Abstract
Vinyl chloride reductases (VC-RDase) are the key enzymes for complete microbial reductive dehalogenation of chloroethenes, including the groundwater pollutants tetrachloroethene and trichloroethene. Analysis of the codon usage of the VC-RDase genes vcrA and bvcA showed that these genes are highly unusual and are characterized by a low G+C fraction at the third position. The third position of codons in VC-RDase genes is biased toward the nucleotide T, even though available Dehalococcoides genome sequences indicate the absence of any tRNAs matching codons that end in T. The comparatively high level of abnormality in the codon usage of VC-RDase genes suggests an evolutionary history that is different from that of most other Dehalococcoides genes.
The discovery of microbial growth coupled to reductive dehalogenation (dehalorespiration) of chloroethenes has led to development of microbial bioremediation as an important strategy for remediation of chloroethene-contaminated sites (14). The microorganisms capable of dehalorespiration of perchloroethene and trichloroethene are phylogenetically diverse and ubiquitous in many soil environments (22). Thus far, only members of the genus Dehalococcoides are known to dehalorespire dichloroethenes and the most toxic congener, vinyl chloride, to harmless ethene (22). The energy metabolism of Dehalococcoides species appears to be confined to reductive dehalogenation with hydrogen as an electron donor, thereby defining the unique biology of this group of microorganisms (9, 20).
The enzymes responsible for reductive dehalogenation in Dehalococcoides have been identified as reductive dehalogenases (RDases) (8, 12, 13, 15, 16). Previous biochemical work demonstrated the activity and substrate range of the RDase responsible for trichloroethene reduction, TceA, in Dehalococcoides strain 195 (12), as well as the vinyl chloride reductase (VC-RDase), VcrA, in Dehalococcoides strain VS (16). Indirect evidence identifying the perchloroethene RDase, PceA, in Dehalococcoides strain 195 (12, 15) and the putative VC-RDase gene (bvcA) in Dehalococcoides strain BAV1 (8) has also been obtained. A combination of molecular tools and whole-genome sequencing projects has led to a database consisting of approximately 90 putative RDase genes for Dehalococcoides (5, 9, 12, 16, 19, 20, 23). Whole-genome sequencing of isolated Dehalococcoides strains (195, CBDB1, and BAV1 draft) (9, 20; http://genome.ornl.gov/microbial/deha_bav1/; http://www.jgi.doe.gov/) has also revealed that many RDase genes are located in or near putative integrated mobile genetic elements, indicating that these genes were potentially acquired through horizontal gene transfer (9, 18, 20).
Here we describe a computational analysis of codon usage in RDase genes that was performed as a means of obtaining further insight into a gene family that has been described as both essential to the biology of Dehalococcoides and associated with signatures for lateral gene transfer. Unless otherwise noted, the computational analysis was performed using the R software language for statistics (version 2.4.1) and the contributed packages seqinR and ade4 (3, 4, 17). The R software is available through the CRAN archives network using the appropriate mirror (http://cran.r-project.org/mirrors.html). Computational analysis of codon usage bias has been used extensively in bacteria to estimate relative expression levels and to identify genes acquired through recent horizontal transfer (1, 2, 6, 7, 10). We will describe elsewhere a correspondence analysis of synonymous codon usage in Dehalococcoides RDase genes which revealed that the G+C fraction at the third position (GC3) is a major discriminating statistic for RDase genes (P. J. McMurdie and S. Holmes, unpublished data).
