Abstract
Genetic instability and genome renewal may cause loss of heterozygosity (LOH) in homothallic wine yeasts (Saccharomyces cerevisiae), leading to the elimination of the recessive lethal or deleterious alleles that decrease yeast fitness. LOH was not detected in genetically stable wine yeasts during must fermentation. However, after sporulation, the heterozygosity of the new yeast population decreased during must fermentation. The frequency of mating between just-germinated haploid cells from different tetrads was very low, and the mating of haploid cells from the same ascus was favored because of the physical proximity. Also, mating restriction between haploid cells from the same ascus was found, leading to a very low frequency of self spore clone mating. This mating restriction slowed down the LOH process of the yeast population, maintaining the heterozygote frequency higher than would be expected assuming a fully random mating of the haploid yeasts or according to the Mortimer genome renewal proposal. The observed LOH occurs because of the linkage of the locus MAT to the chromosome III centromere, without the necessity for self spore clone mating or the high frequency of gene conversion and rapid asymmetric LOH observed in genetically unstable yeasts. This phenomenon is enough in itself to explain the high level of homozygosis found in natural populations of wine yeasts. The LOH process for centromere-linked markers would be slower than that for the nonlinked markers, because the linkage decreases the frequency of newly originated heterozygous yeasts after each round of sporulation and mating. This phenomenon is interesting in yeast evolution and may cause important sudden phenotype changes in genetically stable wine yeasts.
Most wild strains of Saccharomyces cerevisiae carry recessive deleterious alleles in the heterozygous state (4, 5, 7, 10, 13), but most of the loci contain homozygous alleles (10, 13). It has been proposed that this is because the wild homothallic strains of S. cerevisiae tend to be homozygous for most of the genes by genome renewal (Mortimer's proposal [10]). This phenomenon is based on the capacity of the homothallic haploid cells to switch mating type and to conjugate with identical cells from the same single-spore colony (self spore clone mating). This causes loss of heterozygosity (LOH) and should eliminate the recessive lethal or deleterious alleles that decrease yeast fitness leading to slower growth, lower fermentation rate, reduced spore viability, etc. By the same phenomenon, the new homozygous diploids bearing new recessive alleles that increase fitness may replace the parental heterozygous strains (10). The sporulation needed for genome renewal in wine yeasts can take place every year at the end of the vintage. Furthermore, some homothallic yeasts can sporulate in rich media (10, 13), allowing the possibility of genome renewal occuring continuously even during the vegetative growth of the population. This strategy, which probably occurs in nature, has been applied in the laboratory to obtain new fitness-improved wine yeasts which are more suitable for industrial fermentation (13).
In S. cerevisiae, genetic instability is associated with a high rate of LOH (3, 9, 14). High genetic instability and LOH in natural wine yeasts during laboratory propagation under nonselective conditions, but not in the common laboratory strains of S. cerevisiae, have been described recently (3, 14). Also, a high LOH has been reported at the URA3 locus in a transgenic wine yeast strain during must fermentation (12). Therefore, in genetically unstable yeasts, the elimination of recessive lethal or deleterious alleles that decrease yeast fitness could occur rapidly in the absence of sporulation. It has even been suggested, therefore, that sporulation is not significant in terms of the evolution of the yeast genome (12).
Even considering that all these phenomena may cause LOH and some “genome renewal,” it seems likely that Mortimer's proposal (10) is the major mechanism for LOH in wild populations of genetically stable wine yeasts. In this paper we analyze the occurrence of genome renewal by self spore clone mating during must fermentation. In the study, we used new wine yeast strains with good fermentation performance, high rates of sporulation and spore viability, and appropriate genetic markers to analyze the frequency of mating between the different yeasts living in the same fermenting must.
MATERIALS AND METHODS
Yeast strains, culture media, and phenotype tests.
