Abstract
Expression of 14-3-3 σ (σ) is induced in response to DNA damage, and causes cells to arrest in G2. By SAGE (serial analysis of gene expression) analysis, we identified σ as a gene whose expression is 7-fold lower in breast carcinoma cells than in normal breast epithelium. We verified this finding by Northern blot analysis. Remarkably, σ mRNA was undetectable in 45 of 48 primary breast carcinomas. Genetic alterations at σ such as loss of heterozygosity were rare (1/20 informative cases), and no mutations were detected (0/34). On the other hand, hypermethylation of CpG islands in the σ gene was detected in 91% (75/82) of breast tumors and was associated with lack of gene expression. Hypermethylation of σ is functionally important, because treatment of σ-non-expressing breast cancer cell lines with the drug 5-aza-2′-deoxycytidine resulted in demethylation of the gene and synthesis of σ mRNA. Breast cancer cells lacking σ expression showed increased number of chromosomal breaks and gaps when exposed to γ-irradiation. Therefore, it is possible that loss of σ expression contributes to malignant transformation by impairing the G2 cell cycle checkpoint function, thus allowing an accumulation of genetic defects. Hypermethylation and loss of σ expression are the most consistent molecular alterations in breast cancer identified so far.
Although many studies have identified critical genetic and epigenetic changes that mark the transformation of cells in tissues such as colon, pancreas, and lung, similar studies in breast cancer have met with limited success. In this paper we report the identification of a gene, 14-3-3 σ (σ), whose expression is undetectable in 94% (45/48) of breast tumors.
σ was originally identified as an epithelial-specific marker, HME1, which was down-regulated in a few breast cancer cell lines but not in cancer cell lines derived from other tissue types (1). Later studies showed that σ protein (also called stratifin) was abundant in differentiated squamous epithelial cells, but decreased by 95% in simian virus 40-transformed epithelial cells and in primary bladder tumors (2–4).
We investigated the molecular mechanism underlying the low expression of σ in breast cancers. We find that genetic alterations such as loss of heterozygosity (LOH) and intragenic mutations are not major contributing mechanisms for lack of σ expression. Instead, we show that hypermethylation of the CpG-rich region in the σ gene is associated with its transcriptional silencing in the majority of breast tumors. Treatment of breast cancer cell lines that do not express σ with the DNA methyltransferase inhibitor, 5-aza-2′-deoxycytidine (5-aza-dC), leads to partial demethylation of this CpG island and synthesis of σ mRNA. Thus, hypermethylation appears to be responsible for silencing of σ gene expression.
Recent studies have shed light on the function of σ. It was originally identified as a p53-inducible gene that is responsive to DNA damaging agents (5). σ apparently sequesters the mitotic initiation complex, cdc2–cyclin B1, in the cytoplasm after DNA damage (6). This prevents cdc2–cyclin B1 from entering the nucleus where the protein complex would normally initiate mitosis. In this manner, σ induces G2 arrest and allows the repair of damaged DNA (5, 6). Of note, we find that breast cancer cells that do not express σ accumulate significantly more G2-type chromosomal aberrations than cells that express σ. These results suggest that σ participates in the G2 checkpoint control in breast cells. We propose that loss of σ gene expression plays a significant role in breast cancer, as it may facilitate the accumulation of genetic damage conducive to malignant transformation.
Materials and Methods
Cell Lines and Tissues.
The breast cancer cell lines Hs578T, MDA-MB-231, MDA-MB-435, and MCF-7 and the human mammary epithelial cell (HMEC) lines MCF-10A and HBL-100 were obtained and maintained according to instructions (American Type Culture Collection). The two matched tumor cell lines, 21PT and 21MT were propagated as described (7). Cultured normal finite life span HMEC strains, 161, 184, 172, and 48, and the conditionally and fully immortal HMEC lines, 184A1 (passage 15 and 99), and 184B5 were grown as described (http://www.lbl.gov/LBL-Programs/mrgs/review.html). Two additional finite life span HMEC strains (no. 04372 and no. 16637) were grown according to specifications (Clonetics, San Diego). Primary breast tumor tissues were obtained immediately after surgical resection at the Johns Hopkins University or Duke University and stored frozen at −80°C. Microscopic examination of representative tissue sections from each tumor revealed that these samples contained >50% tumor cells. Histopathologically, all of the breast cancer samples from Johns Hopkins were invasive ductal carcinomas (not otherwise specified; NOS). Eighteen of 20 samples from Duke University were also invasive ductal carcinomas (NOS), whereas 2 of 20 were lobular carcinomas. Microdissection of primary tumor cryosections was performed by using a laser capture microscope (8) or by manually scraping the cells with a 20G needle under ×40 magnification (9).
Northern Blot Analysis.
Total RNA was isolated from primary tumor tissues by using Trizol Reagent (Life Technologies). Five micrograms were resolved on 1.5% agarose/formaldehyde gels and transferred to a nylon filter by using standard methods (Gene Screen; DuPont). A 375-bp σ-specific probe was generated by using MCF-10A cDNA as a template and the primers 5′-ACAGGGGAACTTTATTGAGAGG-3′ and 5′-AAGGGCTCCGTGGAGAGGG-3′ (5). Hybridizations were done in Quikhyb (Stratagene) according to the manufacturer's instructions. Filters were exposed to autoradiographic film for up to 5 days. To test for uniform loading of the samples, blots were stripped and reprobed with a 1.5-kb DNA fragment specific for 18S rRNA (ATCC clone HHCSA65).
