Abstract
The occurrence and activation of glutathione S-transferase (GST) and the GST activities in biofilms in cold sulfidic spring waters were compared to the occurrence and activation of GST and the GST activities of the aquatic fungal strains EH5 and EH7 of Mucor hiemalis isolated for the first time from such waters. Using fluorescently labeled polyclonal anti-GST antibodies and GST activity measurements, we demonstrated that a high level of GST occurred in situ in natural biofilms and pure cultures of strain EH5. Measurement of microsomal and cytosolic soluble GST activities using different xenobiotic substrates, including 1-chloro-2,4-dinitrobenzene (CDNB), 1,2-dichloro-4-nitrobenzene, 1,2-epoxy-3-(4-nitrophenoxy)propane, 1-iodo-2,4-dinitrobenzene, and fluorodifen, showed that the overall biotransforming abilities of biofilms were at least sixfold greater than that of strain EH5 alone. Increasing the level of sodium thiosulfate (STS) in the medium stimulated the microsomal and cytosolic GST activities with CDNB of strain EH5 about 44- and 94-fold, respectively, compared to the activities in the control. The induction of microsomal GST activity with fluorodifen by STS was strongly linear, but the initial strong linear increase in cytosolic GST activity with fluorodifen showed saturation-like effects at STS concentrations higher than approximately 1 mM. Using laser scanning confocal and conventional fluorescence microscopy, abundant fluorescently labeled GST proteins were identified in germinating sporangiospores of strain EH5 after activation by STS. High-performance size exclusion chromatography and sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed the presence of at least two main GSTs (∼27.8- and ∼25.6-kDa subunits) in the cytosol of EH5, whereas the major 27.8-kDa subunit was the only GST in microsomes. We suggest that differential cellular GST expression takes place in strain EH5 depending on spore and hyphal development. Our results may contribute to our understanding of induction of GST by sulfurous compounds, as well as to the immunofluorescence visualization of GST in aquatic fungus and fungus-bacterium biofilms.
The glutathione S-transferases (GSTs) (EC 2.5.1.18) are a family of multifunctional cytosolic (38) and membrane-associated microsomal (10, 36) proteins that can catalyze the conjugation of many electrophilic endogenous and xenobiotic compounds to glutathione. Most of our knowledge of the function of GSTs has come from studies of animals and plants, and there have been relatively few studies of microbes (34, 40, 50). The resultant glutathione conjugates are less toxic and more polar than the parent compounds and can be stored in vacuoles or excreted from the biological system, helping the organism regain normal cellular functions and reducing the cellular toxicity of reactive parent or organic intermediates. The existence of a variety of different GST isoenzymes suggests that there is a wide range of substrate specificity (17). GSTs are grouped into seven distinct classes, including the α (basic), μ (near neutral), π (acidic), θ, δ, ξ, and β classes, based on distinct but broad overlapping substrate specificities and N-terminal amino acid sequences (6, 29, 50). With the exception of the θ class enzymes, all GST classes are involved in conjugation of 1-chloro-2,4-dinitrobenzene (CDNB) (16). 1,2-Dichloro-4-nitrobenzene (DCNB) is a preferential substrate of the μ class GST isoenzyme (29), whereas θ class GSTs exhibit high levels of activity with 1,2-epoxy-3-(4-nitrophenoxy)propane (EPNP) (11).
As GST is an S-transferase that is capable of transferring the SH group of glutathione to electrophilic xenobiotic compounds and natural substrates, it is likely that GST turnover is influenced mainly by S stress in aquatic ecosystems. The effects of sulfur stress (e.g., thiosulfate and sulfide stress) on the activity and functions of GST, as well as on in situ immunofluorescence imaging of GST in aquatic microorganisms and biofilms, especially aquatic fungi, have been poorly described. In particular, research on the role of GST in aquatic environments during stress deserves attention (i) because GST may be a universal biomarker as it occurs in many organisms in diverse phyla, (ii) because GST biosynthesis can be stimulated by a diverse range of biotic stress factors (e.g., pathogen invasion) and abiotic stress factors (e.g., heat shock, ozone, ethylene, heavy metals, and xenobiotic compounds), as well as by some inducers or chemoprotective substances (18), (iii) because this multifunctional GST enzyme family is essential for conferring cellular resistance to pollutants, mutagens/carcinogens, drugs, and oxidative stress (9, 18), and (iv) because the ecological role of GST in aquatic biofilms and microorganisms in response to stress (e.g., sulfur stress) is largely unknown. Thus, GST is an interesting biomarker for investigating stress-related processes in microbial communities in spring water and groundwater.