SMR10-11D (MATα/MATa HO/HO SMRR/SMRR [k2+]) is a killer wine yeast (1). SMR10-11DNK (MATα/MATa HO/HO SMRR/SMRR [k20]) is a nonkiller yeast from SMR10-11D. The haploid laboratory yeast YMR107w (mata ho his3 leu2 met15 ura3 ymr107::G418R) was obtained from EUROSCARF (European Saccharomyces cerevisiae Archive for Functional Analysis). E339 (MATα/MATa HO/HO ura3-52/ura3-52 ymr107Δ::G418R/ymr107Δ::G418R [k20]) is a nonkiller homozygous spore clone from the genetic cross SMR10-11DNK × YMR107w. H77 (MATα/MATa HO/HO URA3/ura3Δ0 SMRR/smrS ymr107Δ::G418R/ILV2 [k20]) is a nonkiller heterozygous hybrid from the cross SMR10-11DNK × E339. All these wine S. cerevisiae strains were developed to provide good fermentation performance, high rates of sporulation and spore viability, and appropriate genetic markers to analyze the frequency of mating between the different yeasts present in the same fermenting must.
Standard culture media were used for yeast growth and phenotype tests (6). YEPD agar contained 1% Bacto yeast extract, 2% Bacto peptone, 2% glucose, and 2% Bacto agar. YEPD+G418 is YEPD agar supplemented with G418 (which is the antibiotic Geneticin [Sigma, catalogue number G7034], presented as a concentrated water solution) to a final concentration of 200 μg/ml. Synthetic minimal medium (SD) contained 0.67% yeast nitrogen base (without amino acids but with ammonium sulfate; Difco, Detroit, MI), 2% glucose, and 2% Bacto agar. Uracil (20 mg/liter), l-leucine (30 mg/liter), l-histidine-HCl (20 mg/liter), and l-methionine (20 mg/liter) were added when necessary. SD+SMR is standard SD agar supplemented with sulfometuron (SMR) to a 100-μg/ml final concentration. SMR was prepared in a concentrated dimethyl sulfoxide solution (1%) and added to the medium just before it was poured into petri dishes.
Standard yeast genetic procedures were used for sporulation of cultures and dissection of asci (8). Cells were grown on YEPD plates for 2 days at 30°C, transferred to sporulation plates (1% potassium acetate, 0.1% Bacto yeast extract, 0.05% glucose, 2% Bacto agar), and incubated for 7 to 20 days at 25°C until more than 80% of the cells had sporulated. Twenty-four asci from each yeast were dissected on YEPD plates and incubated for 5 days at 30°C to determine the percentage of viable spores.
Grape must fermentation was performed in 5 ml of sterile white Pardina juice (23°Bx, pH 3.5) supplemented with uracil (20 mg/liter) to facilitate the growth of newly originated homozygous ura3Δ0/ura3Δ0 yeasts. Fermentations were conducted at 25°C for up to 20 days without agitation. The degree Brix values were monitored each day to follow the fermentation kinetics. T15 is the time needed to ferment 15% of the total sugars present in the must, and T100 is the time needed to ferment 100% of the total sugars (13). Suitably diluted samples from each fermentation were spread onto YEPD plates to obtain isolated colonies after 2 days at 30°C. The amount of viable yeast (CFU) was determined by colony counting. SMRR and G418R phenotypes were determined by replica plating on SD+SMR and YEPD+G418 media (1, 2).
Analysis of mating of haploid homothallic yeasts from different asci.
SMR10-11DNK and E339 yeasts were used. Forty intact tetrads from each of the two yeasts were placed together on a YEPD plate and mixed with the needle of the micromanipulator. Rapidly, a small piece of the YEPD agar containing the 80-tetrad mix was inoculated into the sterile grape juice. A must fermentation control, inoculated with a mix of 50 μl of SMR10-11DNK and 50 μl of E339 2-day YEPD broth cultures (vegetative cells), was done in parallel. The yeasts with both SMRR and G418R phenotypes must result from mating of haploid homothallic yeasts from different asci.
Analysis of mating of haploid homothallic yeasts from the same spore clone.
The H77 heterozygous hybrid was used (SMRR and G418R genetic markers are strongly linked in trans configuration, i.e., repulsion linkage phase, the distance between the two markers being only 700 bp). Forty intact tetrads from this hybrid were placed together on a YEPD plate and mixed with the needle of the micromanipulator. Rapidly, a small piece of the YEPD agar containing the 40 tetrads was inoculated into the sterile grape juice. A must fermentation control, inoculated with a mix of 50 μl of H77 2-day YEPD broth culture, was done in parallel. The yeasts with both SMRR and G418R phenotypes must result from mating of haploid homothallic yeasts from different spore clones (from the same or different tetrads).