LOH Studies.
A TG repeat sequence in the 3′ untranslated region of σ was amplified by using: 5′-GAGGAGTGTCCCGCCTTGTGG-3′ (sense) and 5′- GTCTCGGTCTTGCACTGGC-3′ (antisense) primers, which yields a product of 117 bp. The 25-μl reactions contained 50 ng of template DNA (10), 17 mM NH4SO4, 67 mM TrisCl (pH 8.8), 6.7 mM MgCl2, 1% DMSO, 1.5 mM dNTP, 20 ng of each primer, 2 ng of [γ-32P]-labeled sense primer, and 0.5 μl Taq polymerase. PCR conditions were as follows: 1 cycle at 94°C for 90s; 35 cycles at 94°C for 1 min, 57°C for 30s, and 72°C for 30s; and 1 cycle at 72°C for 5 min. PCR products were fractionated on a sequencing gel, which was exposed to autoradiographic film overnight (10).
Mutation Analysis.
A 1.2-kb PCR product, encompassing the entire σ coding sequence, was generated by using two primers, 5′-GTGTGTCCCCAGAGCCATGG-3′ (sense) and 5′-GTCTCGGTCTTGCACTGGCG-3′ (antisense). The PCR contained 50 ng of DNA, 6.4% DMSO, 1.5 mM dNTPs, 100 ng of each primer, and 0.5 μl Taq polymerase in a 50 μl reaction volume. [α-33P]-Labeled cycle sequencing was performed by using the Amplicycle sequencing kit (Perkin-Elmer). Four different [α-33P]-labeled primers were used to sequence the entire σ coding sequence: 5′-CACCTTCTCCCGGTACTCACG-3′ (antisense), 5′-GAGCTCTCCTGCGAAGAG-3′ (sense), 5′-GAGGAGGCCATCCTCTCTGGC-3′ (sense), and 5′-TCCACAGTGTCAGGTTGTCTCG-3′ (antisense).
Transfection of Human Breast Cancer Cell Lines.
A total of 1.5 × 105 cells of MCF-7, MDA-MB-231, and Hs578T, or 2.5 × 105 cells of MDA-MB-435 breast cancer cells were seeded in six-well plates. The following day, transfections were performed by using Trans IT-LT1 (Mirus) as per the manufacturer's instructions. Plasmids used in the transient transfections include: KKH luciferase, containing 4 kb of the σ promoter linked to the luciferase gene in the pGL3-Basic vector (Promega); pCMV-β-gal (Clontech), which was used to correct for the efficiency of transfection; and pGL3-Basic (Promega), which was used as a negative vector control against which KKH luciferase activities were compared. Two micrograms of luciferase reporter plasmid or the pGL3-Basic vector control and 0.5 μg of CMV-β-gal reporter plasmid were used for each transfection.
Luciferase and β-Galactosidase Assays.
Cell lysates were made ≈48 h posttransfection as per the manufacturer's instructions (Promega, Luciferase Assay System). Luciferase and β-galactosidase activities were quantitated by using the luciferase assay system (Promega) and the Aurora GAL-XE reporter gene assay (ICN Pharmaceuticals), respectively. Experiments were done in triplicate, and results of at least two independent experiments are shown. Luciferase activity was first normalized for efficiency of transfection by using the ratio of luciferase to β-galactosidase activity. For each transfected cell line, the results were compared with the mean of pGL3 vector control levels and expressed as fold elevated expression above pGL3. The means and standard deviations of the results of all experiments were calculated and shown as error bars.
Sodium Bisulfite DNA Sequencing.
Genomic DNA was subjected to sodium bisulfite modification as previously described (11). Bisulfite-converted DNA was amplified, as described above, using primers that encompass the first exon of the σ gene: 5′-GAGAGAGTTAGTTTGATTTAGAAG-3′ (sense primer with start at nt 8641) and 5′-CTTACTAATATCCATAACCTCC-3′ (antisense primer with start at nt 9114) which generated a 474-bp PCR product. Conditions for PCR were as follows: 1 cycle at 95°C for 5 min; 35 cycles at 95°C for 45 s, 55°C for 45 s and 72°C for 60 s; and 1 cycle at 72°C for 4 min. The product was purified by using a Qiagen PCR purification kit (Qiagen, Chatsworth, CA) and sequenced by using the sense primer with an Applied Biosystems automated fluorescent sequencer according to the manufacturer's instructions.
Methylation-Specific PCR (MSP).
One microgram of genomic DNA was treated with sodium bisulfite as described (11) and was analyzed by MSP by using a primer set that covered CG dinucleotide numbers 3, 4, 8, and 9 (see Fig. 4). Primers specific for methylated DNA [5′-TGGTAGTTTTTATGAAAGGCGTC-3′ (sense) and 5′-CCTCTAACCGCCCACCACG-3′ (antisense)], and primers specific for unmethylated DNA [5′-ATGGTAGTTTTTATGAAAGGTGTT-3′ (sense) and 5′-CCCTCTAACCACCCACCACA-3′ (antisense)] yielded a 105- to 107-bp PCR product. The PCR conditions were as follows: 1 cycle of 95°C for 5 min; 31 cycles of 95°C for 45 s, 56°C for 30 s and 72°C for 30 s; and 1 cycle of 72°C for 4 min.
