Abstract
A TaqMan format real-time PCR probe was developed against the internal transcribed spacer 2 ribosomal DNA region for the specific detection and quantification of Cryptoperidiniopsis brodyi in environmental samples. The assay specificity was confirmed by testing against related dinoflagellates and verified by sequencing PCR amplicons from natural water samples. Phylogenetic analysis of the sequenced environmental samples also showed that this assay is specific to C. brodyi. The C. brodyi-specific assay was used in conjunction with Pfiesteria piscicida- and Pfiesteria shumwayae-specific real-time PCR assays to investigate the temporal variations of C. brodyi, P. piscicida, and P. shumwayae abundance in the Derwent estuary, Tasmania. The 18-month field survey from November 2004 to April 2006 revealed that C. brodyi occurred in all seasons at very low densities, mostly below 25 cells liter−1, with higher abundance (maximum, 112 cells liter−1) in April and May. P. piscicida was detected only once, in May 2005 at 60 cells liter−1. P. shumwayae was not detected during the survey.
Cryptoperidiniopsis brodyi Steidinger et Litaker is a Pfiesteria-like dinoflagellate (PLD) that is indistinguishable from Pfiesteria piscicida Steidinger et Burkholder by light microscopy (LM). However, it differs from the latter genetically and ultrastructually (19, 37). C. brodyi often co-occurs with Pfiesteria piscicida Steidinger et Burkholder, Pfiesteria shumwayae Glasgow et Burkholder, Karlodinium veneficum (Leadbeater et Dodge) J. Larsen, and other morphologically similar dinoflagellates (22, 38). Since the coexistence of C. brodyi and Pfiesteria species in estuaries makes it difficult to identify these species in water samples by LM, their abundances and seasonal occurrences are poorly understood.
Pfiesteria and PLDs are morphologically defined by their distinct Kofoidian thecal plate formulae (37). The morphological characterization of C. brodyi and Pfiesteria species requires thecal plate analysis by scanning electron microscopy (SEM) on cultured strains in combination with a membrane stripping process (24) that is not suitable for rapid sample processing. While fluorescent in situ hybridization might be a useful alternative, this method is labor intensive and time consuming and suffers from interference by nonspecific binding in complex mixtures (32).
Real-time quantitative PCR that incorporates fluorogenic 5′ nuclease chemistry (TaqMan) is a technique that offers highly sensitive, rapid, and quantitative analysis. The TaqMan system uses sequence-based fluorogenic probes to detect amplified products during PCR cycling, and the detection of PCR amplicons is monitored by measuring the increase in fluorescence. This approach has been used successfully to detect small-subunit ribosomal DNA (SSU rDNA) of P. piscicida in mixed algal cultures; however, there is a major concern with regard to reliable quantification of real-time PCR assays from environmental water samples. Environmental samples often contain organic and inorganic substances with the potential to inhibit PCR assays (18, 42). Thus, the effective removal of PCR inhibitors from samples has to be considered for any field-based survey using real-time PCR assays.
The present study developed a C. brodyi-specific real-time PCR using a TaqMan probe based on internal transcribed spacer 2 (ITS2) of the rDNA. For detection of P. piscicida and P. shumwayae, existing P. piscicida-and P. shumwayae-specific real-time PCR assays (3, 42) were used and tested against different genotypes from Australia. Methods for overcoming PCR-inhibitory substances in environmental samples were examined for application of the real-time PCR protocol with water samples. Temporal occurrences of P. piscicida, P. shumwayae, and C. brodyi were characterized during an 18-month field survey using species-specific real-time PCR assays in a eutrophic Tasmanian estuary.
MATERIALS AND METHODS
Cultures.
The cultures used in this study are listed in Table 1. Nine C. brodyi strains (CBWA11, CBWA12, CBSA4, CBHU1, CBHU2, CBDE1, CBDE2, CBDE10, and CBDE14) identified by SEM and DNA sequence analyses were used as positive controls for C. brodyi-specific real-time PCR. P. piscicida (CCMP1036) and P. shumwayae (CCMP2357) were obtained from CCMP, Bigelow Laboratory for Ocean Sciences, West Boothbay Harbor, ME. Heterotrophic dinoflagellates were grown in 15‰ f/2 medium (11a) at 20°C in the dark. Rhodomonas salina (CS-24; obtained from CSIRO Marine Laboratories, Hobart, Australia) was supplied to C. brodyi and Pfiesteria species as food. All autotrophic cultures listed in Table 1 were obtained from the University of Tasmania's collection of microalgae. These were grown in 28‰ f/2 medium at 20°C with cool white fluorescent lamps on a 12:12-h light:dark cycle. An additional eight heterotrophic strains (CBCM1 to CBCM8, unidentified Pfiesteria look-alike cultures) were collected by Wasele Hosja from the Chapman River, Western Australia, during September 2005, and their identity was assessed by P. piscicida-, P. shumwayae-, and C. brodyi-specific real-time PCR assays.
TABLE 1.
Cultures used in this study
| Culture code | Date of isolation | Locality | Identification |
|---|---|---|---|
| PPSB25 | May 2003 | Surabaya, Indonesia, ballast water | Pfiesteria piscicida |
| CCMP1036 | North Carolina | Pfiesteria piscicida | |
| CCMP2357 | December 2002 | North Carolina | Pfiesteria shumwayae |
| CBWA11 | February 2003 | Brunswick River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBWA12 | February 2003 | Brunswick River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBSA4 | April 2003 | Port Lincoln, South Australia, Australia | Cryptoperidiniopsis brodyi |
| CBHU1 | February 2003 | Huon River, Tasmania, Australia | Cryptoperidiniopsis brodyi |
| CBHU2 | February 2003 | Huon River, Tasmania, Australia | Cryptoperidiniopsis brodyi |
| CBDE1 | March 2004 | Sullivan's Cove, Derwent River, Tasmania, Australia | Cryptoperidiniopsis brodyi |
| CBDE2 | March 2004 | Sullivan's Cove, Derwent River, Tasmania, Australia | Cryptoperidiniopsis brodyi |
| CBDE10 | March 2003 | Sandy Bay, Derwent River, Tasmania, Australia | Cryptoperidiniopsis brodyi |
| CBDE14 | November 2004 | Sandy Bay, Derwent River, Tasmania, Australia | Cryptoperidiniopsis brodyi |
| CBCM1 | September 2005 | Chapman River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBCM2 | September 2005 | Chapman River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBCM3 | September 2005 | Chapman River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBCM4 | September 2005 | Chapman River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBCM5 | September 2005 | Chapman River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBCM6 | September 2005 | Chapman River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBCM7 | September 2005 | Chapman River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CBCM8 | September 2005 | Chapman River, Western Australia, Australia | Cryptoperidiniopsis brodyi |
| CS-24 | Derwent River, Tasmania, Australia | Rhodomonas salina | |
| GCPL01 | April 1996 | Port Lincoln, South Australia, Australia | Gymnodinium catenatum |
| ASDB01 | Diana's Basin, Tasmania, Australia | Akashiwo sanguinea | |
| GFPL01 | July 2001 | Port Lincoln, South Australia, Australia | Gymnodinium falcatum |
| CAWD63 | Waimangu, New Zealand | Karenia mikimotoi | |
| KDVSR01 | July 2001 | Swan River, Perth, Australia | Karlodinium veneficum |
| ACTRA02 | October 1997 | Triabunna, Tasmania, Australia | Alexandrium catenella |
| HAWL169 | Port River, Adelaide, South Australia, Australia | Heterosigma akashiwo | |
| PRMED01 | March 2003 | Wonboyn Lakes, New South Wales, Australia | Prorocentrum minimum |
| APMLV01 | December 1999 | Louisville Point, Tasmania, Australia | Amphidinium massartii |
Collection and processing of environmental samples.