RESULTS
Mating of haploid homothallic yeasts from different asci.
The analysis of the frequency of mating between haploid yeasts from two diploid homothallic strains after spore germination was done by inoculating sterile grape must with a mix (1:1) of tetrads from SMR10-11DNK (SMRR/SMRR) and E339 (ymr107Δ::G418R/ymr107Δ::G418R). This mating will yield heterozygous SMRR/G418R diploid strains. Both strains sporulated very well, over 80% of tetrads after 7 days in sporulation media, and the spore viability was higher than 91%. As the control, the same sterile must was inoculated with a mix (1:1) of vegetative cells of the same strains. The fermentation started earlier in the must inoculated with vegetative cells (T15 = 2.6 days) than in that inoculated with the spores (T15 = 4.75) because of the time needed for spore germination, haploid cell mating, and yeast growth. Despite this, both fermentations were properly completed by day 15. No heterozygous SMRR/G418R diploid yeast was detected in the control fermentation, i.e., neither sporulation and mating (genome renewal) nor rare mating was detected. In the tetrad-inoculated fermentation, only 1.5% of the total population corresponded to heterozygous SMRR/G418R diploid yeasts, which appeared the third day after inoculation (Table 1). The frequency of mating between haploid yeasts from spores belonging to different tetrads was thus very low.
TABLE 1.
Analysis of the frequency of genetic markers during the fermentation of must inoculated with tetrads and vegetative cells of SMR10-11DNK (SMRR) and E339 (G418R) yeastsa
| Day | Tetrads (T15 = 4.75, T100 = 15)
|
Vegetative cells (T15 = 2.6, T100 = 15)
|
||||||
|---|---|---|---|---|---|---|---|---|
| CFU/ml | % with phenotype:
|
CFU/ml | % with phenotype:
|
|||||
| G418R | SMRR | G418R + SMRR | G418R | SMRR | G418R + SMRR | |||
| 2 | 4 × 104 ± 5 × 103 | 18 ± 1.5 | 82 ± 1.5 | 0 ± 0 | 3.6 × 107 ± 1 × 106 | 30 ± 2 | 70 ± 2 | 0 ± 0 |
| 3 | 5.4 × 105 ± 3 × 104 | 4.6 ± 1.7 | 93 ± 2 | 1.5 ± 0.3 | 2.5 × 107 ± 1 × 106 | 20 ± 1.7 | 80 ± 1.7 | 0 ± 0 |
| 8 | 1.1 × 108 ± 2 × 107 | 0 ± 0 | 100 ± 0 | 0 ± 0 | 2 × 108 ± 2 × 106 | 1 ± 0.6 | 99 ± 0.6 | 0 ± 0 |
G418R, resistant to G418; SMRR, resistant to SMR; G418R + SMRR, resistant to both G418 and SMR. The data are the mean values (percentages) of two independent experiments and standard errors. One hundred to 300 colonies were analyzed from each sample. The first sample was taken on the second day of fermentation because of the time needed for spore germination, mating, and yeast population growth.
Mating of haploid homothallic yeasts from the same spore clone.