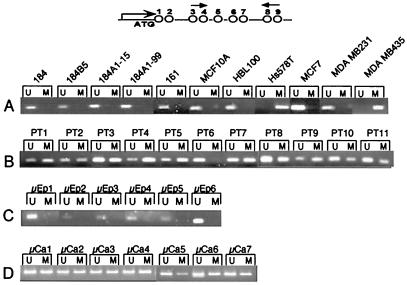
Figure 4.
MSP analysis of DNA from cell line and primary tumors. (A) HMEC finite life span strains (184 and 161) and immortalized HMEC lines (184B5, 184A1-15, 184A1-99, MCF-10A, and HBL-100), and breast cancer cell lines, Hs578T, MCF-7, MDA-MB-231, and MDA-MB-435. (B) PT, primary breast tumors. (C) Ep, microdissected epithelial ducts from six normal breast tissues. (D) Ca, microdissected carcinoma cells from seven primary breast tumors. The CpGs in σ spanned by the primers are shown above.
Treatment of Cells with 5-aza-dC.
Cells were seeded at a density of 2 × 106 cells/100-mm plate. Twenty-four hours later cells were treated with 0.75 μM 5-aza-dC (Sigma) (12). Total cellular RNA and genomic DNA were isolated from the cells at 0 and 3 days after addition of 5-aza-dC as described above.
RT-PCR.
RNA was treated with RNase-free DNase (Boehringer Mannheim) (1 μg/μl) for 2 h at 37°C, followed by heat inactivation at 65°C for 10 min. RT reactions contained 1 μg DNase-treated RNA, 0.25 μg/μl pdN6 random primers (Pharmacia), 1× first strand buffer (GIBCO-BRL), 0.5 mM dNTP (Pharmacia), and 200 units MMLV (Moloney murine leukemia virus)-RT (GIBCO-BRL), and were incubated for 1 h at 37°C. PCR was performed by using the σ-specific primers 5′-GTGTGTCCCCAGAGCCATGG-3′ and 5′-ACCTTCTCCCGGTACTCACG-3′ by using buffer conditions described above. The PCR conditions were as follows: 1 cycle of 95°C for 5 min; 30 cycles of 60°C for 45 s, 72°C for 45 s and 95°C for 45 s. The PCR samples were resolved by electrophoresis in a 2% agarose gel.
Assay for G1 and G2 Checkpoint and Chromosomal Aberrations.
The G1 cell cycle checkpoint and chromosomal aberrations in mitosis were assessed as described previously (13). Specifically, cells in plateau phase were irradiated with 3 Gy, subcultured after 24 h, and metaphases were collected. G1 type aberrations were examined at metaphase. All categories of asymmetric chromosome aberrations were scored: dicentrics, centric rings, interstitial deletions/acentric rings, and terminal deletions.
The efficiency of G2 checkpoint control was evaluated by measuring the proportion of cells in metaphase after irradiation. Chromosomal aberrations at mitosis were assessed by counting chromatid breaks and gaps per metaphase as described recently (14). Specifically, cells in exponential growth phase were irradiated with 1 Gy. Metaphases were harvested 45 and 90 min following irradiation and examined for chromatid type breaks and gaps. Fifty metaphases each were scored for G1 and G2 types of chromosomal aberrations. Mitotic index was also determined by a procedure described previously (13, 14).
Introduction of σ into the σ-Negative Breast Cancer Cell Line MDA-MB-435 by Adenoviral Infection.
Cells were seeded and grown to 50% confluency. Adenovirus encoding either σ or β-galactosidase (5) was added to the culture at a multiplicity of infection of 5000:1, and infection was allowed to take place overnight. The cells were harvested, fixed, and stained with Hoechst dye and subjected to fluorescence-activated cell sorter analysis (5).
Results
σ Expression in Normal, Immortalized, and Tumorigenic Breast Epithelial Cells.
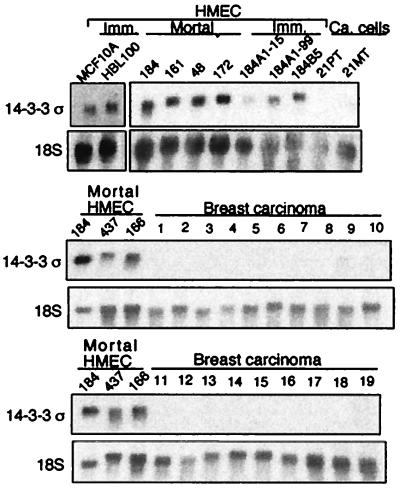
By SAGE (serial analysis of gene expression) analysis, σ was found to be expressed at an average of 7-fold lower levels in three human breast cancer cell lines 21PT, 21MT, and MDA-MB-468 than in two populations of normal finite life span HMECs (38). Northern blot analysis was performed to confirm this finding in other breast cancer cell lines and in primary breast tumors. No expression of σ was detected in 45 of 48 (94%) primary tumors. The results from 19 representative primary tumors are shown in Fig. 1. In contrast, σ was expressed at easily detectable levels in all six finite life span HMEC strains and five immortalized but nontumorigenic HMECs (Fig. 1). These results indicate that loss of σ gene expression is a frequent event in human breast cancer.