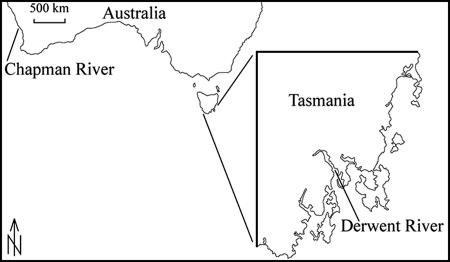
Water samples were obtained from Sullivan's Cove and Sandy Bay in the Derwent River, Tasmania, Australia (Fig. 1), at 2-week intervals from November 2004 to April 2006. The presence of C. brodyi at both sampling sites (2- to 3-m depth) was confirmed in 2003 to 2004. A low cell abundance of Pfiesteria or Pfiesteria-like organisms was observed by LM in the samples from the Derwent River during 2003 to 2004. To quantify cells and DNA yield, large-volume water samples (10 liter) were collected from the surface with a 10-liter bucket, poured through phytoplankton net (10-μm mesh size; 23-cm mouth diameter × 40-cm length; but the effective open mesh area was confirmed to be 5 μm), and concentrated in a 50-ml cod end container. The concentrated water samples from Sullivan's Cove and Sandy Bay were mixed in a glass bottle. A subsample of 50 ml from the glass bottle was centrifuged at 4,000 × g for 10 min at room temperature, and the cell pellet was stored at −70°C. To prevent degradation of the target DNA, samples were processed within 1 h of collection.
FIG. 1.
Locations where water samples were collected for this study.
DNA extraction.
After the cell pellets of the environmental samples stored at −70°C had defrosted, DNA was extracted from the cell pellets using a DNeasy tissue kit, following the manufacturer's protocol (QIAGEN, Hilden, Germany). As Pfiesteria and PLD are armored dinoflagellates, and extraction buffers may not be fully effective at lysing the cells, the standard DNeasy tissue kit method was compared to an alternative DNA-extracting method using a sonicator probe (Thomas Optical & Scientific Co. Pty. Ltd.). Three aliquots of 6,000 C. brodyi (CBWA12) cells each were incubated in ATL (tissue lysis buffer; QIAGEN) and sonicated for 40 s, and subsequently, the manufacturer's protocol for purification of DNA was followed. The threshold cycle (CT) values of the sonicated samples were measured using real-time PCR and compared to those obtained with the nonmodified DNeasy method. When the threshold line was set to a fluorescence value of 0.02 for 6-carboxyfluorescein (FAM), similar CT values were obtained by the two methods (18.34 ± 0.05 for no sonication and 18.53 ± 0.19 for sonication; n = 3; P > 0.05 by Student's t test). The DNA extraction method using a sonicator probe takes more time than the nonmodified DNA extraction method and can increase the risk of cross-contamination during the processing of a large number of DNA samples. Thus, the nonmodified DNeasy method was chosen for extracting DNA for constructing standard curves of real-time PCR assays and for processing environmental samples. DNA extracts (n = 6) from C. brodyi (CBWA12; 240,000 cells) were also quantified using the PicoGreen double-stranded DNA (dsDNA) quantification kit following the manufacturer's protocol (Molecular Probes, Eugene, Oregon).
PCR inhibitor removal from environmental samples.
Three different methods were compared for their capacity to remove PCR inhibitors from environmental samples: PVPP (polyvinylpolypyrrolidone), PVP360 (polyvinylpyrrolidone), and a 100-fold dilution of the DNA extract, using nontreated samples as a control. Water samples collected on 15 February 2005 were tested with real-time PCR to confirm the absence of C. brodyi and then used to detect PCR inhibitors. The following treatments were applied to water samples without natural C. brodyi, spiked with 1 μl of CBDE10 DNA (1.2 ng μl−1): (i) serial DNA dilutions of water sample spiked with CBDE10 DNA, (ii) water sample spiked with CBDE10 DNA for treatment with PVPP, and (iii) water sample spiked with CBDE10 DNA for treatment with PVP360. DNA was extracted from the cell pellets using a DNeasy tissue kit. ATL (180 μl) and proteinase K (20 μl) were added and thoroughly mixed by vortexing. Following incubation of the cell lysates for 1 h at 55°C, PVPP or PVP360 was added to a final concentration of 2% and the cell lysates were incubated for 30 min at 55°C. The cell lysates were then processed according to the manufacturer's instructions. C. brodyi-specific real-time PCR was performed with DNA from these treatments. The absence of PCR inhibitors in the 100-fold DNA dilution of all three treatments was confirmed. The absence of PCR inhibitors in the 100-fold DNA dilution was also confirmed with 10 other water samples that were randomly chosen among the field samples. The results of the 100-fold dilution of the DNA were calculated into cell estimates for the quantitative component of the assay. When large volumes of cell pellets (over 10 mg) were processed for DNA extraction, the DNeasy kit reagent volume was increased twice.
Design of TaqMan probe and primer set for detection of C. brodyi.
C. brodyi and related dinoflagellate rDNA sequences available from GenBank were aligned using the program ClustalX (40). Manual searches of the alignments were conducted to determine unique sequences and to develop a C. brodyi-specific real-time PCR assay. The sequences for the primer-probe set were conserved within C. brodyi strains, but discriminated between C. brodyi and other dinoflagellates. To minimize the variation in assay sensitivity caused by intraspecies variations within C. brodyi strains, mixed-base degenerate primer codes were used (Table 2). The primer and probe sequences of the target species were analyzed with Primer 3 (Whitehead Institute and Howard Hughes Medical Institute, Maryland) and Oligo analyzer 3.0 (Integrated DNA Technologies, Inc., Iowa) software for optimal melting temperature and secondary structure, and subsequently, the primers and probe were synthesized by Geneworks Pty. Ltd. (Hindmarsh, South Australia, Australia). The probe was dual-labeled with the fluorescent dyes FAM and 6-carboxytetramethyl-rhodamine (TAMRA) at the 5′ and 3′ ends, respectively.