Sterile must was inoculated with a homothallic heterozygous SMRR/G418R diploid strain, H77, to analyze the frequency of self spore clone mating after spore germination and mating type switch, which should increase the frequency of homozygous G418R/G418R and SMRR/SMRR diploid strains. H77 sporulated very well, over 82% of tetrads after 7 days in sporulation medium, and the spore viability was greater than 88%. No relationship was found between the nonviability of the spores and the presence or absence of either of the two genetic markers. As a control, the same sterile must was inoculated with vegetative cells of the same strain. The fermentation was higher in the must inoculated with vegetative cells (T15 = 2.25; T100 = 16) than in that inoculated with the asci (T15 = 3.6; T100 = 18.7) because of the time needed for spore germination, haploid cell mating, and yeast growth. Both fermentations were properly completed before day 19. No homozygous G418R/G418R or SMRR/SMRR diploid yeast was detected in the control fermentation, i.e., we did not detect any LOH by sporulation and mating, gene conversion, mitotic recombination, or chromosome loss and endoreduplication. Homozygous diploid yeasts were detected from the start in the must inoculated with H77 asci (15% G418R/G418R and 13% SMRR/SMRR, Table 2, day 2). These frequencies were lower than would be expected if the mating of the haploid yeast had been fully random (25% for each homozygote) and much lower than would be expected with frequent self spore clone mating after a mating type switch (higher than 25% for each homozygote, fast genome renewal). In contrast, the frequency of heterozygous G418R/SMRR (72%) was higher than would be expected (50%) if there had been fully random mating of the haploid yeast. The frequency of each yeast genotype fluctuated over the course of the must fermentation, with the frequency of heterozygous G418R/SMRR decreasing from 72% to 51%, probably because of the different fitnesses of the new yeasts that arise after sporulation and mating and the differing resistance to the stressing fermentation conditions, mostly low pH and increasing alcohol concentration.
TABLE 2.
Analysis of the frequency of genetic markers during the first fermentation of must inoculated with tetrads and vegetative cells of the H77 [ymr107Δ::G418R/ymr107 ILV2(SMRS)/ILV2(SMRR)] hybrida
| Day | Tetrads (T15 = 3.6, T100 = 18.7)
|
Vegetative cells (T15 = 2.25, T100 = 16)
|
||||||
|---|---|---|---|---|---|---|---|---|
| CFU/ml | % with phenotype:
|
CFU/ml | % with phenotype:
|
|||||
| G418R | SMRR | G418R + SMRR | G418R | SMRR | G418R + SMRR | |||
| 2 | 3.5 × 105 ± 1 × 103 | 15 ± 0.6 | 13 ± 1.1 | 72 ± 0.6 | 3 × 106 ± 2 × 105 | 0 ± 0 | 0 ± 0 | 100 ± 0 |
| 4 | 2.1 × 107 ± 2 × 106 | 15 ± 1.7 | 30 ± 0.5 | 55 ± 1.1 | 1 × 107 ± 8 × 105 | 0 ± 0 | 0 ± 0 | 100 ± 0 |
| 8 | 1.9 × 107 ± 9 × 105 | 13 ± 0.4 | 35 ± 1.1 | 52 ± 1 | 1.5 × 107 ± 2 × 106 | 0 ± 0 | 0 ± 0 | 100 ± 0 |
| 11 | 2.8 × 107 ± 1 × 106 | 26 ± 0.4 | 23 ± 0.4 | 51 ± 0.7 | 2.2 × 107 ± 6 × 105 | 0 ± 0 | 0 ± 0 | 100 ± 0 |
G418R, resistant to G418; SMRR, resistant to SMR; G418R + SMRR, resistant to both G418 and SMR. The data are the mean values (percentages) of two independent experiments and standard errors. One hundred to 300 colonies were analyzed from each sample. The first sample was taken on the second day of fermentation because of the time needed for spore germination, mating, and yeast population growth.
A second similar experiment was done by inoculating the sterile must with tetrads from a sporulated culture of the yeasts from day 2 (low alcohol concentration, still-healthy growing yeast culture) of the first tetrad fermentation (15% G418R/G418R, 13% SMRR/SMRR, and 72% G418R/SMRR; Table 2). The yeast sporulated very well, over 79% of tetrads after 7 days in sporulation medium, and the spore viability was greater than 86%. Again, no homozygous G418R/G418R or SMRR/SMRR diploid yeast was detected in the control fermentation inoculated with vegetative cells of H77, and a lower-than-expected frequency was found in the tetrad-inoculated fermentation, 20% G418R/G418R and 26% SMRR/SMRR (Table 3). In contrast, the frequency of heterozygous G418R/SMRR (54%) was again higher than would be expected if there had been fully random mating of the haploid yeasts (0.724 × 0.5 = 0.36). The frequency of each yeast genotype fluctuated over the course of the must fermentations, but this time the frequency of G418R/SMRR heterozygotes increased, indicating that the fitness and resistance to the stressing fermentation conditions of the new yeasts arising after sporulation may change in different ways.
