (P < 0.005; n = 41) and did not correlate significantly with any of the OTU abundance values (P > 0.05; n = 10) or the total bacterial abundance (P > 0.05; n = 39).
FIG. 5.
Glycolate concentrations for each 24-h sampling period. The data for three consecutive days in week 5 (5-1, 5-2, and 5-3) are indicated separately by the dashed lines and open symbols. Sample times were normalized to the sunrise time for the day. The error bars indicate the standard deviations for triplicate measurements of one sample per time point.
DISCUSSION
glcD diversity and abundance.
Marine bacteria with the genetic potential to use glycolate (based on the presence of glcD) are phylogenetically diverse and yet appear to be restricted to specific clades. Across different phyla, many of the marine glcD sequences formed clades that grouped more closely with each other than with sequences belonging to nonmarine bacteria. Moreover, two clades comprised phylotypes observed previously in Pacific Northwest waters, as well as in an Atlantic Gulf Stream Ring. In Pacific Northwest waters, five phylotypes (OTUs 20, 26, 28, 62, and 68) found in Dabob Bay in 2004 had been observed previously in Parks Bay and San Juan Channel in Washington in 2000. Three of these resampled OTUs (OTUs 26, 62, and 68) represented relatively more abundant members of the glycolate-utilizing bacterial component in Dabob Bay in 2004. The presence of these OTUs across time and space suggests that they are important and stable members of bacterial communities in the coastal waters of the Pacific Northwest.
The diversity of bacteria with the genetic potential for glycolate utilization decreased over the course of the bloom period. In addition to this decrease, shifts in the relative abundance of different phylotypes were detected. Specifically, three different patterns were observed: early-bloom phylotypes (OTUs 18, 26, 37, and 52) that were associated with the onset of the phytoplankton bloom, a late-bloom phylotype (OTU 68) whose abundance increased as the phytoplankton bloom progressed, and an abundant phylotype (OTU 62) that was present throughout the phytoplankton bloom. Artifacts in PCR amplification can result in either over- or undersampling of OTUs in clone libraries (Table 4) and can skew the evenness and number of the species observed. The decreasing trend in glcD diversity for both Shannon and Simpson diversity indices, which weigh evenness and species richness differently (20), and the general agreement in seasonal trends between qPCR results and clone library frequencies indicate that PCR amplification bias was probably not the cause of the trends observed. The different trends in abundance among the different glycolate-utilizing phylotypes probably reflected real differences in response to the phytoplankton community and to glycolate availability.
Changes in diversity associated with different phytoplankton-derived dissolved organic carbon compounds have been observed using 16S rRNA genes (10, 22, 36, 43). In these studies, however, the workers monitored the whole bacterial community using 16S rRNA analyses, which integrate all processes affecting the bacterial community, and consequently could detect only general trends or changes in a few dominant phylotypes. Our results indicate that bulk community analyses (such as analyses with 16S rRNA genes) would not have provided the sensitivity necessary to detect the decrease in diversity, as well as the succession among the glycolate-utilizing bacteria.
glcD transcript levels and glycolate utilization.
The glcD mRNA levels of all of the four phylotypes whose RNA transcripts were detectable in the environmental samples exhibited a diel pattern. This diel pattern of glcD mRNA levels corresponded to the known diel periodicity of glycolate production and subsequent release by phytoplankton (9, 27, 28, 34, 35) and implied that the bacteria were actively using glycolate during the day.
Because the majority of bacterial mRNA have half-lives on the order of minutes (4, 42), the differences in transcript abundance between night and day likely reflected the “immediate” response of bacteria to glycolate availability. Thus, the diel pattern of glcD transcription levels was probably caused by differential accumulation of transcripts associated with upregulation during the day due to a response to increased glycolate availability and downregulation at night due to a response to a halt in glycolate production and excretion by phytoplankton at night. Increased mRNA degradation at night or synchronous division at night, while possible, was probably not the cause of the diel patterns observed.