TABLE 2.
Primers and TaqMan probes for species-specific real-time PCR
| Dinoflagellate | Primer or probe | Sequence (5′→3′) | Reference |
|---|---|---|---|
| Cryptoperidiniopsis brodyi | Forward primer CBITSF | TTGACACGTTGAAGTGAWGGA | This study |
| Reverse primer CBITSR | ACAGCCAATGAAAGAGTKATGACAA | ||
| Probe CBITSP | FAM-CATCTCATCGCTCGCCGTCGAT-TAMRA | ||
| Pfiesteria piscicida | Forward primer 107 | CAGTTAGATTGTCTTTGGTGGTCAA | 3 |
| Reverse primer 320 | TGATAGGTCAGAAAGTGATATGGTA | ||
| Probe | FAM-CATGCACCAAAGCCCGACTTCTCG-TAMRA | ||
| Pfiesteria shumwayae | Forward primer PSMTF2 | TGACTTTCTAACTTCTAACTTCTTTACATC | 42 |
| Reverse primer PSCOBR1 | AACACCATCCATAGAATATTTCTCTCATG |
Specificity of TaqMan probe and primers.
The sequences of the probe and primers were checked against published sequences in GenBank by BLAST homology search. Prior to real-time PCR, the specificity of the forward and reverse primers (Table 2) was tested by standard real-time PCR using SYBR green, which does not require a dual-labeled probe. The specificity testing was performed with 11 other dinoflagellates (Table 1) and R. salina. The SYBR green system measures any dsDNA, which may include nonspecific real-time PCR products such as primer dimers (12). The absence of primer dimers or nonspecific products was tested by inspecting melting curves and analyzing the amplicons of the target DNA (128 bp) in a 2% agarose gel stained with ethidium bromide. Subsequently, real-time PCR using a TaqMan probe was performed, and the specificity was confirmed with 11 dinoflagellates and R. salina.
Optimization of primer and probe concentrations.
To achieve optimal performance, a dilution series of primers and probe concentrations was tested by real-time PCR assay. The testing primer and probe concentrations ranged from 100 to 500 nM and 100 to 300 nM, respectively. The primer and probe concentrations that provided the lowest CT value and the highest total increase of fluorescence compared to the background were selected. The primer and probe concentrations were 0.2 μM and 0.15 μM, respectively.
Real-time PCR assay conditions.
PCR assays using TaqMan probes were performed on a Rotor-Gene RG-3000A (Corbett Research, New South Wales, Australia). The following reagents were added for a 20-μl reaction mixture: 10 μl of platinum quantitative PCR supermix-UDG (Invitrogen, New South Wales, Australia), forward and reverse primers each at a final concentration of 0.2 μM, fluorogenic probe at a final concentration of 0.15 μM, 2 μl of template DNA, and PCR-grade water to a final volume of 20 μl. For PCR assays using SYBR green, the following components were added for a 20-μl reaction mixture: 10 μl of platinum SYBR green real-time PCR supermix-UDG (Invitrogen) and concentrations of forward and reverse primers, template DNA, and PCR-grade water as described above. The thermal cycling conditions consisted of 2 min at 50°C and 2 min at 95°C followed by 45 cycles (50 cycles for SYBR green) of 10 s at 95°C and 45 s (40 s for SYBR green) at 60°C. Fluorescence data were collected at the end of each cycle, and determination of the cycle threshold line was carried out automatically by the instrument. PCR amplicons that were positive with SYBR green real-time PCR were further analyzed by gel electrophoresis through 2% agarose gels to confirm the amplicon size.
Construction of standard curves.
Standard curves using cell numbers were constructed from C. brodyi (CBWA12) and P. piscicida (PPSB25) cultures. Cell numbers (55,000 cells for C. brodyi and 11,000 cells for P. piscicida; n = 1) were estimated by LM in a hemocytometer (Blau Brand, Germany) before harvesting the cells, and genomic DNA was extracted. Tenfold serial dilutions of the DNA extracts were used to construct the standard curve (duplicate measurements by real-time PCR). The cell number of the target species in field samples was determined from CT values from comparison with the standard curve. To evaluate the accuracy and precision of the standard curves, known concentrations of C. brodyi cells (CBWA12; 480, 4,800, and 48,000 cells; n = 3) were spiked with sterile, filtered, environmental samples (0.2-μm membrane filter; MFS, California) followed by DNA extraction, 100-fold dilution of the DNA extracts, and real-time PCR. The results of the spiked experiments were then compared to the standard curves.
Species verification of PCR amplicons from environmental samples by sequence analysis.
PCR amplicons obtained in November 2004 to May 2005 that gave positive real-time PCR results (seven PCR products with C. brodyi-specific primers and one product with P. piscicida-specific primers) were visualized on an agarose gel stained with ethidium bromide. The band on the agarose gel was reamplified by use of the “band stab” approach (2). The band was stabbed with a stainless steel syringe needle, and the needle was dipped into the PCR mixture. PCR assays were carried out immediately, and the PCR products were purified. The purified PCR amplicons were cloned using the pGEM-T vector system (Promega, Madison, WI), and then one clone of each PCR product was sequenced with pUC/M13 primers (Promega, Madison, WI) on a Beckman-Coulter CEQ2000 capillary electrophoresis sequencer.
Phylogenetic analysis.
Partial ITS and SSU rDNA sequences of environmental samples were aligned with sequences of closely related organisms available on GenBank by using ClustalX (40). The dinoflagellate Karlodinium veneficum was chosen as the outgroup taxon, and the final alignment was refined manually. Bayesian analysis (likelihood) was carried out using the computer program MrBayes v. 3.12 (13). Nucleotides were treated as independent unordered characters of equal weight, and gaps were interpreted as missing data. The Bayesian analysis was run for 150,000 generations, using a 4-by-4 nucleotide-substitution, general time-reversible model with gamma-distributed among-site rate variation and a proportion of invariable sites (GTR+G+I). The resulting tree was sampled every 100 generations.
Microscopic observation.
The presence of Pfiesteria or Pfiesteria-like dinoflagellates in environmental samples collected every 2 weeks was checked by LM before the samples were stored at −70°C. Microscopic observations were compared to the real-time PCR results.
Nucleotide sequence accession numbers.
The sequences of field samples DE250505, DE281104, DE190105, DE140305, DE280305, DE120405, DE270405, and DE160505 have been deposited in GenBank under accession numbers DQ991364 to DQ991371, respectively. Sample codes: DE, Derwent River in Tasmania; the code for the sampling station (DE) is followed by a number indicating the day, month, and year of the sampling event (e.g., DE250505, Derwent River sampled on 25 May 2005).
RESULTS
Specificity of primers and a probe for real-time PCR assay.