Cold sulfidic spring waters can contain a diverse range of microorganisms, especially bacteria, spirally ordered filaments, and fungus-like filaments living at relatively low temperatures (10 to 11°C) and high sulfide concentrations (20), which may produce various GST isoenzymes in response to environmental stress. Stress factors such as sulfides, heavy metals, and nitrate are commonly found in sulfidic spring waters (20). In this investigation, our objective was to study the effects of sulfur stress on the induction of GSTs in biofilms and the aquatic fungus Mucor hiemalis strain EH5 in a cold sulfidic spring, as well as on the immunofluorescence localization and characterization of GSTs, especially during fungal spore germination. Such studies should increase our knowledge of the underlying mechanisms of GST-mediated xenobiotic compound biotransformation in response to sulfur compounds.
MATERIALS AND METHODS
Location and hydrogeochemistry of the Irnsing-H2S and Teugn springs.
The Irnsing-H2S (20) and Teugn springs are located in the Karst area of Franconian Alb near Irnsing and Teugn, respectively, within 25 km of each other (Bavaria, Germany). A chemical analysis of the spring waters was performed by ion chromatography, using a Dionex DX 100 chromatography system equipped with a precolumn and an analytical column. For anions an AG 12A precolumn (53 by 4 mm) was connected to an AS 42A analytical column (255 by 4 mm), and for cations a CG 12A precolumn (53 by 4 mm) was connected to a CS 12A analytical column (255 by 4 mm). The anions were eluted with an aqueous mobile phase containing 2.7 mM Na2CO3 and 0.3 mM NaHCO3, whereas the cations were eluted with 20 mM H2SO4 at a flow rate of 1 ml/min. The ions were detected with an electrical conductivity detector. The heavy metals after complexation with EDTA were analyzed with a Dionex 500 system (53). The dissolved organic carbon content was determined by using a Shimadzu TOC 5.000A system according to the manufacturer's protocol. Total sulfide contents were determined by titration using a Microquant sulfide test kit (Merck, Germany) and were also estimated by CdS precipitation (45). The dissolved H2S content in spring water was determined with an H2S electrode (model H2S-Mikrosensor; ATM Analysentechnik, Rostock, Germany). The oxygen content of spring water was recorded with an Oximeter OXI 197-S using a Cell Ox 325 electrode (WTW, Weilheim, Germany), the electrical conductivity was determined with a detector (model Konduktometer LF 196 with a TetraCon 96-1.5 electrode; WTW), and the temperature, redox potential, and pH were determined using a detector-pH meter (pH 196) equipped with TA 197-pH, Pt 4805, and E50-pH-Einstabmesskette sensors (WTW), respectively. A pesticide analysis was performed at a public water supply facility and quality control center (Wasserwirtschaftsamt, Schweinfurt, Germany) using a thermostat-controlled (40°C) reversed-phase Dionex high-performance liquid chromatography (HPLC) system (Hypersil octyldecyl silane column [250 by 4 mm; particle size, 3 μm]) and a mobile phase consisting of acetonitrile with 0.15 g/liter ammonium acetate buffer at a flow rate of 0.9 ml/min, and the pesticides were detected at selected UV wavelengths (210, 220, 245, and 275 nm) for quantification; in addition, the UV spectra (210 to 360 nm) were recorded for identification using a photodiode array detector (model UVD 340 S). The column was calibrated with 54 different pesticides.
Characterization of microbial community in biofilm.