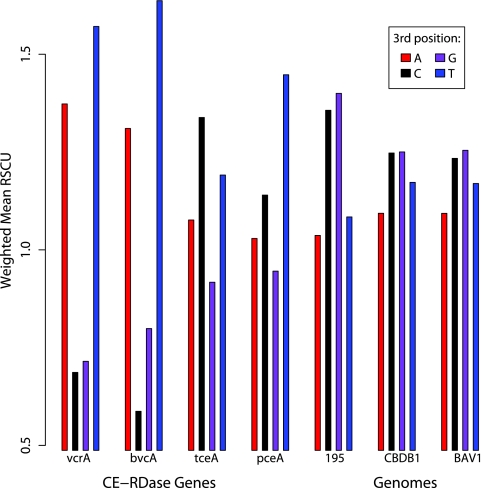
The codon usage bias (CUB) of chloroethene reductase (CE-RDase) genes was summarized using a weighted mean of relative synonymous codon usage (RSCU) (21) values for each nucleotide at the third position, as shown in Fig. 1. The average values for all open reading frames (ORFs) from the three available genome sequences were also included for comparison. As Fig. 1 shows, the third-position nucleotide usage in VC-RDase genes was different from the third-position nucleotide usage in other CE-RDase genes and different from the third-position nucleotide usage in the average Dehalococcoides gene. A localized scan of third-position usage for the length of CE-RDase genes did not reveal any bias due to the position in the gene (data not shown).
FIG. 1.
Summary of RSCU by third-position nucleotide in CE-RDase genes: mean RSCU values (21) for codons having the same nucleotide at the third position, weighted by the number of codons. Values for vcrA, bvcA, tceA, and pceA are shown individually along with values for the complete genome sequences of D. ethenogenes 195, strain CBDB1, and strain BAV1.
To rigorously evaluate the total differences in synonymous codon usage between genes or groups of genes, we employed the following formula, first proposed by Karlin et al. (7):
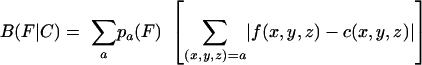 |
(1) |
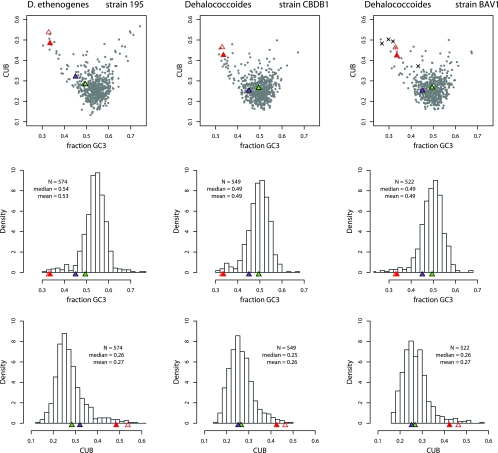
where f(x,y,z) and c(x,y,z) are the normalized frequencies of codon xyz in gene(s) F and C, respectively, and pa(F) is the normalized frequency of amino acid a in gene(s) F. Equation 1 yields a single value, B, representing the total difference in synonymous codon usage between F and C, where F and C can be genes or groups of genes (7). When C is defined as all codons from all ORFs in a genome sequence, the resulting B(F|all) is referred to as the CUB of gene F relative to the average codon usage of the genes in genome C. In this way, CUB was calculated for all available Dehalococcoides RDase genes, as well as for every ORF from the three available Dehalococcoides genome sequences. These values were plotted against the GC3 and are shown in Fig. 2 (top row). As this figure shows, the VC-RDase genes were unusual compared to all Dehalococcoides genomes, having both an unusually low GC3 and a CUB of approximately 0.5, which Karlin et al. described as “rare” for comparable gene groups (7). The fringe status of the VC-RDase genes was in contrast to the status of tceA, pceA, or most Dehalococcoides RDase genes, which appear to be bounded by the “cloud” of genomic ORFs. For better resolution, each axis is also displayed as a histogram in Fig. 2 (middle and bottom rows), and the positions of known CE-RDase genes are indicated.
FIG. 2.
CUB of CE-RDase genes relative to the whole genome. (Top row) CUB of individual genes from the complete genomes of D. ethenogenes 195, CBDB1, and BAV1 plotted against GC3. The vcrA and bvcA genes are indicated by solid and open red triangles, while the tceA and pceA genes are indicated by green and purple triangles, respectively. ORFs from the genomes are indicated by gray circles. ORFs less than 300 codons long are not shown to mitigate the representation of sampling effects. ×, ORFs in the immediate vicinity of bvcA in the strain BAV1 genome. (Middle row) Histograms for GC3 of all genomic ORFs from the genome of each strain. N is the number of ORFs included in the plot. The relative positions of CE-RDase genes are indicated by colored triangles, as described above. (Bottom row) Histograms for CUB of all ORFs from the genome of each strain. Colored triangles indicate the locations of CE-RDase genes, as described above.