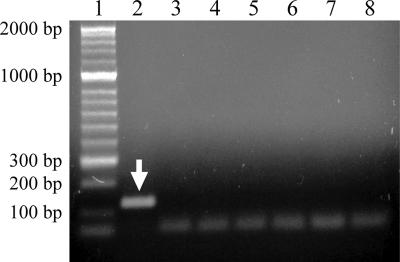
The sequences of the primer-probe combination had at least 31-bp mismatches with sequences of related organisms. BLAST searches showed that the sequences of the selected primers and probe matched only with the sequences of C. brodyi. In the real-time PCR using SYBR green, C. brodyi gave a strong positive fluorescent signal after 12 cycles, while closely related species produced faint signals only after 37 cycles (results not shown). Melting-curve analysis of the PCR products showed that the melting temperatures of C. brodyi and closely related species were 86°C and 75 to 77°C, respectively, indicating that the weak signals observed after 37 cycles were likely to be primer dimers (results not shown). Agarose gel analysis of the PCR products showed only one amplicon of the expected size (128 bp) for C. brodyi and no products for other species (Fig. 2). Real-time PCR experiments showed that the primer-probe set was specific to the target for which it was designed and did not react with unrelated organisms (results not shown), consistent with the predictions from sequence alignments. Sequence analyses indicated that other related species unavailable for experimental analysis in this study would also not be detected.
FIG. 2.
Agarose gel analysis showing C. brodyi-selective real-time PCR products. The real-time PCR was carried out using a SYBR green-based system with the primers CBITSF and CBITSR. The arrow indicates a positive real-time PCR product of 128 bp. Lanes: 1, 2-kbp-ladder molecular size marker; 2, C. brodyi; 3, P. piscicida; 4, P. shumwayae; 5, Karlodinium veneficum (=micrum); 6, Karenia mikimotoi; 7, Prorocentrum minimum; 8, no-template control.
For additional confirmation, heterotrophic dinoflagellates were isolated from a water sample collected on 28 November 2004 that showed a positive PCR result for C. brodyi. These isolates were established as clonal cultures. One of the cultures was examined by SEM and rDNA sequence analyses, and it corresponded with C. brodyi.
The specificities of the P. piscicida and P. shumwayae assays (SSU rDNA-based primers and mitochondrial cytochrome b-based primers, respectively) were tested with Australian genotypes of C. brodyi. No nonspecific reactions were produced by these primers. An environmental sample from the Derwent River samples assayed with P. piscicida-specific primers was also sequenced and was identical to the documented P. piscicida sequence.
Phylogenetic analysis.
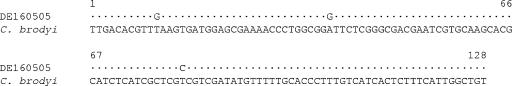
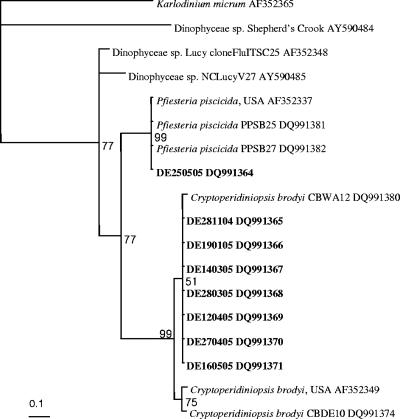
PCR amplicons generated from 7 environmental samples with C. brodyi-specific primers were identical, or differed by only 1 bp from each other, and differed from C. brodyi CBWA12 by 2.3 to 3.1%. The sequence comparison between C. brodyi and one of the field samples is shown in Fig. 3. Bayesian analysis of environmental samples and other related species showed that the sequences of the PCR amplicons clustered in a clade with C. brodyi at the 51 to 99% bootstrap values (Fig. 4). C. brodyi, P. piscicida, and other related dinoflagellates were rooted as distinct clades.
FIG. 3.
Alignment of partial ITS2 rDNA sequences from environmental sample and C. brodyi (CBWA12). The field sample DE160505 was sampled from the Derwent River on 16 May 2005 and was detected by C. brodyi-specific real-time PCR. Dots indicate nucleotides identical to those of C. brodyi.
FIG. 4.
Phylogram based on Bayesian analysis of partial ITS2 rDNA sequences (128 bp) of environmental samples and closely related organisms. The environmental samples DE281104 to DE160505 were sampled from the Derwent River from 28 November 2004 to 16 May 2005. These samples were detected by C. brodyi-specific real-time PCR and were sequenced. The sample DE250505 was collected on 25 May 2005 and amplified with P. piscicida-specific real-time PCR. The numbers adjacent to each node represent probabilities which were derived from 150,000 generations using a general time-reversible evolution model with gamma-distributed (0.248) among-site rate variation. The strains sequenced in the present study are shown in bold. Consistency index, 0.837; rescaled consistency index, 0.696; retention index, 0.831; homoplasy index, 0.162.
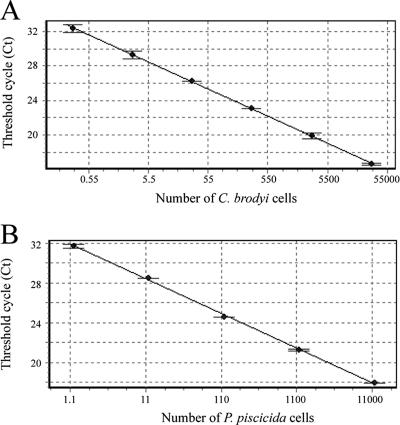
Standard curve and detection limit.
Standard curves were constructed with 10-fold serial dilutions of extracted DNA from C. brodyi and P. piscicida cultures. A strong linear relationship between the CT values and the log of the starting cell number was established, with a correlation coefficient (r2) of ≥0.997 (Fig. 5). These assays could detect less than one cell of the target species in a reaction. When 100-fold dilutions of DNA extracts from C. brodyi cultures (480, 4,800, and 48,000 cells) spiked with environmental samples were compared to the standard curves, concentrations of the spiked cells (4.0 ± 0.9, 41 ± 10, and 564 ± 99 cells, respectively; n = 3; P > 0.05 by Student's t test) did not significantly differ from those of standard curves (4.8, 48, and 480 cells). The DNA content in C. brodyi cells was also measured by using a PicoGreen dsDNA quantification kit, and the amount of DNA was 7,274 ± 1,254 fg (mean ± standard deviation) per cell.
FIG. 5.
Linear relationship between the CT values and the cell numbers for C. brodyi (r2 = 0.997) (A) and for P. piscicida (r2 = 0.998) (B). The standard errors from two measurements are shown as error bars.
PCR inhibitors from environmental water samples.