TABLE 3.
Analysis of the frequency of genetic markers during the second fermentation of must inoculated with tetrads (from a sporulated culture of the yeasts from day 2 of the first fermentation) and vegetative cells of the H77 hybrida
| Day | Tetrads (T15 = 3, T100 = 15)
|
Vegetative cells (T15 = 1.5, T100 = 11)
|
||||||
|---|---|---|---|---|---|---|---|---|
| CFU/ml | % with phenotype:
|
CFU/ml | % with phenotype:
|
|||||
| G418R | SMRR | G418R + SMRR | G418R | SMRR | G418R + SMRR | |||
| 2 | 4.3 × 105 ± 2 × 103 | 20 ± 0.5 | 26 ± 0.9 | 54 ± 0.5 | 1.3 × 105 ± 8 × 103 | 0 ± 0 | 0 ± 0 | 100 ± 0 |
| 7 | 6.2 × 107 ± 2 × 105 | 13 ± 0.9 | 18 ± 0.6 | 69 ± 1.4 | 2 × 106 ± 1 × 105 | 0 ± 0 | 0 ± 0 | 100 ± 0 |
| 11 | 6.2 × 107 ± 8 × 105 | 19 ± 0.9 | 17 ± 0.1 | 64 ± 0.8 | 7.4 × 107 ± 7 × 105 | 0 ± 0 | 0 ± 0 | 100 ± 0 |
| 21 | 6.3 × 107 ± 1 × 105 | 21 ± 0.7 | 7.4 ± 1.3 | 71 ± 1 | 9.4 × 107 ± 2 × 105 | 0 ± 0 | 0 ± 0 | 100 ± 0 |
G418R, resistant to G418; SMRR, resistant to SMR; G418R + SMRR, resistant to both G418 and SMR. The data are the mean values (percentages) of two independent experiments and standard errors. One hundred to 300 colonies were analyzed from each sample. The first sample was taken on the second day of fermentation because of the time needed for spore germination, mating, and yeast population growth.
DISCUSSION
S. cervisiae spores remain inside the ascus until germination occurs in favorable environmental conditions. In the absence of germination the ascus wall has to be artificially destroyed to disseminate the spores (8). Without continuous shaking, as is the case for most vineyard or winery musts before tumultuous fermentation, this circumstance should restrict the mating between just-germinated haploid cells from different tetrads, favoring the mating of haploid cells from the same ascus because of the physical proximity. This explains our finding that the frequency of mating between haploid yeasts from two diploid homothallic strains, SMR10-11DNK and E339, after spore germination in the grape must was 1.5% (Table 1). As the two strains were inoculated in equal amounts (1:1), and mating between haploid yeasts from different asci of the same strain is also possible and undetectable, we estimate a frequency of only 3% for different-ascus yeast mating under our working conditions.