Figure 1.
Northern blot analysis of σ gene expression in HMECs and primary breast tumors. Data from 6 finite life span HMEC strains, 184, 161, 48, 172, 437, and 166 (Clonetics HMECs, 04372 and 16637); 5 immortal HMEC lines, 184B5, 184A1 (passage 15), 184A1 (passage 99), MCF-10A, and HBL-100; breast cancer cell lines 21PT and 21MT; 19 representative breast carcinomas are shown.
Genetic Alterations Within the σ Gene.
Possible causes for loss of σ gene expression in breast tumors include deletion of the chromosomal region containing the gene or intragenic mutations that lead to decreased mRNA stability. σ localizes to chromosome 1p35, an arm that has been extensively studied for LOH in breast cancer (5, 15). LOH has been observed for the 1p32–36 region at a frequency of 15–25%. However, it is not known whether the region lost in these tumors includes σ (16–19). Therefore, we looked for loss of σ by using a TG repeat sequence within the 3′ untranslated region of the σ gene itself. By using primers that span the TG repeats, we amplified the locus in 45 sets of normal and tumor DNA pairs. Twenty of 45 (44%) of the patients were found to be heterozygous with respect to the length of the PCR-product. Only 1 of the 20 tumor specimens exhibited LOH (Table 1). Eleven of these 20 samples were tested by Northern blot analysis, and no σ transcripts were detectable (data not shown). These results led us to examine whether there were smaller genetic changes within the coding region of σ. We PCR-amplified and sequenced the entire 1190-bp coding region from σ-non-expresssing (σ-negative) breast cancer cell lines, MDA-MB-435 and Hs578T and seven primary tumor tissues. No mutations were found (data not shown). In addition, 25 primary tumor DNA samples were analyzed by single-stranded conformation polymorphism, and no abnormalities were detected (data not shown). These results suggest that genetic alterations within σ is not a primary mechanism for loss of gene expression.
Table 1.
Incidence of σ alterations in breast cancer
| Sample | σ expression, Northern blot analysis | No. with methylated σ/total
|
No. with LOH/total, TG repeat PCR | No. with mutation/total
|
||
|---|---|---|---|---|---|---|
| Sequencing | MSP | Sequencing | SSCP | |||
| Normal breast | ||||||
| Mortal HMEC strains | 6 /6 | 0 /1 | 0 /3 | |||
| Immortal HMEC lines | 5 /5 | 0 /1 | 0 /5 | |||
| Reduction mammoplasty, microdissected epithelium | 0 /6 | 0 /1 | ||||
| Breast cancer | ||||||
| Cell lines | ||||||
| MCF-7 | + | − | − | |||
| MDA-MB-231 | + | − | − | |||
| MDA-MB-435 | − | + | + | − | ||
| Hs 578t | − | + | + | − | ||
| Primary tumors | 2 /45 | 10 /10 | 43 /50 | 1 /20 | 0 /7 | 0 /25 |
| Microdissected carcinoma | 32 /32 | |||||
Epigenetic Alterations of the σ Gene.
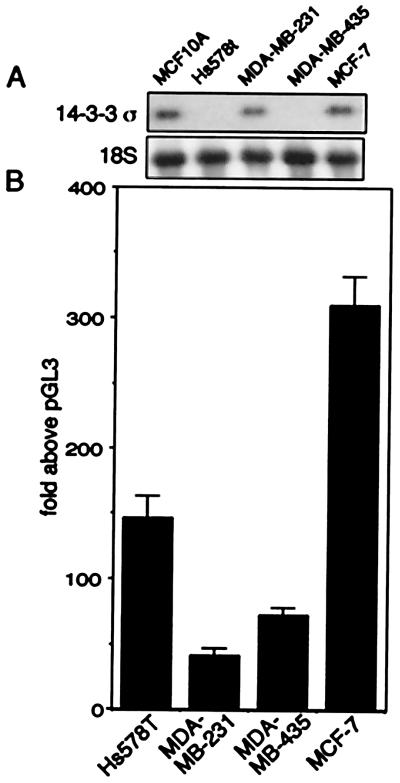
We next tested whether the lack of σ mRNA was due to deficiencies in factors required for σ transcription. The two breast cancer cell lines, MDA-MB-435 and Hs578T, served as model systems for σ-negative primary tumors that harbored wild-type σ alleles, whereas the two breast cancer cell lines, MCF-7 and MDA-MB-231, served as σ-positive controls because they both express detectable levels of σ (Fig. 2A). The plasmid KKH-luciferase contains 4 kb of sequence upstream of the transcriptional start site of σ linked to the luciferase reporter gene; this upstream region contains the sequences necessary for p53 and γ-irradiation-inducible transcription of σ (5). Following transient transfection of the four cell lines with the reporter plasmid, we observed high levels of expression (70- to 300-fold above the promoterless parental vector) in both σ-negative and σ-positive breast cancer cell lines (Fig. 2B). These results indicate that the σ-negative breast cancer cells, like the σ-positive cells, were able to support transcription from the σ promoter equally well, and contained factors required for transcription.