Environmental water samples may potentially include inhibitors affecting DNA extraction or subsequent PCR amplification (41). For example, the DNA extracts obtained from Derwent River samples completely inhibited real-time PCR (Table 3). For removal of the PCR inhibitors, two chemical treatments (PVPP and PVP360) were attempted. The DNA extracts from water samples were completely inhibitory even in the presence of PVPP and PVP360. Serial dilutions of the DNA extracts were quantified as well, and it was possible to detect 10-fold dilutions of the DNA extracts. Tenfold dilution along with PVPP treatment was still inhibitory, but it was more effective at reducing inhibition than PVP or nontreatment. Diluted 100-fold, the DNA extracts effectively reduced the effect of inhibitors and there was no significant difference between treatments (P > 0.05 by Student's t test) (Table 3). Accordingly, DNA extracted from Derwent River samples was diluted 100-fold and then used for real-time PCR.
TABLE 3.
Real-time PCR efficiencies for Derwent River samples (15 February 2005) with PVPP or PVP360 and different dilutions of the samples
| Sample dilution | Sample and treatment | CTa |
|---|---|---|
| None | C. brodyi DNA | 23.24 ± 0.15 |
| C. brodyi DNA + field sample | 0 | |
| C. brodyi DNA + field sample + PVPP | 0 | |
| C. brodyi DNA + field sample + PVP360 | 0 | |
| 10-fold | C. brodyi DNA | 26.93 ± 0.23 |
| C. brodyi DNA + field sample | 35.91 ± 0.36 | |
| C. brodyi DNA + field sample + PVPP | 29.26 ± 0.41 | |
| C. brodyi DNA + field sample + PVP360 | 30.46 ± 0.20 | |
| 100-fold | C. brodyi DNA | 30.18 ± 0.33 |
| C. brodyi DNA + field sample | 30.36 ± 0.19 | |
| C. brodyi DNA + field sample + PVPP | 30.76 ± 0.46 | |
| C. brodyi DNA + field sample + PVP360 | 30.66 ± 0.58 |
Mean±standard deviation of triplicate wells.
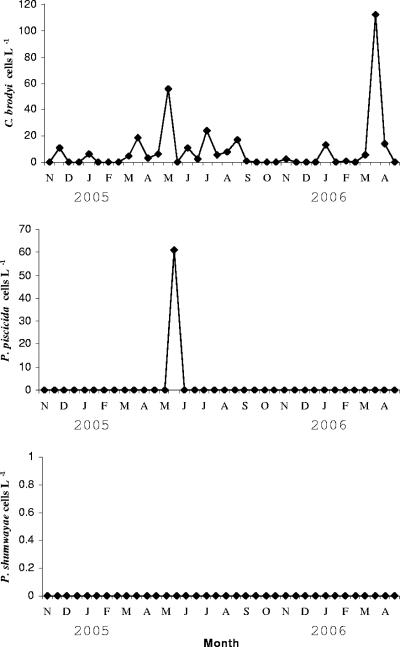
Seasonal changes in biomass of C. brodyi, P. piscicida, and P. shumwayae in Derwent River.
The 18-month field survey conducted in the Derwent River showed that C. brodyi cell concentrations were generally low (Fig. 6; Table 4). The DNA of C. brodyi was detected in all seasons during the survey period. C. brodyi abundance ranged from 1 to 24 cells liter−1 in most months, and cell numbers increased to 55 to 112 cells liter−1 in April and May. C. brodyi was more abundant in late summer to autumn in the Derwent River. In contrast, P. piscicida occurred only once, in May 2005, at 60 cells liter−1, and P. shumwayae was never detected. From the microscopic analysis, very low cell abundances of C. brodyi or Pfiesteria-like species were evident during the survey and they were occasionally observed (below quantification level) only in samples that tested positive by real-time PCR assay (Table 4). DNAs extracted from eight cultures from the Chapman River, Western Australia, were identified by using P. piscicida-, P. shumwayae-, and C. brodyi-specific real-time PCR assays. They exhibited increased signals in fluorescence detected by C. brodyi-specific real-time PCR.
FIG. 6.
Temporal variations in C. brodyi, P. piscicida, and P. shumwayae abundances in the Derwent River, Tasmania, during 2004 to 2006, quantified by species-specific real-time PCR. The values are the means of triplicate wells.
TABLE 4.
C. brodyi abundance in Derwent River from November 2004 to April 2006a
| Date | Cells liter−1 | LM result |
|---|---|---|
| 11/14/04 | 0 | − |
| 11/28/04 | 10.56 ± 7.14 | + |
| 12/11/04 | 0 | − |
| 12/22/04 | 0 | − |
| 1/19/05 | 5.89 ± 3.00 | + |
| 1/27/05 | 0 | − |
| 2/15/05 | 0 | − |
| 2/26/05 | 0 | − |
| 3/14/05 | 4.86 ± 4.02 | − |
| 3/28/05 | 18.70 ± 7.05 | − |
| 4/12/05 | 3.09 ± 2.40 | − |
| 4/27/05 | 6.04 ± 5.52 | − |
| 5/16/05 | 55.81 ± 3.17 | + |
| 5/25/05 | 0 | − |
| 6/10/05 | 10.59 ± 6.17 | − |
| 6/26/05 | 2.57 ± 1.50 | − |
| 7/17/05 | 24.00 ± 7.09 | − |
| 7/29/05 | 5.65 ± 3.96 | − |
| 8/15/05 | 7.70 ± 6.52 | − |
| 8/28/05 | 16.84 ± 5.11 | − |
| 9/11/05 | 1.26 ± 1.12 | − |
| 9/29/05 | 0 | − |
| 10/17/05 | 0 | − |
| 10/25/05 | 0 | − |
| 11/16/05 | 2.46 ± 1.50 | − |
| 11/30/05 | 0 | − |
| 12/12/05 | 0 | − |
| 12/21/05 | 0 | − |
| 1/15/06 | 13.23 ± 6.50 | + |
| 1/30/06 | 0 | − |
| 2/14/06 | 1.21 ± 0.96 | − |
| 2/27/06 | 0 | − |
| 3/18/06 | 5.35 ± 3.65 | − |
| 3/27/06 | 112.27 ± 2.50 | + |
| 4/10/06 | 14.02 ± 7.02 | − |
| 4/29/06 | 0 | − |
Cells liter−1 estimated using C. brodyi-specific real-time PCR. Values are the mean ± standard deviation of triplicate wells. Date, mo/day/yr; +, positive detection of C. brodyi or Pfiesteria-like species by LM analysis; −, negative detection.
DISCUSSION
Real-time specific PCR for ecological study.
In order to detect and enumerate C. brodyi from environmental samples, a C. brodyi-specific real-time PCR was developed from ITS2 rDNA. Currently, there are two formally described Pfiesteria species, P. piscicida and P. shumwayae. The number of C. brodyi in the field is difficult to quantify by microscopic methods, because C. brodyi superficially looks very similar to Pfiesteria spp. and numerous other small dinoflagellates. Real-time PCR with species-specific primers can provide high sensitivity for quantification of individual heterotrophic species (18, 42). Its application would contribute to understanding the temporal and geographic distributions of small estuarine dinoflagellates in the environment. The real-time PCR assay developed in this study provided specific and sensitive detection of C. brodyi in environmental samples. The detection limit for the real-time PCR assay was below one cell per reaction, similar to the results reported in other studies (3, 11, 18, 42).