Fluorescent in situ hybridization (FISH) was performed using known fluorescently labeled (Texas Red, Pacific blue, or Cy3 or fluorescein label at the 5′ position) oligonucleotide probes (ARCH915 for archaea [46]; CREN499 for the domain Crenarchaeota and EURY498 for the domain Euryarchaeota [7]; ALF1b for α-Proteobacteria, BET42a for β-Proteobacteria, and GAM42a for γ-Proteobacteria [31]; DELTA495a,b,c for δ-Proteobacteria [30]; CF for cytophaga and flavobacteria [51]; HGC for high-G+C-content bacteria [42]; EUB338 for eubacteria [2]; MH1 for Mucor sp. [see below]; UNIV1392 for all microbes [47]). The labeled oligonucleotide probes were obtained as salt-free, highly purified preparations from MWG (Germany). Briefly, formaldehyde-fixed biofilm samples (10 μl) were suspended in wells of a slide carrier (Marienfeld, Germany), passed quickly through a Bunsen burner flame, and then dehydrated using an ethanol gradient (50, 80, and 100%) and hybridized in situ at 46°C separately using selected oligonucleotide probes (see above and below) (4, 32, 43) with the NaCl concentration in the wash buffer adjusted based on optimized formamide concentrations (10 to 30%) in hybridization buffers.
Isolation and purification of M. hiemalis strains EH5 and EH7.
Intact floating biofilms were collected from the Irnsing-H2S and Teugn sulfidic springs in sterile 50-ml Falcon tubes and transported to the laboratory on ice. The biofilms were concentrated by centrifugation (4,000 × g, 10 min) at 4°C and were purified by three washing and centrifugation cycles with 50 ml of phosphate-buffered saline (PBS) (20 mM phosphate buffer, 150 mM NaCl) (pH 7.4). After another centrifugation, the pellet was weighed, and aliquots of the pellet were used either for isolation and purification of the fungus or for other experiments (see below). An aliquot of the pellet (5 to 10 mg [wet weight]) was streaked on solid fungal malt extract agar growth medium (3) usually supplemented with 100 ppm streptomycin sulfate (Sigma-Aldrich, Germany). Strains EH5 and EH7 from sulfidic spring water biofilms were isolated and purified by repeated inoculation-growth cycles. A final check of strain purity was performed after staining with a 3% safranin solution (bioMérieux, France) with 2.5% glycerin and 1.65% potassium hydroxide by using phase-contrast microscopy (Axiolab; Carl Zeiss).
Identification of strains EH5 and EH7.
Microscopic examination of strains EH5 (Irnsing-H2S spring) and EH7 (Teugn spring) grown on malt extract agar plates revealed a mycelium morphology that allowed tentative identification of the fungi as M. hiemalis strains. The identities of the strains were then verified by crossing experiments with plus and minus strands of M. hiemalis f. hiemalis (plus-type strain DSM 2655 and minus-type strain DSM 2656; DSMZ, Braunschweig, Germany) (see Results). Additionally, the presence of strains EH5 and EH7 in sulfidic spring water was investigated by using phase-contrast microscopy.
RNA extraction, RT-PCR, and phylogenetic classification of EH5.
Total RNA was extracted from fungal samples using an RNA isolation kit (QIAGEN, Germany) and was quantified by spectrophotometry (A260). The 18S rRNA (0.5 to 1 μg) region was transcribed and amplified to obtain cDNA by reverse transcription (RT)-PCR using primers MUC1(f) and MUC2(r) (Table 1); four replications were performed with a Titan one-tube RT-PCR system kit (Boehringer, Germany). The RT-PCR program consisted of 50°C for 30 min, 94°C for 4 min, and 25 cycles of 94°C for 30 s, 50°C for 45 s, and 68°C for 4 min, followed by 68°C for 10 min and a pause at 4°C to obtain cDNA. Amplification of the target region was checked by electrophoresis on a 1% agarose gel containing 1 μl toluidine blue. The cDNA products were subjected to PCR (100 μl) using a mixture containing 81.3 μl of sterile distilled water, 2 μl of RT-PCR product, either 2 μl of 10 μM MUC1(f) and 2 μl of 10 μM MUC3(r) or 2 μl of 10 μM MUC4(f) and 2 μl 10 μM MUC2(r), 2 μl of a solution containing each deoxynucleoside triphosphate at a concentration of 10 mM, 10 μl of buffer, and 0.7 μl (3.5 U) of Taq polymerase (Boehringer, Germany). The PCR program consisted of 94°C for 3 min and 30 cycles of 94°C for 45 s, 55°C for 1 min, and 72°C for 2 min, followed by 72°C for 10 min and incubation at 4°C to obtain double-stranded DNA. The double-stranded DNA products obtained were purified by HPLC using a linear gradient (0 to 100%) of 1 M NaCl and 20 mM Tris (pH 9) (solution A) with 0.5 M NaCl and 20 mM Tris (pH 9) (solution B) on a TSKgel DNA-NPR-S column (particle size, 2.5 μm; 75 by 4.6 mm; ToshoHaas, Germany) at a flow rate of 0.5 ml/min (8), desalted, and subjected to sequence analysis.