Figure 2.
σ-positive and σ-negative cells support transcription from an exogenous promoter-reporter construct. (A) Northern blot analysis of σ expression in breast cancer cell lines. HMEC MCF-10A, positive control. Blots were reprobed with 18S rRNA. (B) σ promoter linked-luciferase activity in σ-positive and σ-negative cells following transient transfection.
σ has a CpG-rich region (CpG island) within its first and only exon that begins near the transcription initiation site and ends ≈800 bp downstream. To explore a role of hypermethylation in silencing σ gene expression, we first determined the nucleotide sequence of this region after treating the DNA with sodium bisulfite (20). PCR primers were designed to amplify a region spanning 27 CpG dinucleotides within the CpG island. No significant methylation was observed by using DNAs from four σ-positive cells including the finite life span HMEC 184, immortal MCF-10A, and two tumorigenic breast cancer cell lines, MCF-7 and MDA-MB-231. In contrast, DNAs from two σ-negative breast cancer cell lines, Hs578T and MDA-MB-435, were fully methylated at all of the CpG sites (Fig. 3). Because there was a strong correlation between σ methylation status and mRNA expression in all of the cell lines examined, we also analyzed 10 σ-negative primary breast tumors (Table 1). All of the tumor DNAs exhibited partial or complete methylation of the 27 CpG dinucleotides (Fig. 3).
Figure 3.
Nucleotide sequencing of σ-DNA following NaHSO3-treatment of normal and breast tumor samples. ○, Unmethylated; ●, completely methylated; ◐, partially methylated cytosines.
Next, an MSP assay was used to detect methylation of the CpG island by using primers spanning the region between CpG dinucleotides 3 and 9 within the σ gene (Fig. 4). Primers were designed that take advantage of the nucleotide sequence differences between methylated and unmethylated DNA as a result of bisulfite modification. By this method, 5/5 σ-positive finite life span HMEC strains were completely unmethylated (Fig. 4A). In addition, DNAs from the σ-positive immortalized breast epithelial cells (MCF-10A, HBL-100) and breast cancer cell lines (MCF-7 and MDA-MB-231), were also unmethylated at the sites examined (Fig. 4A). In contrast, DNAs from the σ-negative breast cancer cell lines, Hs578T and MDA-MB-435, were fully methylated (Fig. 4A). Similarly, 43 of 50 samples from primary breast tumors were partially or completely methylated (representative results shown in Fig. 4B). Of these 43 tumors, 26 were examined by Northern blot analysis, and all 26 lacked detectable σ gene expression (Fig. 1B and data not shown). Three of the seven unmethylated breast tumor samples also lacked σ transcripts (data not shown); the expression pattern for the remainder was not tested. These results suggest that aberrant methylation of σ is a frequent event in breast cancer, but that other mechanisms are responsible for silencing the gene in a small fraction of breast tumors.
Previous reports indicate that σ gene expression is restricted to differentiated epithelial cells (1–4). To clearly ascertain the cellular origin of methylated DNA, normal and tumor tissues were microdissected and analyzed for σ methylation by MSP. All six DNA samples of microdissected mammary epithelial cells obtained from reduction mammoplasty specimens were unmethylated (Fig. 4C). In contrast, all 32 samples of DNA from microdissected breast carcinomas were methylated within the σ CpG island (representative cases shown in Fig. 4D). These results indicate that hypermethylation of the σ gene is associated with loss of gene expression in the majority of primary breast tumors. The data from gene expression, genetic, and epigenetic studies are summarized in Table 1.
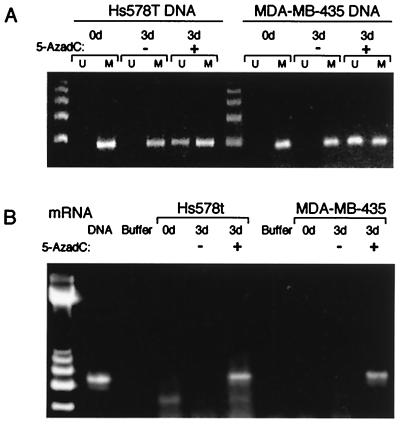
To determine the effect of methylation on σ gene expression, we treated the two fully methylated, σ-negative cell lines, Hs578T and MDA-MB-435, with the DNA methyltransferase inhibitor, 5-aza-dC. Treatment of cells with 0.75 μM 5-aza-dC for 3 days led to demethylation of the CpG rich region encompassed by the MSP primers (Fig. 5A). Moreover, 5-aza-dC treatment resulted in reactivation of gene expression, as demonstrated by RT-PCR (Fig. 5B). These results indicate that methylation is at least partially responsible for loss of σ transcription in breast cancer cells.
Figure 5.
Treatment with 5-aza-dC triggers re-expression of σ. (A) MSP analysis of DNA from breast cancer cell lines MDA-MB-435 and Hs578T before and after treatment for 3 days with 5-aza-dC. (B) RT-PCR of cells from A shows that σ mRNA is re-expressed in both cell lines following treatment for 3 days with 5-aza-dC.
Functional Consequences of Loss of σ in Breast Cancer Cells.