When environmental samples are examined to obtain quantitative results, one main problem is false-negative results caused by real-time PCR inhibitors coextracted with target DNA. Surface water contains phenolic compounds, humic acids, and heavy metals, all known to be PCR inhibitors (41). The efficiency of removal of PCR inhibitors is generally dependent on the sample purity and volume. PVPP and PVP are known to remove humic compounds from DNA (10). In the present study, a dilution step and chemical treatments to remove these real-time PCR inhibitors were tested. The chemical treatment might not be sufficient to remove impurities from 10-liter Derwent River samples that might interfere with the real-time PCR assay. Processing low sample volumes and increasing PCR volumes may help to increase the DNA extraction efficiency and reduce concentrations of PCR inhibitors, so that overall sensitivity may be increased. However, where target organisms are present in low abundance in environmental water samples, stochastic effects on cell quantification become significant, by reducing the amount of the water sample being processed. Therefore, it may be a better strategy to sacrifice assay sensitivity than to process smaller volumes of water. However, an acceptable compromise should be decided by the objective of the study and potential inhibition should be checked by parallel analysis of natural samples with and without spiked C. brodyi cells.
Specificity of real-time PCR assay.
Previous studies have demonstrated that a primer set used in the SYBR green system can be used in a standard PCR assay to determine the presence or absence of target DNA (1). The primer set used in this study may also be applied in standard PCR assay format. The SYBR green and TaqMan probe systems have been known to have similar assay sensitivities, but with different levels of specificity (12, 16). In this study, no non-C. brodyi strain produced a signal in either the SYBR green-based or the TaqMan probe-based real-time PCR. However, when real-time PCR is applied to field samples, uncertainty in the assay specificity can result from the potential presence of unknown related species with similar sequences to the primer sites. Therefore, sequencing analysis of positive real-time PCR amplicons is required as further confirmation of assay specificity. More-variable DNA sequences, such as ITS rDNA regions, are useful genetic markers for the development of highly accurate species-specific real-time PCR. Homology searches of a nucleotide database revealed that the ITS2 rDNA region contains sequences unique to the target molecule, to which a primer-probe set of the real-time PCR was designed. Given the higher variability of the ITS regions compared to the SSU, LSU, and 5.8S genes, ITS sequence data have been used to analyze other dinoflagellates at the interspecies level (8, 21, 27). The higher divergence means that designing real-time PCR primer-probes based on ITS regions may reduce false positives.
Sequence and phylogenetic analyses.
One copy of an ITS2 rDNA fragment from each water sample was sequenced for confirmation of assay specificity. Three nucleotides differed from C. brodyi (CBWA12) DNA, and the sequenced samples were all identified as identical to C. brodyi by phylogenetic analysis. The sequence variation in Fig. 3 could be a result of intraclonal variation between rDNA loci in single C. brodyi cells, because only one clone was sequenced and the sequences were not obtained by consensus sequencing. Phylogenetic analyses of C. brodyi and other related dinoflagellates using ITS-5.8S rDNA sequences were previously described (23). When the partial ITS-based phylogram from this study was compared with the entire ITS-5.8S-based phylogram, the order of divergences among this dinoflagellate grouping differed slightly. Although the partial ITS-based phylogram clearly clustered the Derwent River samples (GenBank no. DQ991365 to DQ991371) with C. brodyi, the phylogram from this study may not resolve the phylogeny of C. brodyi and related dinoflagellates, because sort sequences (128 bp) were used to generate this phylogenetic tree.
Seasonal changes in C. brodyi, P. piscicida, and P. shumwayae in Derwent River, Tasmania.
Quantification of cells by real-time PCR could be conducted either by using the absolute number of gene copies in the target species or by relative concentrations of the target species. A single P. piscicida cell has been reported to have 100 to 200 rDNA copies (34). However, the number of rDNA copies in C. brodyi and P. shumwayae is not yet known. Therefore, the abundances of these species in environmental samples were calculated by comparisons with a standard curve generated from serial dilutions of DNA. The rDNA copy number in the natural populations was assumed to be the same as in the culture used as the standard.
The 18-month survey from the Derwent River showed that vegetative cells of C. brodyi were regularly present at very low abundance (mostly below 25 cells liter−1) but were slightly more abundant (55 to 112 cells liter−1) in April and May. Accurate quantification of C. brodyi in field samples has not been previously reported, due to lack of a quantitative molecular tool. Similarly low abundances were also shown for Pfiesteria species, which were detected by real-time PCR or PCR-fluorescent fragment detection techniques (9, 18, 42).
Zoospore sizes of C. brodyi range from 6 to 11 μm in length and from 5 to 10 μm in width (38). P. piscicida has been reported to have vegetative cell sizes ranging from 8.5 to 12 μm (20). In this study, a phytoplankton net (5-μm open mesh area) was used to concentrate large volumes (10 liters) of environmental samples for rapid sample processing. The mesh size was close to the size of the zoospores, and thus, it may allow some C. brodyi and P. piscicida cells to pass through the mesh. Centrifugation of the cells at 4,000 × g for 10 min also might be somewhat insufficient to settle all the cells. This could have affected the seasonal abundance results presented in Fig. 6 and underestimated the cell numbers, although this is unlikely to alter the conclusion of low abundance of C. brodyi and P. piscicida. Processing water samples through filtration with glass microfiber filters could minimize the potential loss of the organisms during sample collection.
Prey-dependent growth has been shown for C. brodyi and related dinoflagellates (6, 17, 26, 35). These dinoflagellates are known to feed on various microalgae, bacteria, protozoan ciliates, rotifers, tissues of fish, and planktonic shellfish larvae (5, 7, 35, 36). When preferred prey, such as cryptophytes, including R. salina, are supplied at high rates, higher cell densities can be obtained for these dinoflagellates (17, 26). Size, shape, and/or chemical cues of prey may affect food preferences of heterotrophic dinoflagellates (14, 25). Field experiments showed that the relationship between P. piscicida and total chlorophyll a concentrations may be influenced by appropriate prey rather than the abundance of total phytoplankton (9, 18). The seasonal changes in C. brodyi in the Derwent River thus may be associated with availability of suitable prey at times, implying that prey co-occurred in all seasons and prey cell densities increased during April and May, when Tasmanian coastal seawater reaches its annual temperature maximum. A positive correlation between a heterotrophic dinoflagellate and prey abundances has been shown in a previous study (15). When cell densities of Stoeckeria algicida (morphologically similar to Shepherd's Crook) peaked at 17,400 cells ml−1 in July, being the warmer season in Korea, cell densities of Heterosigma akashiwo (algal prey for S. algicida) decreased, indicating that S. algicida populations may have considerable grazing impact on H. akashiwo and contribute to the decline of H. akashiwo blooms (15). However, the microscopic counts used in that study were inadequate for discriminating morphologically similar species and may erroneously have included cells that resemble S. algicida, such as C. brodyi, Pfiesteria spp., and Karlodinium veneficum. As the protocol developed in this study allowed for the estimation of C. brodyi abundances from environmental samples, the use of real-time PCR will provide higher detection accuracies of the marine dinoflagellates in field-based studies than traditional methods.