We used a Monte Carlo approach to assess the statistical significance of differences in codon usage between individual genes and a whole genome. The CUB was calculated for each of 105 simulated genes, which were created by a random sampling of codons from all ORFs in the whole genome. We defined a P value for each Dehalococcoides RDase gene as the fraction of simulated genes with a CUB value larger than the CUB value of the RDase gene:
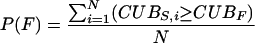 |
(2) |
where P(F) is the P value defined for RDase gene F, N is the total number of simulated genes, CUBS,i is the CUB value for simulated gene i, and CUBF is the CUB value for gene F. Unusual Dehalococcoides RDase genes are shown in Table S1 in the supplemental material. Unusual ORFs in the complete genome sequences are shown in Table S2 in the supplemental material. In 20 repeated Monte Carlo trials for each genome sequence, an average of less than one simulated gene per trial had a CUB value larger than the value observed for either VC-RDase gene, corresponding to a P value of <10−5. We concluded that it is highly unlikely (P < 10−5) that the difference in codon usage between VC-RDase genes and typical Dehalococcoides genes is a result of random variability in synonymous codon usage in Dehalococcoides genes.
For some bacteria unusual synonymous codon usage occurs in highly expressed genes as a consequence of strong selective forces (7). For example, unusually high levels of expression have been correlated with a specific unusual codon usage observed in ribosomal protein genes in Escherichia coli (7). However, a high level of expression of VC-RDase genes does not explain their unusual codon usage because their codon usage is distinct from that of any other Dehalococcoides genes that might be expected to be expressed at high levels in Dehalococcoides (data not shown). Furthermore, recent proteomic work revealed that TceA, Fdh, Hup, the product of the DET1407 gene, and subunits of ATP synthase were the most abundant proteins in Dehalococcoides ethenogenes strain 195 under chloroethene-dechlorinating conditions (15). The codon usage of the corresponding genes clustered well within the “cloud” of genes in Fig. 2 and was significantly different from that of the VC-RDase genes.
The codon usage in VC-RDase genes was also unusual because it is not consistent with tRNAs available in the Dehalococcoides genome. While codons having T at the third position were highly favored in VC-RDase genes (Fig. 1), not a single tRNA anticodon predicted by Dehalococcoides genome analyses matches codons ending in T (9, 20; http://genome.ornl.gov/microbial/deha_bav1/; http://www.jgi.doe.gov/). These anticodon predictions were independently confirmed here using the tRNAscan-SE analysis available from the genomic tRNA database (11). The anticodon predictions were also consistent with the genome-wide trend to favor codons with G or C at the third position (Fig. 1). However, codons ending with T are present throughout the Dehalococcoides genomes at levels that appear to be neither favored nor disfavored (RSCU value, ∼1.0). The significant bias in favor of codons ending in T, despite the absence of matching tRNAs in Dehalococcoides, further supports the notion that VC-RDase genes have an unusual evolutionary history.
The codon bias of VC-RDase genes was among the most severely divergent codon biases of the available Dehalococcoides genes. The observation that codon usage in most other RDase genes was not unusual compared to the codon usage in the Dehalococcoides genome was consistent with the strong patterns of operon orientation, strand bias, and localization near the origin of replication shown previously for many Dehalococcoides RDase genes (9, 20). It was also consistent with the hypothesis that there has been rampant movement of RDase genes within and between the closely related genomes of different Dehalococcoides strains, which Kube et al. suggested as a possible explanation for the association between many RDase genes and mobile genetic elements (9). Interestingly, genes in the immediate vicinity of bvcA in the strain BAV1 genome are characterized by a similarly unusual codon usage (Fig. 2); these genes include a putative phage integrase gene adjacent to bvcA, which is consistent with the hypothesis that bvcA was acquired by strain BAV1 through phage-associated horizontal transfer. While this analysis could not demonstrate the cause of the unusual codon usage, the extraordinary difference in codon usage in VC-RDase genes and the poor adaptability of the codon usage to available tRNAs are consistent with the notion that VC-RDase genes were horizontally acquired from a previously unknown microorganism. This prediction has important implications for the evolution of reductive dehalogenation in the environment.