The function of human σ has been analyzed in human colon carcinoma cells (5, 6). These studies demonstrated that following ionizing irradiation, σ sequesters cdc2–cyclin B1 complexes in the cytoplasm, thus arresting the cells in G2 (6). These actions prevent the cell from initiating mitosis before repair of its damaged DNA. Colon carcinoma cells lacking σ can still initiate, but do not maintain, G2 arrest, leading to mitotic catastrophe and cell death.
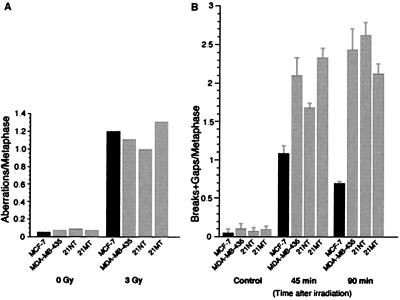
In an attempt to determine the effects of loss of σ gene expression on cell cycle regulation in breast cancer cells, we tested the effects of γ-irradiation on the σ-negative breast cancer cell lines, MDA-MB-435, 21NT, and 21MT, and the σ-positive breast cancer cell line, MCF-7. First, G1 type chromosomal aberrations were examined 24 h after cells were exposed to 3 Gy of γ-irradiation. All categories of G1-type chromosomal aberrations were scored at metaphase; their frequency was identical in the two cell types (Fig. 6). These results indicate that the examined cell lines have similar G1 cell cycle checkpoint control responses to ionizing radiation.
Figure 6.
σ-negative cells have a defective G2 cell cycle checkpoint and have an increased number of G2 type chromosomal aberrations. (A) The frequency of G1 type chromosomal aberrations in the σ-positive cell line, MCF-7, and σ-negative cancer cell lines, MDA-MB-435, 21MT, and 21NT. (B) The frequency of G2 type chromosome aberrations in MCF-7, MDA-MB-435, 21MT, and 21NT. Results of two experiments are shown.
Next, we evaluated the G2 checkpoint function in the four cell lines. Cells in exponential growth phase were γ-irradiated with 1 Gy and, and metaphases were examined for chromatid type breaks and gaps. Defective G2 arrest will increase these values. The results showed a striking difference in the ability of σ-negative cells and σ-positive cells to repair their damaged DNA (Fig. 6). Forty-five minutes after irradiation, σ-negative cells exhibited up to twice as many G2 type chromosomal aberrations as MCF-7 cells. This number increased to 3-fold by 90 min (Fig. 5). These are highly reproducible and statistically significant differences, similar to those reported for cells from cancer prone individuals (14, 21), well known for their defective G2 check points. Moreover, although repair of DNA damage was evident in the MCF-7 cells, as seen by a decrease in the number of G2 type aberrations between 45 and 90 min, no decrease was seen in σ-negative cells (Fig. 6). The mitotic index (MI) was also determined (13, 14) at the same time points. It varied from 5.7 to 7.3 for the four cell lines. Following irradiation, the mitotic index declined significantly in MCF-7 cells (MI = 7.0 to 2.5) at 90 min after irradiation, whereas there was no significant decline (range, 0–0.7) in the other cell lines, suggesting that G2 delay occurs in σ-positive MCF-7 cells, but not in the three σ-negative breast cancer cell lines.
Finally, to further demonstrate the role of σ in G2 checkpoint function in breast cells, we overexpressed a cloned copy of the gene in the σ-negative cell line MDA-MB-435 as well as in normal breast epithelial cells by using the adenovirus expression system used to express σ in colon cancer cells (5). Overexpression of σ in these breast epithelial cells led to a rapid and permanent G2 arrest, whereas the control virus-infected cells showed no effect (data not shown).
In summary, these results suggest that although the σ-negative cell lines have a functional G1 cell cycle checkpoint, they accumulate more genetic damage following irradiation which is consistent with its failure to arrest in G2 in response to DNA damage.
Discussion
Despite years of intense study, no molecular alterations common to the vast majority of breast cancers have yet been found. For instance, only about 30% of breast cancers overexpress c-erb2, epidermal growth factor receptor, cyclin D1, or c-myc (reviewed in Ref. 22). We report that in striking contrast to normal breast tissue, >90% of breast cancers lack detectable expression of σ (Fig. 1). In fact, loss of σ gene expression is the most common molecular event in primary breast cancers that has been reported thus far.
σ is a member of a superfamily that is responsible for instituting the G2 cell cycle checkpoint in response to DNA damage in human cells (5, 6). Four kilobases of σ upstream regulatory sequence directed the expression of a linked reporter gene in both σ-negative and σ-positive cell lines, suggesting that the normal trans-acting factors required for σ expression are active in both cell types (Fig. 2). However, several lines of evidence demonstrated that hypermethylation of σ occurs in 91% of primary breast cancers and is strongly associated with loss of σ gene expression in these tumors (Figs. 3 and 4; Table 1).
A number of studies have found both tumor and metastasis suppressor genes to be abnormally methylated in breast cancer (11, 23–28). Among these, E-cadherin and MDGI (mammary derived growth factor inhibitor) are the most frequently hypermethylated genes, with ≈50% of breast tumors exhibiting these epigenetic alterations (23, 24). In comparison, the exceptionally high incidence of σ gene methylation raises the possibility that it may play an important role in breast cancer.