TABLE 1.
Nucleotide sequences of the RT-PCR and PCR primers designed for amplification of 18S rRNA and cDNA, respectively, selected based on the homology of the Mucor racemosus 16S-like rRNA gene sequence with the Mucor mucedo 18S rRNA gene sequence
| Primer | Sequence (5′-3′) | Position | Type |
|---|---|---|---|
| MUC1(f) | TGT GAA ACT GCG AAT GGC TC | 81-100 | Forward |
| MUC2(r) | CTA CTG ATC CCA TAT GCT CAC TAT AAC C | 1678-1651 | Reverse |
| MUC3(r) | CAT TAT TCC ATG CTA ACA CAT TCA AGC | 824-798 | Reverse |
| MUC4(f) | AGT TGT TGC AGT TAA AAC GTC CG | 611-633 | Forward |
A sequence analysis of the PCR products (∼750 to 1,100 bp; concentration, 30 ng/μl) was carried out with a sequencer (model 373; Applied Biosystems, United States) using cycle sequencing Big-Dye terminator chemistry. The 18S rRNA gene sequence was assembled by alignment of partial sequences from PCR products using the Lasergene DNA Star (v.6) software and was compared with the 18S rRNA gene data (NCBI) for identification using the BLASTN 2.0.11 software (1). Reference 18S rRNA gene sequences representing broad fungal diversity were downloaded from the NCBI database and stored in a fungal 18S rRNA gene database. Multiple sequence alignment and calculation of phylogenetic trees for the 18S rRNA gene sequence of the aquatic M. hiemalis strains and other fungal 18S rRNA gene sequences along with sequences of outgroup taxa (Aspergillus fumigatus [accession no. M55626], Boletus satanas [M94337], Candida albicans [X53497], Kluyveromyces lactis [X51830], Leucostoma personii [M83259], Penicillium chrysogenum [M55628], and Russula compacta [U59093]) (44) were performed using the ClustalW software (version 1.8.3; http://www.ebi.ac.uk/clustalw/), and data were exported in the Nexus format for further processing with the PAUP 4.0b10 software (Sinauer Associates, United States). A phylogenetic tree was constructed using optimality criterion parsimony and bootstrap analysis with 1,000 replicates. The phylogenetic tree constructed was visualized and exported as an enhanced metafile by using the TreeView software (version 1.6.6; http://taxonomy.zoology.gla.ac.uk/).
FISH of EH5.