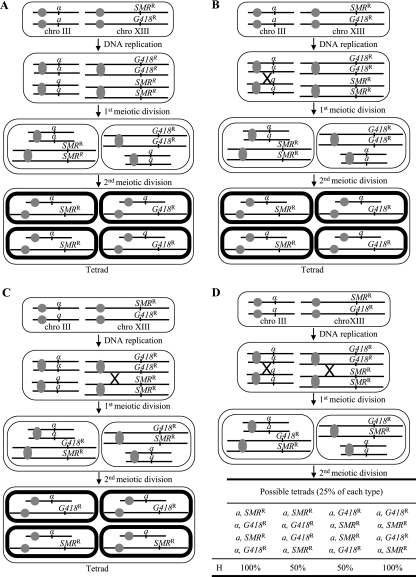
Given this mating restriction, it is interesting to ask what the mating behavior of homothallic haploid yeasts form the same ascus was. The frequencies of homozygous diploid yeasts from H77 asci (15% G418R/G418R and 13% SMRR/SMRR; Table 2) were lower than would be expected if the mating of the haploid yeast had been fully random (25% for each type) and much lower than would be expected if any frequent self spore clone mating had occurred. This result indicates that there is some restriction on mating between haploid cells from the same ascus, together with a very low frequency of self spore clone mating. This restriction decreases the LOH of the yeast population, maintaining the frequency of heterozygous G418R/SMRR (72%) higher than would be expected assuming fully random mating of the haploid yeasts (50%) or according to the genome renewal proposal (10). We propose that this mating restriction is just the result of chromosome segregation during meiosis and can be explained for H77 as follows. The G418R and SMRR (ILV2) markers are strongly trans linked and far enough away from the chromosome XIII centromere, 63 cM (11), to be considered non-centromere linked (the maximum distance for being considered centromere linked is 50 cM). The locus MAT, which determines the mating type and hence the mating restriction of just-germinated haploid yeasts, is located on chromosome III and is centromere linked, 30 cM (11). In the absence of meiotic crossover between G418R (or SMRR) or MAT and its corresponding centromere, the marker segregations would be as shown in Fig. 1A. As a result, 100% of the new yeasts arising from spore germination and haploid yeast mating would be heterozygous G418R/SMRR. This situation would occur at a frequency of 1 minus the other three possible situations to be explained below, i.e., 1 − [(0.5 × 0.7) + (0.5 × 0.3) + (0.5 × 0.3)] = 0.35. If a single crossover occurs only between G418R (or SMRR) and the centromere, the segregations would be as shown in Fig. 1C. As a result, 50% of the new yeasts would be heterozygous G418R/SMRR. This situation would occur at a frequency of 0.5 × 0.7 = 0.35 (frequency of recombination between G418R or SMRR and the centromere) × (frequency of no recombination between MAT and the centromere). Hence, the heterozygous frequency in this situation would be 0.175. If a single crossover occurs only between MAT and the centromere, the segregations would be as shown in Fig. 1B. As a result, 50% of the new yeasts would be heterozygous G418R/SMRR. This situation would occur at a frequency of 0.3 × 0.5 = 0.15 (frequency of recombination between MAT and the centromere) × (frequency of no recombination between G418R or SMRR and the centromere). Hence, the heterozygous frequency in this situation would be 0.075. And finally, if a double crossover occurs between G418R (or SMRR) and MAT and their corresponding centromeres, the segregations would be as shown in Fig. 1D. As a result, either 50% or 100% of the new yeasts would be heterozygous G418R/SMRR, depending on which of the four types of tetrad arises. This situation would occur at a frequency of 0.5 × 0.3 = 0.15 (frequency of recombination between MAT and the centromere) × (frequency of recombination between G418R or SMRR and the centromere). Hence, the heterozygous frequency would be 0.1125. Therefore, the expected total frequency for heterozygous G418R/SMRR is 0.7125, which is very close to the frequency of 0.72 found at the beginning of the first tetrad-inoculated must fermentation (Table 2). Also, the expected frequency of heterozygous yeasts at the beginning of the second must fermentation, inoculated with tetrads from a yeast population containing 15% G418R/G418R, 13% SMRR/SMRR, and 72% G418R/SMRR, is 0.72 × 0.7125 = 0.513, which is very close to the frequency found, 0.54 (day 2, Table 3). That is, LOH in homothallic wine yeast occurs because of the linkage of the locus MAT to the chromosome III centromere, without the need for self spore clone mating (10), mitotic gene conversion (12), or rapid asymmetric LOH due to genetic instability (3, 14). This phenomenon is enough in itself to explain the high level of homozygosity found in natural populations of wine yeasts (10, 13). It will also lead to genome renewal in the population without the need for self spore clone mating. The LOH would be slower for centromere-linked markers than for the non-centromere-linked markers, because the linkage decreases the frequency of newly originated heterozygous yeasts after each round of sporulation and mating (Fig. 1B and C).
FIG. 1.
H77 tetrad formation and spore genotype in the absence of genetic crossover between the locus ILV2 (G418R and SMRR) and the chromosome XIII centromere (A), after a single meiotic crossover between the locus MAT (a and α) and the chromosome III centromere (B), after a single genetic crossover between the locus ILV2 and the chromosome XIII centromere (C), and after double meiotic crossover between the locus ILV2 and the chromosome XIII centromere and between the locus MAT and the chromosome III centromere (D). Chro, chromosome; H, frequency of hybrids.