Eight heterotrophic cultures identified by C. brodyi-specific real-time PCR.
Identification of eight cultures from the Chapman River, Western Australia, was confirmed by species-specific real-time PCR. This assay was completed in approximately 1.5 h. Therefore, this assay would be useful for the rapid and sensitive identification of clonal cultures, as well as environmental samples. However, for confirmation of the morphological and genetic characters of these cultures, SEM and sequencing analyses are required. The finding of C. brodyi in the Chapman River confirms that this species is widely distributed and commonly present in Australia. P. shumwayae has been detected previously on every continent by using molecular probes based on SSU rDNA, suggesting that P. shumwayae is a cosmopolitan species (4, 28, 29, 30, 31, 32, 33, 39). In Australia, this species was detected in various locations in Queensland, South Australia, Victoria, Western Australia, and Tasmania (P. A. Rublee, personal communication). However, no P. shumwayae was detected by mitochondrial cytochrome b-based probes during this study. The discrepancy between previous findings and this study raises a question about whether this is due to the limited sampling area of this study or false-positive reactions by the SSU rDNA-based probes. Companion work testing the specificity of the P. shumwayae-selective probes on C. brodyi and related species will be published elsewhere (25a).
In conclusion, the real-time PCR assay developed in this study demonstrated high sensitivity and specificity to detect and quantify C. brodyi from environmental samples. A real-time PCR assay was successfully implemented for the ecological study of this dinoflagellate. Our real-time PCR approach readily detected low abundances of C. brodyi (1 to 112 cells liter−1), revealing that C. brodyi is generally present as a minor component of phytoplankton in the Derwent River and that Pfiesteria species are rare in Tasmania. The detection of C. brodyi and P. piscicida at such low concentrations also suggests that real-time PCR, with appropriate adaptation of sample extraction, is an effective tool for investigating the distributions and seasonal dynamics of dinoflagellates in the marine environment.
Acknowledgments
We thank Lucy Harlow for technical assistance with real-time PCR and for valuable advice and suggestions. We also thank Vasele Hosja of the Water Ways Commission for providing Chapman River samples.
This work was supported by a grant from the Internal Research Grant Scheme (IRGS), University of Tasmania.
Footnotes
Published ahead of print on 23 February 2007.
REFERENCES
- 1.Audemard, C., S. R. Kimberly, and E. M. Burreson. 2004. Real-time PCR detection and quantification of the protistan parasite Perkinsus marinus in environmental waters. Appl. Environ. Microbiol. 70:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolch, C. J. S. 2001. PCR protocol for genetic identification of dinoflagellates directly from single cysts and plankton cells. Phycologia 40:162-168. [Google Scholar]
- 3.Bowers, H. A., T. Tengs, H. B. Glasgow, J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers, H. A., T. M. Trice, R. E. Magien, D. M. Goshorn, B. Michael, E. F. Schaefer, P. A. Rublee, and D. W. Oldach. 2006. Detection of Pfiesteria spp. by PCR in surface sediments collected from Chesapeake Bay tributaries (Maryland). Harmful Algae 5:342-351. [Google Scholar]
- 5.Burkholder, J. M., and H. B. Glasgow. 1995. Interactions of a toxic estuarine dinoflagellate with microbial predations and prey. Arch. Protistenkd. 145:177-188. [Google Scholar]
- 6.Burkholder, J. M., and H. B. Glasgow. 1997. Pfiesteria piscicida and other Pfiesteria-like dinoflagellates: behavior, impacts, and environmental controls. Limnol. Oceanogr. 42:1052-1075. [Google Scholar]
- 7.Burkholder, J. M., H. B. Glasgow, and N. Deamer-Melia. 2001. Overview and present status of the toxic Pfiesteria complex (Dinophyceae). Phycologia 40:186-214. [Google Scholar]
- 8.Connell, L. M. 2001. Rapid identification of marine algae (Raphidophyceae) using three-primer PCR amplification of nuclear internal transcribed spacer (ITS) regions from and archived material. Phycologia 41:15-21. [Google Scholar]
- 9.Coyne, K. J., D. A. Hutchins, C. E. Hare, and S. C. Cary. 2001. Assessing temporal and spatial variability in Pfiesteria piscicida distribution using molecular probing techniques. Aquat. Microb. Ecol. 24:275-285. [Google Scholar]
- 10.Cullen, D. W., and P. R. Hirsch. 1998. Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biol. Biochem. 30:983-993. [Google Scholar]
- 11.Galluzzi, L., A. Penna, E. Bertozzini, M. Vila, E. Garcés, and M. Magnani. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Guillard, R. R. L., and J. H. Ryther. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea cleve. Can. J. Microbiol. 8:229-239. [DOI] [PubMed] [Google Scholar]
- 12.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time PCR and application of this technique for examination of cheeses. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 14.Jeong, H. J., and M. I. Latz. 1994. Growth and grazing rates of the heterotrophic dinoflagellates Protoperidinium spp. on red tide dinoflagellates. Mar. Ecol. Prog. Ser. 106:173-185. [Google Scholar]
- 15.Jeong, H. J., J. S. Kim, J. Y. Park, J. H. Kim, S. Kim, I. H. Lee, S. H. Lee, J. H. Ha, and W. H. Yih. 2005. Stoeckeria algicida n. gen., n. sp. (Dinophyceae) from the coastal water off southern Korea: morphology and small subunit ribosomal DNA gene sequence. J. Eukaryot. Microbiol. 52:382-390. [DOI] [PubMed] [Google Scholar]
- 16.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, S., M. R. Mulholland, H. Zhang, T. N. Feinstein, F. J. Jochem, and E. J. Carpenter. 2004. Intense grazing and prey-dependent growth of Pfiesteria piscicida (Dinophyceae). J. Phycol. 40:1062-1073. [Google Scholar]
- 18.Lin, S., H. Zhang, and A. Dubois. 2006. Wide distribution and low abundance of Pfiesteria piscicida as detected by mtDNA-18S rDNA real-time PCR. J. Plankton Res. 28:667-681. [Google Scholar]