Hypermethylation of the σ gene occurs in a CpG-rich region that extends from the transcriptional initiation site to the middle of the coding region. Bisulfite genomic sequencing of this 500-bp region showed that is consistently and densely methylated in σ-negative cell lines and primary breast tumors (Figs. 3 and 4). Several studies have clearly documented that gene activity correlates inversely with the density of gene-specific CpG island methylation, but is less dependent on the position and distance of the methylated DNA sequences from the transcriptional initiation site (29–31). With respect to σ, dense methylation just downstream of its transcriptional start site is strongly associated with gene silencing. Furthermore, in σ-negative cell lines, 5-aza-dC-induced demethylation of the CpG island leads to reactivation of gene expression, indicating that hypermethylation plays a causal role in σ gene inactivity (Fig. 5 A and B).
Hypermethylated DNA is known to interact with at least one methyl-CpG binding protein, MeCP2, that forms a transcriptionally repressive complex with the histone deacetylase and transcriptional corepressor, SIN3A (32, 33). We also found that treatment of σ-negative cells with the histone deacetylase inhibitor, trichostatin A, leads to reactivation of the σ gene (our unpublished findings). Together, these results suggest that methylation-mediated chromatin condensation is responsible for suppressing σ transcription in breast cancer cells.
σ is a member of the 14-3-3 superfamily that is responsible for G2 cell cycle checkpoint control in response to DNA damage in human cells (5, 6). In addition to any growth advantage resulting from a loosening of this checkpoint control mechanism, loss of σ function is predicted to cause an increase in DNA damage in response to γ-irradiation (6). Accordingly, irradiated, σ-negative breast cancer cells accumulated significantly more chromosomal breaks and gaps than the σ-positive cells, MCF-7. It is intriguing to speculate that, in response to irradiation and following exposure to DNA damaging drugs, σ-negative primary breast cancer cells may accumulate lethal amounts of DNA damage, inducing their death by a process called mitotic catastrophe (6, 34, 35). Primary breast cancers are sensitive to adjuvant radiation therapy (36, 37). Although difficult, a prospective study could be undertaken to determine σ expression in material obtained at biopsy to examine whether the response to radiation is altered by the σ status of the tumor. Similarly, the small number of tumors that recur after adjuvant radiation could be examined for σ gene expression.
In summary, CpG island methylation is an epigenetic change that is largely responsible for silencing of the σ gene and occurs in a majority of breast cancers. Loss of σ may play a role in determining the sensitivity of breast cancers to radiation therapy. Further evaluation of σ gene expression in tissue samples such as fine-needle biopsies and premalignant lesions like ductal carcinoma in situ (DCIS) may provide the foundation for its development as a marker for early detection.
Acknowledgments
We thank Drs. Bert Vogelstein and Vimla Band for generously sharing reagents and thank Sonu Dhar for expert technical assistance. We thank Sharyl Nass, Nancy Davidson, Alan Rein, and Bert Vogelstein for critically reviewing the manuscript. This work was supported by grants from the Susan G. Komen Foundation, U.S. Public HealthService Grant CA48943, and the Breast Cancer Research Foundation to S.S.; Grant T32 CA09630 to A.T.F., Grant CA-24844 and Contract DE-AC03-76SF00098 to M.R.S., and Grant NS34746 to T.K.P.
Abbreviations
- σ
14-3-3 σ
- SAGE
serial analysis of gene expression
- 5-aza-dC
5-aza-2′deoxycytidine
- LOH
loss of heterozygosity
- MSP
methylation-specific PCR
- HMEC
human mammary epithelial cell
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100566997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100566997
References
- 1.Prasad G L, Valverius E M, McDuffe E, Cooper H L. Cell Growth Differ. 1992;3:507–513. [PubMed] [Google Scholar]
- 2.Leffers H, Madsen P, Rasmussen H H, Honore B, Andersen A H, Walbum E, Vandekerckhove J, Celis J E. J Mol Biol. 1993;231:982–998. doi: 10.1006/jmbi.1993.1346. [DOI] [PubMed] [Google Scholar]
- 3.Velluci V F, Germino F J, Reiss M. Gene. 1995;166:213–220. doi: 10.1016/0378-1119(95)00543-9. [DOI] [PubMed] [Google Scholar]
- 4.Ostergaard M, Rasmussen H H, Nielsen H V, Vorum H, Orntoft T F, Wolf H, Celis J E. Cancer Res. 1997;57:4111–41117. [PubMed] [Google Scholar]