The observed diel patterns most likely represented a tight coupling between heterotrophic bacterial utilization and phytoplankton glycolate excretion. The glycolate concentrations during the spring phytoplankton bloom in Dabob Bay in 2004 did not vary on a diel cycle or change dramatically over the bloom period, although phytoplankton excrete different amounts of glycolate over a diel cycle and under different nutrient regimes, which occur during a phytoplankton bloom (34, 35). Previous work by Fuhrman (16) and Fuhrman and Ferguson (18) on bacterial uptake of dissolved free amino acids, which are also highly labile compounds, showed that the measurable levels of dissolved free amino acids in seawater were low (nanomolar range) and changed very slowly, but the uptake rates were high, on the same order as the production rates. Thus, these workers concluded that there was a tight coupling between release and uptake of these compounds. In our study, rather than using radioactive tracers to track the flow of glycolate, we used the bacterial transcription of glcD, a gene specifically required for utilization of phytoplankton-derived glycolate, as a proxy for bacterial uptake. Our results suggest that there was a similar, direct, and specific coupling between bacterial uptake and phytoplankton excretion of glycolate.
The diel pattern of glcD transcript abundance detected in this study is the first such pattern described for a gene related to heterotrophy. Diel patterns of mRNA transcription of the nifH gene for nitrogen fixation have been detected in cyanobacteria in situ (5). Different diel patterns (peaks at different times of the day and night) were detected for five phylotypes of cyanobacteria, which likely resulted from a response to light that was also coupled to the specific physiology for nitrogen fixation in each phylotype. However, no diel pattern of nifH transcription was evident for the γ-proteobacterial (and presumably heterotrophic) phylotype (5).
The transcriptional abundance (RNA/DNA ratio) of glcD was always less than 0.1, suggesting that at the moment of sampling only a portion of the bacteria in each phylotype was actively responding to glycolate. The short half-lives of bacterial mRNA may partially explain this ratio. The filtration time was limited to 20 min, but the collection time was longer; altogether, these times represented several half-lives for the average bacterial mRNA. Under the most conservative scenario, in which transcription stops immediately when water is collected, if we assume that the sample collection time was on the order of 5 to 6 mRNA half-lives, the adjusted RNA/DNA ratio would have been around 2 to 3. For the nifH gene, upregulation resulted in an RNA/DNA ratio of 102 to 103 (5), so sample collection time may not fully explain the glcD ratio. This ratio may also occur if only a fraction of the cells in each bacterial phylotype actively transcribe glcD at any one time or if only a subset in each phylotype is metabolically active. Studies of the activities of the overall bacterial community have shown repeatedly that only a fraction of a marine bacterial community is metabolically active at any given time (23, 55). Whether this partial activity is attributable to different phylotypes being active or to different fractions of any phylotype being active at a given time is not known; however, the RNA transcription results for the two subphylotypes of OTU 68 suggest that it is possible for different fractions of a phylotype to behave differently.
Another explanation for this RNA transcription pattern may be the spatial distribution of bacteria relative to the phytoplankton cells. The bacteria sampled in this study were unlikely to be attached to the phytoplankton as the dominant phytoplankton species present were larger than 10 μm (21) and the bacterial size sampled was less than 10 μm. The observed glcD RNA/DNA ratio may reflect the fact that only some of the (free-living) glycolate-utilizing bacteria at any given moment were in the phycosphere containing concentrations of glycolate that were high enough to induce glcD transcription.
Implications of diel and bloom period patterns of abundance and activity of glycolate utilizers.
The decrease and shift in glycolate-utilizing diversity during the phytoplankton bloom suggests that there was competition or selection among the glycolate-utilizing bacteria. Competition among glycolate utilizers may result from different physiologies (e.g., uptake rates and assimilation efficiency) for glycolate utilization that may be coupled to utilization of other organic compounds, such as serine and glycine, that are also produced through photorespiration. Alternatively, the change in diversity may reflect specific interactions between specific lineages of bacteria and phytoplankton. The dominant phytoplankton species changed over the course of the phytoplankton bloom. The change in the diversity of the glycolate-utilizing bacteria may have reflected tight coupling between the bacterial and phytoplankton lineages that dominated at different stages during the phytoplankton bloom and deserves further study. Regardless of the mechanism, the community dynamics of glycolate-utilizing bacteria has important implications for nutrient cycling in marine environments.