- 19.Litaker, R. W., P. A. Tester, A. Colomi, M. G. Levy, and E. J. Noga. 1999. The phylogenetic relationship of Pfiesteria piscicida, cryptoperidiniopsoid sp., Amyloodinium ocellatum and a Pfiesteria-like dinoflagellate to other dinoflagellates and apicomplexans. J. Phycol. 35:1379-1389. [Google Scholar]
- 20.Litaker, R. W., M. W. Vandersea, S. R. Kibler, V. J. Madden, E. J. Noga, and P. A. Tester. 2002. Life cycle of the heterotrophic dinoflagellate Pfiesteria piscicida (Dinophyceae). J. Phycol. 38:442-463. [Google Scholar]
- 21.Litaker, R. W., M. W. Vandersea, S. R. Kibler, K. S. Reece, N. A. Stokes, K. A. Steidinger, D. F. Millie, B. J. Bendis, R. J. Pigg, and P. A. Tester. 2003. Identification of Pfiesteria piscicida (Dinophyceae) and Pfiesteria-like organisms using internal transcribed spacer-specific PCR assays. J. Phycol. 39:754-761. [Google Scholar]
- 22.Marshall, H. G., D. Seaborn, and J. Wolny. 1999. Monitoring result for Pfiesteria piscicida and Pfiesteria-like organisms from Virginia waters in 1998. Virginia J. Sci. 50:287-298. [Google Scholar]
- 23.Marshall, H. G., P. E. Hargraves, J. M. Burkholder, M. W. Parrow, M. Elbrächter, E. H. Allen, V. M. Knowlton, P. A. Rublee, W. L. Hynes, T. A. Egerton, D. L. Remington, K. B. Wyatt, A. J. Lewitus, and V. C. Henrich. 2006. Taxonomy of Pfiesteria (Dinophyceae). Harmful Algae 5:481-496. [Google Scholar]
- 24.Mason, P. L., W. K. Vogelbein, L. W. Haas, and J. D. Shields. 2003. An improved stripping technique for lightly armored dinoflagellates. J. Phycol. 39:253-258. [Google Scholar]
- 25.Naustvoll, L. J. 2000. Prey size spectra and food preferences in thecate heterotrophic dinoflagellates. Phycologia 39:187-198. [Google Scholar]
- 25a.Park, T. G., C. J. S. Bolch, and G. M. Hallegraeff. Morphological and molecular genetic characterization of Cryptoperidiniopsis brodyi (Dinophyceae) from Australia-wide isolates. Harmful Algae, in press.
- 26.Parrow, M. W., and J. M. Burkholder. 2003. Estuarine heterotrophic Cryptoperidiniopsoids (Dinophyceae): life cycle and culture studies. J. Phycol. 39:678-696. [Google Scholar]
- 27.Penna, A., and M. Magnani. 1999. Identification of Alexandrium (Dinophyceae) species using PCR and rDNA-targeted probes. J. Phycol. 35:615-621. [Google Scholar]
- 28.Rhodes, L. L., J. M. Burkholder, H. B. Glasgow, P. A. Rublee, C. Allen, and J. E. Adamson. 2002. Pfiesteria shumwayae (Pfiesteriaceae) in New Zealand. N. Z. J. Mar. Freshw. Res. 36:621-630. [Google Scholar]
- 29.Rhodes, L. L., J. E. Adamson, P. A. Rublee, and E. Schaefer. 2006. Geographic distribution of Pfiesteria spp. (Pfiesteriaceae) in Tasman Bay and Canterbury, New Zealand (2002-03). N. Z. J. Mar. Freshw. Res. 40:211-220. [Google Scholar]
- 30.Rublee, P. A., J. W. Kempton, E. F. Schaefer, C. Allen, J. Harris, D. W. Oldach, H. Bowers, T. Tengs, J. M. Burkholder, and H. B. Glasgow. 2001. Use of molecular probes to assess geographic distribution of Pfiesteria species. Environ. Health Perspect. 109:765-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rublee, P. A., C. Allen, E. Schaefer, L. Rhodes, J. Adamson, C. Lapworth, J. Burkholder, and H. Glasgow. 2004. Global distribution of toxic Pfiesteria complex species detected by PCR assay, p. 320-322. In K. A. Steidinger, J. H. Landsberg, C. R. Tomas, and G. A. Vargo (ed.), Harmful algae, 2002. Florida Fish And Wildlife Conservation Commission, Florida Institute of Oceanography and Intergovernmental Oceanographic Commission of UNESCO, St. Petersburg, FL.
- 32.Rublee, P. A., D. L. Reminfton, E. F. Schaefer, and M. M. Marshall. 2005. Detection of the dinozoans Pfiesteria piscicida and P. shumwayae: a review of detection methods and geographic distribution. J. Eukaryot. Microbiol. 52:83-89. [DOI] [PubMed] [Google Scholar]
- 33.Rublee, P. A., R. Nuzzi, R. Waters, E. F. Schaefer, and J. M. Burkholder. 2006. Pfiesteria piscicida and Pfiesteria shumwayae in coastal waters of Long Island, New York, USA. Harmful Algae 5:374-379. [Google Scholar]
- 34.Saito, K., T. Drgon, J. A. Robledo, D. N. Krupatkina, and G. R. Vasta. 2002. Characterization of the rRNA locus of Pfiesteria piscicida and development of standard and quantitative PCR-based detection assays targeted to the nontranscribed spacer. Appl. Environ. Microbiol. 68:5394-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seaborn, D. W., A. M. Seaborn, W. M. Dunstan, and H. G. Marshall. 1999. Growth and feeding studies on the algal feeding stage of a Pfiesteria-like dinoflagellate. Virginia J. Sci. 50:337-344. [Google Scholar]
- 36.Springer, J. J., S. E. Shumway, J. M. Burkholder, and H. B. Glasgow. 2002. Interaction between the toxic estuarine dinoflagellate Pfiesteria piscicida and two species of bivalve mollusks. Mar. Ecol. Prog. Ser. 245:1-10. [Google Scholar]
- 37.Steidinger, K. A., J. Landsberg, R. W. Richardson, E. Truby, B. Blaskesley, P. Scott, P. Tester, T. Tengs, P. Mason, S. Morton, D. Scaborn, W. Litaker, K. Reece, D. Oldach, L. Haas, and G. Vasta. 2001. Classification and identification of Pfiesteria and Pfiesteria-like species. Environ. Health Perspect. 109:661-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steidinger, K. A., J. H. Landsberg, P. L. Mason, W. K. Vogelbein, P. A. Tester, and R. W. Litaker. 2006. Cryptoperidiniopsis brodyi gen. et sp. nov. (Dinophyceae), a small lightly armored dinoflagellate in the Pfiesteriaceae. J. Phycol. 42:951-961. [Google Scholar]
- 39.Tango, P., R. Magnien, D. Goshorn, H. Bowers, B. Michael, R. Karrh, and D. Oldach. 2006. Associations between fish health and Pfiesteria spp. in Chesapeake Bay and mid-Atlantic estuaries. Harmful Algae 5:352-362. [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewnaik, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, H., and S. Lin. 2005. Development of a cob-18S rRNA gene real-time PCR assay for quantifying Pfiesteria shumwayae in the natural environment. Appl. Environ. Microbiol. 71:7053-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]