- 5.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 6.Chan T A, Hermeking H, Langauer C, Kinzler K W, Vogelstein B. Nature (London) 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 7.Band V, Zajchowski D, Swisshelm K, Trask D, Kulesa V, Cohen C, Connolly J, Sager R. Cancer Res. 1990;50:7351–7357. [PubMed] [Google Scholar]
- 8.Schutze K, Lahr G. Nat Biotechnol. 1998;16:737–742. doi: 10.1038/nbt0898-737. [DOI] [PubMed] [Google Scholar]
- 9.Umbricht C B, Sherman M E, Dome J, Carey L A, Marks J, Kim N, Sukumar S. Oncogene. 1999;18:3407–3414. doi: 10.1038/sj.onc.1202714. [DOI] [PubMed] [Google Scholar]
- 10.Evron E, Cairns P, Halachmi N, Ahrendt S A, Reed A L, Sidransky D. Cancer Res. 1997;57:2888–2889. [PubMed] [Google Scholar]
- 11.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson A T, Lapidus R G, Baylin S B, Davidson N E. Cancer Res. 1995;55:2279–2283. [PubMed] [Google Scholar]
- 13.Pandita T K, Hall E J, Hei T K, Piatyszek M A, Wright W E, Piao C Q, Pandita R K, Willey J C, Geard C R, et al. Oncogene. 1996;13:1423–1430. [PubMed] [Google Scholar]
- 14.Morgan S E, Lovly C, Pandita T K, Shiloh Y, Kastan M B. Mol Cell Biol. 1997;17:2020–2029. doi: 10.1128/mcb.17.4.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bieche I, Lidereau R. Genes Chromosomes Cancer. 1995;14:227–251. doi: 10.1002/gcc.2870140402. [DOI] [PubMed] [Google Scholar]
- 16.Genuardi M, Tsihira H, Anderson D E, Saunders G F. Am J Hum Genet. 1989;45:73–82. [PMC free article] [PubMed] [Google Scholar]
- 17.Trent J, Yang J M, Emerson J, Dalton W, McGee D, Massey K, Thompson F, Villar H. Genes Chromosomes Cancer. 1993;7:194–203. doi: 10.1002/gcc.2870070403. [DOI] [PubMed] [Google Scholar]
- 18.Nagai H, Negrini M, Carter S L, Gillum D R, Rosenberg A L, Schwartz G F, Croce C M. Cancer Res. 1995;55:1752–1757. [PubMed] [Google Scholar]
- 19.Tsukamoto K, Noriko I, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G, Nakamura Y, Emi M. Cancer. 1998;82:317–322. [PubMed] [Google Scholar]
- 20.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parshad R, Sanford K, Jones G M. Proc Natl Acad Sci USA. 1983;80:5612–5616. doi: 10.1073/pnas.80.18.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch D R, Wei L L. Endocrine-Related Cancer. 1998;5:155–197. [Google Scholar]
- 23.Huynh H, Alpert L, Pollak M. Cancer Res. 1996;56:4865–4870. [PubMed] [Google Scholar]
- 24.Graff J R, Herman J G, Lapidus R G, Chopra H, Xu R, Jarrard D F, Isaacs W B, Pitha P M, Davidson N E, Baylin S B. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 25.Bachman K E, Herman J G, Corn P G, Merlo A, Costello J F, Cavenee W K, Baylin S B, Graff J R. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 26.Hakkarainen M, Wahlfors J, Myohanen S, Hiltunen M O, Eskelinen M, Johansson R, Janne J. Int J Cancer. 1996;69:471–474. doi: 10.1002/(SICI)1097-0215(19961220)69:6<471::AID-IJC9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 27.Lapidus R G, Ferguson A T, Ottaviano Y L, Parl F F, Smith H S, Weitzman S A, Baylin S B, Issa J P, Davidson N E. Clin Cancer Res. 1996;2:805–810. [PubMed] [Google Scholar]
- 28.Lapidus R G, Nass S J, Butash K A, Parl F F, Weitzman S A, Graff J G, Herman J G, Davidson N E. Cancer Res. 1998;58:2515–2519. [PubMed] [Google Scholar]
- 29.Hsieh C-L. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kass S U, Goddard J P, Adams R L P. Mol Cell Biol. 1993;13:7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rideout W M I, Versole-Cire P E, Spruck III C H, Hustad C M, Coetzee G A, Gonzales F A, Jones P A. Mol Cell Biol. 1994;14:6143–6152. doi: 10.1128/mcb.14.9.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 33.Wade P A, Jones P L, Vermaak D, Veenstra G J, Imhof A, Sera T, Tse C, Ge H, Shi Y B, Hansen J C, Wolffe A P. Cold Spring Harbor Symp Quant Biol. 1998;63:435–445. doi: 10.1101/sqb.1998.63.435. [DOI] [PubMed] [Google Scholar]
- 34.Heald R, McLoughlin M, McKeon F. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- 35.Pines J. Nature (London) 1999;397:104–105. doi: 10.1038/16344. [DOI] [PubMed] [Google Scholar]
- 36.Clark R M, Whelan T, Levine M, Roberts R, Willan A, McCulloch P, Lipa M, Wilkinson R H, Mahoney L J. J Natl Cancer Inst. 1996;88:1659–1664. doi: 10.1093/jnci/88.22.1659. [DOI] [PubMed] [Google Scholar]
- 37.Fisher B, Dignam J, Wolmark N, Mamounas E, Costantino J, Poller W, Fisher E R, Wickerham D L, Deutsch M, Margolese R, et al. J Clin Oncol. 1998;16:441–452. doi: 10.1200/JCO.1998.16.2.441. [DOI] [PubMed] [Google Scholar]
- 38.Nacht M, Ferguson A T, Zhang W, Petroziello J M, Cook B P, Gao Y H, Maguire S, Riley D, Coppola G, Landes G M, et al. Cancer Res. 1999;59:5464–5470. [PubMed] [Google Scholar]