The relative abundance of the six OTUs quantified exhibited a diel pattern not mirrored in the total bacterial abundance, which may have reflected the activity of these OTUs. Diel patterns of overall bacterial abundance have been observed previously in some systems (17, 25). Generally, the greatest abundance occurred at night and in the early morning, and the abundance decreased during the day (17, 25). This diel pattern has been attributed to more intense bacterial grazing during the day than at night. In one study, no diel pattern was detected for heterotrophic bacteria, but the greatest abundance of Synechococcus was detected at night, again presumably due to grazing during the day (48). The diel pattern for glycolate-utilizing bacterial abundance relative to the abundance of the whole bacterial population may similarly have been due to differential grazing during the day. The patterns of glcD transcription suggest that glycolate-utilizing bacteria were active during the day. Preferential grazing on active bacteria by ciliates and nanoflagellates has been observed (24, 46). Another possibility is increased viral lysis during the day, when bacteria are more active, presumably due to tight coupling with phytoplankton production (51). If glycolate-utilizing bacteria are indeed preferentially grazed or lysed because they are active, glycolate-utilizing bacteria may represent a key component of the bacterial community during phytoplankton blooms for remineralization of phytoplankton exudates and for facilitated transfer of phytoplankton-derived carbon to other trophic levels.
This study is the first study in which the utilization of a specific organic carbon compound by marine bacteria was investigated in detail at the phylotype level and at the mRNA transcript level. By using a specific functional gene (as opposed to the 16S rRNA gene), we were able to monitor bacterial utilization of the highly labile compound glycolate. Traditionally, it has been difficult to quantify bacterial utilization of highly labile dissolved organic compounds because release and uptake are so tightly coupled that changes in seawater concentrations are negligible (16, 18). The results of this study suggest that we can monitor bacterial responses at the molecular level to determine the details of potentially tight coupling between marine bacteria and phytoplankton. In fact, we were able to detect both tight coupling of glycolate-utilizing bacteria to phytoplankton photorespiration on a diel cycle and an effect on the bacterial community structure over a phytoplankton bloom. The results of this study have important ecological implications for how this bacterium-phytoplankton interaction influences the flow of carbon up and down trophic levels and in marine ecosystems in general.
Acknowledgments
We thank J. Pierson, K. Hubbard, E. Lin, R. Marohl, P. von Dassow, and F. McCroskey, as well as R. McQuin and N. Hix of the R/V Clifford A. Barnes, for help with field sampling. We are grateful to A. Riddle, C. Berthiaume, J. Piovia-Scott, S. Carpenter, M. Parker, S. Reda, and N. Ahlgren for invaluable help in the lab and to D. Grunbaum, G. Rocap, J. Baross, and J. Cherrier for insightful discussions concerning this research.
This research was supported by a National Science Foundation Graduate Research Fellowship and a National Oceanographic and Atmospheric Administration Dr. Nancy Foster Scholarship to W. W. Y. Lau; by grant DE-FG03-00ER62982 from the Biotechnological Investigations-Ocean Margins Program, Biological and Environmental Research, U.S. Department of Energy, to E. V. Armbrust and R. G. Keil; and by a Gordon and Betty Moore Foundation Marine Microbiology Investigator Award to E. V. Armbrust.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Albert, D. B., and C. S. Martens. 1997. Determination of low-molecular-weight organic acid concentrations in seawater and pore-water samples via HPLC. Mar. Chem. 56:27-37. [Google Scholar]
- 2.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 3.Barnes, C. A., and C. C. Ebbesmeyer. 1978. Some aspects of Puget Sound's circulation and water properties, p. 209-228. In B. Kjerfve (ed.), Estuarine transport processes. University of South Carolina Press, Belle W. Baruch Library in Marine Science, Columbia.
- 4.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church, M. J., C. M. Short, B. D. Jenkins, D. M. Karl, and J. P. Zehr. 2005. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic north Pacific Ocean. Appl. Environ. Microbiol. 71:5362-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, A. W., M. J. Maguire, and J. R. Coleman. 1981. Mithramycin-2-phenylindole and 4′-6-diamidino-2-phenylindole (DAPI)-DNA staining for fluorescence micro-spectrophotometric measurement of DNA in nuclei, plastids, and virus-particles. J. Histochem. Cytochem. 29:959-968. [DOI] [PubMed] [Google Scholar]
- 7.Colwell, R. K., C. X. Mao, and J. Chang. 2004. Interpolating, extrapolating, and comparing incidence-based species accumulation curves. Ecology (New York) 85:2717-2727. [Google Scholar]
- 8.Cottrell, M. T., D. L. Kirchman, and L. Yu. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coughlan, S. J., and R. H. Al Hasan. 1977. Studies of uptake and turnover of glycolic acid in the Menai Straits North Wales. J. Ecol. 65:731-746. [Google Scholar]
- 10.Covert, J. S., and M. A. Moran. 2001. Molecular characterization of estuarine bacterial communities that use high- and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127-139. [Google Scholar]
- 11.Ducklow, H. W., and C. A. Carlson. 1996. Growth of bacterioplankton and consumption of dissolved organic carbon in Sargasso Sea. Aquat. Microb. Ecol. 10:69-85. [Google Scholar]
- 12.Ducklow, H. W., D. L. Kirchman, H. L. Quinby, C. A. Carlson, and H. G. Dam. 1993. Stocks and dynamics of bacterioplankton carbon during the spring bloom in the eastern North Atlantic Ocean. Deep-Sea Res. Part II Top. Stud. Oceanogr. 40:245-263. [Google Scholar]
- 13.Ebbesmeyer, C. C. 1973. Some observations of medium scale water parcels in a fjord: Dabob Bay, Washington. Ph.D. thesis. University of Washington, Seattle.