A FISH protocol to detect aquatic M. hiemalis was tested by using biofilms from the Irnsing-H2S, Bad Gögging, Marching, Teugn, Sippenauer Moor, Schwandorf, and Künzing cold sulfidic springs (Bavaria) and was verified by isolating pure cultures of M. hiemalis from the biofilms. The FISH analysis was carried out at least in duplicate using object slide wells (Marienfeld, Germany). An oligonucleotide probe containing the specific antisense sequence MH1 (5′-TCG CCG ACC GAA GCC AAC AA-3′) (see Results) of M. hiemalis f. hiemalis was designed using the NCBI data bank to target the complementary 18S rRNA gene sequence, synthesized, and coupled to the fluorescence dye Texas Red (Molecular Probes, United States) at the 5′ position. The oligonucleotide probes were purified by HPLC and desalted (HPSF grade; MWG-Biotech, Germany) prior to use. Water without oligonucleotide probes was used as a negative control in parallel incubations. The biofilm samples were fixed with 5% formaldehyde in PBS (pH 7.4), and an aliquot (20 μl) was transferred to a slide well (Marienfeld, Germany), fixed to the slide by quick passage through a Bunsen burner flame, dehydrated using an ethanol gradient (50, 70, and 100%), washed twice with 50 μl PBS (pH 6.0), incubated with 50 μl chitinase (concentration, 0.5 μg/μl in PBS [pH 6.0]; Sigma, Germany) at 25°C for 60 min, washed twice with 50 μl PBS (pH 7.4) to remove excess reagents, incubated with 50 μl of a proteinase K solution (concentration, 0.1 to 0.5 μg/ml in 5× Tris-HCl-EDTA buffer [pH 7.2]) at 25°C for 5 to 10 min, washed twice with 50 μl PBS (pH 7.4), incubated in 20 μl of hybridization stock solution (see below) containing 1 μl of an oligonucleotide probe (MH1 coupled to Texas Red at the 5′ position) or water at 68°C overnight (see below), and finally washed twice with 50 μl 5× Tris-HCl-EDTA buffer (pH 7.2) prior to fluorescence microscopy. The hybridization stock solution (400 μl) (see above) was prepared by using a modified protocol (48) and consisted of 6.3 μl of 10% sodium dodecyl sulfate (SDS), 50 μl of 0.8% N-lauroyl-sarcosine, 4.5 μl of deionized formamide, 40 μl of diethyl pyrocarbonate-treated water, 100 μl of 20× SSC (1× SSC is 0.15 M sodium chloride and 0.15 M sodium citrate) (pH 7), and 199.2 μl of water.
In situ fluorescence imaging of GST proteins.
Specimens were visualized with a fluorescence microscope (Axiolab; Carl Zeiss, Göttingen, Germany), and excitation and emission wavelengths were set depending on the fluorescence label. Pictures were taken with a Coolpix 995 camera (Nikon).
Laser scanning confocal fluorescence microscopy of GST-anti-GST complexes was performed with laser excitation at 495 nm and emission at 510 to 525 nm (maximum emission, 519 nm) (LSM model 410; Carl Zeiss). The fluorescence z-scanning technique (23, 24) was used to visualize GST proteins (see below).
Visualization of GST proteins in the microbial community.
The GST proteins present in the microbial community of the Irnsing-H2S spring were visualized by labeling with anti-GST Alexa Fluor 488 conjugates (Molecular Probes, United States). A concentrated purified biofilm pellet (ca. 10 mg [wet weight] [see above]) was suspended in 2.5 ml of PBS (pH 7.4), and an aliquot (ca. 2.5 mg [wet weight] in 250 μl) was transferred to an Eppendorf cup. Anti-GST Alexa Fluor 488 conjugates (2 mg/ml) were diluted 1:200 and wetted with the ProTrans reagent (MoBiTec, Germany) (see below). Luria-Bertani medium supplemented with 10 g/liter glucose and liquid C- and N-rich medium (26) were mixed at a ratio of 1:1 (vol/vol). Then 1 ml of the mixture was transferred to the Eppendorf cup. The wetted antibody mixture was added to the cells, gently shaken, and incubated with light protection at ca. 20°C for at least 4 h.
Treatment with STS.
M. hiemalis strain EH5 was cultivated on sodium thiosulfate (STS)-containing malt extract agar plates for ca. 4 weeks. STS (Merck, Darmstadt, Germany) was added to hot (ca. 60°C) liquid malt extract agar (buffered with Tris-HCl, pH 8.3) to final concentrations of 0 to 3.25 mM.
GST enzyme preparation.
Preparation of samples from biofilms and M. hiemalis strains was carried out as described previously (41) usually in triplicate, with some modifications. Freshly harvested, concentrated biofilm pellets (ca. 1 g [wet weight]/sample) after centrifugation (see above) or fungal mycelium (5 g [fresh weight]/sample; ca. 4-week-old cultures) scraped off malt extract agar plates was quickly frozen with liquid nitrogen. Frozen samples were immediately homogenized, purified, and fractionated to obtain microsomal and cytosolic protein fractions as described previously (41).
GST activity assay and protein determination.