The frequency of heterozygous G418R/SMRR yeast changed in opposite directions during different must fermentations. It decreased during the first must fermentation from 72% to 51% (Table 2) and increased during the second fermentation from 54% to 71% (Table 3). This indicates that the relative fitness of the different types of yeasts may change in different ways after each round of sporulation, germination, and mating. However, those relative fitness changes should not affect the progressive LOH of the yeast population since they would occur randomly in either of the two possible senses.
In conclusion, sporulation and mating restrictions during must fermentation lead to LOH in homothallic S. cerevisiae strains. Genetically stable wine yeasts tend to be homozygous for most of the genes simply because of the linkage of the locus MAT to the chromosome III centromere, without the need for self spore clone mating, mitotic gene conversion, or rapid asymmetric LOH. Also, the speed of LOH depends on the centromere linkage of each marker, because the stronger this linkage, the lower the frequency of newly originated heterozygous yeast after each round of sporulation and mating.
Acknowledgments
This work was funded by grants 2PR01B002 and 2PR04B003 from the Extremadura Regional Government, Spain.
Footnotes
Published ahead of print on 23 February 2007.
REFERENCES
- 1.Ambrona, J., M. Maqueda, E. Zamora, and M. Ramírez. 2005. Sulfometuron resistance as genetic marker for yeast populations in wine fermentations. J. Agric. Food Chem. 53:7438-7443. [DOI] [PubMed] [Google Scholar]
- 2.Ambrona, J., A. Vinagre, M. Maqueda, M. L. Álvarez, and M. Ramírez. 2006. Rhodamine-pink as genetic marker for yeast populations in wine fermentations. J. Agric. Food Chem. 54:2977-2984. [DOI] [PubMed] [Google Scholar]
- 3.Ambrona, J., A. Vinagre, and M. Ramírez. 2005. Rapid asymmetric evolution of Saccharomyces cerevisiae wine yeasts under apparently non-selective conditions. Yeast 22:1299-1306. [DOI] [PubMed] [Google Scholar]
- 4.Bakalinsky, A. T., and R. Snow. 1990. Conversion of wine strains of Saccharomyces cerevisiae to heterothallism. Appl. Environ. Microbiol. 56:849-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guijo, S., J. C. Mauricio, J. M. Salmon, and J. M. Ortega. 1997. Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and “flor” film ageing of dry sherry-type wines. Yeast 13:101-117. [DOI] [PubMed] [Google Scholar]
- 6.Guthrie, C., and G. R. Fink (ed.). 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, CA. [PubMed]
- 7.Jiménez, J., and T. Benítez. 1987. Genetic analysis of highly ethanol-tolerant wine yeasts. Curr. Genet. 12:421-428. [Google Scholar]
- 8.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome instability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 10.Mortimer, R. K., P. Romano, G. Suzzi, and M. Polsinelli. 1994. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10:1543-1552. [DOI] [PubMed] [Google Scholar]
- 11.Mortimer, R. K., C. R. Schild, C. R. Contopoulou, and J. A. Kans. 1991. Genetic and physical maps of Saccharomyces cerevisiae. Methods Enzymol. 194:827-863. [DOI] [PubMed] [Google Scholar]
- 12.Puig, S., A. Querol, E. Barrio, and J. E. Pérez-Ortín. 2000. Mitotic recombination and genetic changes in Saccharomyces cerevisiae during wine fermentation. Appl. Environ. Microbiol. 66:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramírez, M., J. A. Regodón, F. Pérez, and J. E. Rebollo. 1999. Wine yeast fermentation vigor may be improved by elimination of recessive growth-retarding alleles. Biotechnol. Bioeng. 65:212-218. [DOI] [PubMed] [Google Scholar]
- 14.Ramírez, M., A. Vinagre, J. Ambrona, F. Molina, M. Maqueda, and J. E. Rebollo. 2004. Genetic instability of heterozygous hybrid populations of natural wine yeasts. Appl. Environ. Microbiol. 70:4686-4691. [DOI] [PMC free article] [PubMed] [Google Scholar]