- 14.Edenborn, H. M., and C. D. Litchfield. 1987. Glycolate turnover in the water column of the New York Bight apex. Mar. Biol. 95:450-467. [Google Scholar]
- 15.Felsenstein, J. 2004. PHYLIP (phylogeny inference package), 3.6 ed. Department of Genome Sciences, University of Washington, Seattle.
- 16.Fuhrman, J. A. 1987. Close coupling between release and uptake of dissolved free amino acids in seawater studied by an isotope dilution approach. Mar. Ecol. Prog. Ser. 37:45-52. [Google Scholar]
- 17.Fuhrman, J. A., R. Eppley, W, A. Hagstrom, and F. Azam. 1985. Diel variations in bacterioplankton, phytoplankton, and related parameters in the Southern California Bight. Mar. Ecol. Prog. Ser. 27:9-20. [Google Scholar]
- 18.Fuhrman, J. A., and R. L. Ferguson. 1986. Nanomolar concentrations and rapid turnover of dissolved free amino acids in seawater: agreeement between chemical and microbiological measurements. Mar. Ecol. Prog. Ser. 33:237-242. [Google Scholar]
- 19.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayek, L. C., and M. A. Buzas. 1996. Surveying natural populations. Columbia University Press, New York, NY.
- 21.Horner, R. A., J. R. Postel, C. Halsband-Lenk, J. J. Pierson, G. Pohnert, and T. Wichard. 2005. Winter-spring phytoplankton blooms in Dabob Bay, Washington. Prog. Oceanogr. 67:286-313. [Google Scholar]
- 22.Janse, I., G. Zwart, M. J. E. C. van der Maarel, and J. C. Gottschal. 2000. Composition of the bacterial community degrading Phaeocystis mucopolysaccharides in enrichment cultures. Aquat. Microb. Ecol. 22:119-133. [Google Scholar]
- 23.Karner, M., and J. A. Fuhrman. 1997. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koton-Czarnecka, M., and R. J. Chrost. 2003. Protozoan prefer large and metabolically active bacteria. Pol. J. Environ. Stud. 12:325-334. [Google Scholar]
- 25.Kuipers, B., G. J. van Noort, J. Vosjan, and G. J. Herndl. 2000. Diel periodicity of bacterioplankton in the euphotic zone of the subtropical Atlantic Ocean. Mar. Ecol. Prog. Ser. 201:13-25. [Google Scholar]
- 26.Lau, W. W. Y., and E. V. Armbrust. 2006. Detection of glycolate oxidase gene glcD diversity among cultured and environmental marine bacteria. Environ. Microbiol. 8:1688-1702. [DOI] [PubMed] [Google Scholar]
- 27.Leboulanger, C., V. Martin-Jezequel, C. Descolas-Gros, A. Sciandra, and H. J. Jupin. 1998. Photorespiration in continuous culture of Dunaliella tertiolecta (Chlorophyta): relationships between serine, glycine, and extracellular glycolate. J. Phycol. 34:651-654. [Google Scholar]
- 28.Leboulanger, C., L. Oriol, H. J. Jupin, and C. Descolas-Gros. 1997. Diel variability of glycolate in eastern tropical Atlantic Ocean. Deep-Sea Res. 44:2131-2139. [Google Scholar]
- 29.Ledyard, K. M., and J. W. H. Dacey. 1996. Microbial cycling of DMSO and DMS in coastal and oligotrophic seawater. Limnol. Oceanogr. 41:33-40. [Google Scholar]
- 30.Lee, P. A., P. A. Saunders, S. J. de Mora, D. Deibel, and M. Levasseur. 2003. Influence of copepod grazing on concentrations of dissolved dimethylsulfoxide and related sulfur compounds in the North Water, northern Baffin Bay. Mar. Ecol. Prog. Ser. 255:235-248. [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malin, G., W. H. Wilson, G. Bratbak, P. S. Liss, and N. H. Mann. 1998. Elevated production of dimethylsulfide resulting from viral infection of cultures of Phaeocystis pouchetii. Limnol. Oceanogr. 43:1389-1393. [Google Scholar]
- 33.Malmstrom, R. R., R. P. Kiene, and D. L. Kirchman. 2004. Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49:597-606. [Google Scholar]