Acknowledgments
This work was supported by a grant from the Western Region Hazardous Substance Research Center and SERDP to A.M.S. S.H. was partially funded by grant NSF-DMS-0241246, and P.J.M. was partially funded by grant FP-91671901-0 from the U.S. Environmental Protection Agency's Science to Achieve Results (STAR) program.
We thank two anonymous reviewers for their useful comments and also the Joint Genome Institute for general public access to the draft genome sequence of Dehalococcoides strain BAV1.
Footnotes
Published ahead of print on 16 February 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Azad, R. K., and J. G. Lawrence. 2005. Use of artificial genomes in assessing methods for atypical gene detection. PLoS Comput. Biol. 1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailly-Bechet, M., A. Danchin, M. Iqbal, M. Marsili, and M. Vergassola. 2006. Codon usage domains over bacterial chromosomes. PLoS Comput. Biol. 2:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charif, D., and J. R. Lobry. seqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences and analysis. In H. R. U. Bastolla, M. Porto, and M. Vendruscolo (ed.), Structural approaches to sequence evolution: molecules, networks, populations, biological and medical physics, biomedical engineering, in press. Springer-Verlag, New York, NY.
- 4.Chessel, D., A. B. Dufour, and J. Thioulouse. 2004. The ade4 package. I. One-table methods. R News 4:5-10. http://CRAN.R-project.org/doc/Rnews/. [Google Scholar]
- 5.Holscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. Von Wintzingerode, H. Gorisch, F. E. Loffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain, R., M. C. Rivera, J. E. Moore, and J. Lake. 2002. Horizontal gene transfer in microbial genome evolution. Theor. Popul. Biol. 61:489-495. [DOI] [PubMed] [Google Scholar]
- 7.Karlin, S., J. Mrazek, and A. M. Campbell. 1998. Codon usages in different gene classes of the Escherichia coli genome. Mol. Microbiol. 29:1341-1355. [DOI] [PubMed] [Google Scholar]
- 8.Krajmalnik-Brown, R., T. Holscher, I. N. Thomson, F. M. Saunders, K. M. Ritalahti, and F. E. Loffler. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kube, M., A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269-1273. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genomes. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowe, T., and S. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty, P. L. 1997. Breathing with chlorinated solvents. Science 276:1521-1522. [DOI] [PubMed] [Google Scholar]
- 15.Morris, R. M., S. Sowell, D. Barofsky, S. Zinder, and R. Richardson. 2006. Transcription and mass-spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes. Environ. Microbiol. 8:1499-1509. [DOI] [PubMed] [Google Scholar]
- 16.Muller, J. A., B. M. Rosner, G. Von Abendroth, G. Meshulam-Simon, P. L. McCarty, and A. M. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Development Core Team. 2005. R: a language and environment for statistical computing. Technical report. R Foundation for Statistical Computing, Vienna, Austria.
- 18.Regeard, C., J. Maillard, C. Dufraigne, P. Deschavanne, and C. Holliger. 2005. Indications for acquisition of reductive dehalogenase genes through horizontal gene transfer by Dehalococcoides ethenogenes strain 195. Appl. Environ. Microbiol. 71:2955-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regeard, C., J. Maillard, and C. Holliger. 2004. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J. Microbiol. Methods 56:107-118. [DOI] [PubMed] [Google Scholar]
- 20.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. E. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 21.Sharp, P. M., T. M. Tuohy, and K. R. Mosurski. 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14:5125-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smidt, H., and W. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 23.Waller, A. S., R. Krajmalnik-Brown, F. E. Loffler, and E. A. Edwards. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]