- 34.Parker, M. S., and E. V. Armbrust. 2005. Synergistic effects of light, temperature and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 41:1142-1153. [Google Scholar]
- 35.Parker, M. S., E. V. Armbrust, J. Piovia-Scott, and R. G. Keil. 2005. Induction of photorespiration by light in the centric diatom Thalassiosira weissflogii (Bacillariophyceae): Molecular characterization and physiological consequences. J. Phycol. 40:557-567. [Google Scholar]
- 36.Pinhassi, J., A. Farooq, J. Hemphala, R. A. Long, J. Martinez, U. L. Zweifel, and A. Hagstrom. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 37.Pinhassi, J., M. M. Sala, H. Havskum, F. Peter, O. Guadayol, A. Malits, and C. Marrase. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinhassi, J., R. Simo, J. M. Gonzalez, M. Vila, L. Alonso-Saez, R. P. Kiene, M. A. Moran, and C. Pedro-Alio. 2005. Dimethylsulfoniopropionate turnover is linked to the composition and dynamics of the bacterioplankton assemblage during a microcosm phytoplankton bloom. Appl. Environ. Microbiol. 71:7650-7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pomeroy, L. R. 1974. The ocean food web, a changing paradigm. BioScience 24:499-504. [Google Scholar]
- 40.Ramakers, C., J. M. Ruijter, R. H. L. Deprez, and A. F. M. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 41.Rappe, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 42.Redon, E., P. Loubiere, and M. Cocaign-Bousquet. 2005. Role of mRNA stability during genome-wide adaptation of Lactococcus lactis to carbon starvation. J. Biol. Chem. 280:36380-36385. [DOI] [PubMed] [Google Scholar]
- 43.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simo, R., S. D. Archer, C. Pedro-Alio, L. Gilpin, and C. E. Stelfox-Widdicombe. 2002. Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 47:53-61. [Google Scholar]
- 45.Sokal, R. R., and F. J. Rohlf. 1995. Biometry: the principles and practice of statistics in biological research. WH Freeman, New York, NY.
- 46.Tadonleke, R. D., D. Planas, and A. Lucotte. 2005. Microbial food webs in boreal humic lakes and reservoirs: ciliates as a major factor related to the dynamics of the most active bacteria. Microb. Ecol. 49:325-341. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai, A. Y., K. P. Chiang, J. Chang, and G. C. Gong. 2005. Seasonal diel variations of picoplankton and nanoplankton in a subtropical western Pacific coastal ecosystem. Limnol. Oceanogr. 50:1221-1231. [Google Scholar]
- 49.van Hannen, E. J., W. Mooij, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vila, M., R. Simo, R. P. Kiene, J. Pinhassi, J. M. Gonzalez, M. A. Moran, and C. Pedro-Alio. 2004. Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl. Environ. Microbiol. 70:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winters, C., G. J. Herndl, and M. G. Weinbauer. 2004. Diel cycles in viral infection of the bacterioplankton in the North Sea. Aquat. Microb. Ecol. 35:207-216. [Google Scholar]
- 52.Wright, R. T., and N. M. Shah. 1975. The trophic role of glycolic-acid in coastal sea water. Part 1. Heterotrophic metabolism in sea water and bacterial cultures. Mar. Biol. 33:175-183. [Google Scholar]
- 53.Wright, R. T., and N. M. Shah. 1977. The trophic role of glycolic-acid in coastal sea water. Part 2. Seasonal changes in concentration and heterotrophic use in Ipswich Bay Massachusetts USA. Mar. Biol. 43:257-264. [Google Scholar]
- 54.Zar, J. H. 1996. Biostatistical analysis. Prentice Hall, Englewood Cliffs, NJ.
- 55.Zweifel, U. L., and A. Hagstrom. 1995. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts). Appl. Environ. Microbiol. 61:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]