The GST activities in the microsomal and soluble cytosolic fractions were determined (14, 19, 41) using 1-iodo-2,4-dinitrobenzene (IDNB) (Sigma-Aldrich), CDNB and fluorodifen (Riedel de Haën, Germany), DCNB (Fluka, Germany), and EPNP (Fisher Scientific, Germany) as substrates. The formation of glutathione conjugates from reduced glutathione (GSH) (Sigma-Aldrich, Germany) and substrates was measured using a double-beam photometer at 340 nm for CDNB-GSH, at 345 nm for DCNB-GSH, at 354 nm for IDNB-GSH, at 360 nm for EPNP-GSH, and at 370 nm for fluorodifen-GSH over a 3-min period. The protein contents of the microsomal and soluble fractions were determined (5) using bovine serum albumin as a standard (Bradford protein kit; Sigma-Aldrich).
HPSEC and test for linearity of the fluorescence signal.
Fluorescently labeled or unlabeled GST proteins (1 to 10 μg) were separated by high-performance size exclusion chromatography (HPSEC) with a Shimadzu biocompatible 10Ai HPLC system equipped with a photodiode array detector (model SPD-M10A VP; Shimadzu) and a Merck-Hitachi fluorescence detector (model F-1050). UV detection was performed at 260 nm, and fluorescence signals were detected at 519 nm (excitation wavelength, 495 nm). Proteins were separated on a GFC 300-8 HPSEC column (300 by 7.7 mm; particle size, 8 μm; Macherey-Nagel, Germany) with a GFC 8P precolumn (50 by 7.7 mm) packed with the same gel (21). For hydrophilic proteins the optimized mobile phase consisted of 20 mM NaH2PO4 and 200 mM NaCl according to the manufacturer's protocol, and the optimum flow rate of the mobile phase was set at 0.3 ml/min. A small amount (0.1%) of methanol was added in some cases at flow rates of 0.3 to 0.5 ml/min to suppress hydrophobic interactions during analysis of stable proteins. The column was calibrated each time with 1 to 2.6 μg of the native protein standards horse ferritin (Mr, 450 × 103), bovine catalase (240 × 103), bovine albumin (67 × 103), egg albumin (45 × 103), chymotrypsinogen A (25 × 103), equine myoglobin (17.8 × 103), and cytochrome c (12.4 × 103) (Serva, Germany). The void volume and total permeation volume at a flow rate of 0.3 ml/min were located at retentions times around 25 min (polystyrene sulfonate, sodium salt; 780 kDa) and 37 min (KNO3), respectively. The linearity of the fluorescence signal of Alexa Fluor-labeled anti-GST was checked by using various concentrations.
The cytosolic protein fractions from strain EH5 and biofilms were investigated without incubation or after incubation (pH 7.4, 1 h, 25°C) with Alexa Fluor-labeled GST antibody (MoBiTec, Germany). For comparison, the cytosolic GSTs from EH5 (5 × 106 germinated spores) were purified by the glutathione affinity-magnetic separation technique (Bio-Nobile, Finland) prior to injection into the HPSEC system.
GST activation and labeling using fluorescence labeled-GST antibodies. (i) Activation of GST in sporangiospores.
GST in strain EH5 growing on solid malt extract agar was activated by addition of 0.82 mM STS. Mycelium mats were scraped off solid media containing no STS (control) and 0.82 mM STS, chopped into 1- to 2-cm pieces, and suspended in 45 ml of sterile PBS (pH 7.4). Each suspension was ultrasonicated (10 min) and sieved (mesh size, 0.5 to 1 mm) by using gentle pressure. Sporangiospores were purified from filtrates by repeated centrifugation (4,000 × g, 10 min) and washing of the pellets with PBS (pH 7.4). The purity and amounts of sporangiospores were determined using a bright-field hemacytometer (Sigma, Germany) and microscopic imaging. About 5 × 107 spores from solid media containing no STS and 0.82 mM STS were transferred into 40 ml of liquid C- and N-enriched media containing no STS and 0.82 mM STS (26), respectively. Control liquid medium without spores was used as a blank. The spores were incubated for 24 h at 30°C with continuous shaking at 150 rpm to induce germination. After the germination stage was checked microscopically, germinated spores were purified three times by washing the pellets with C- and N-enriched medium and by centrifugation (4,000 × g, 5 min) to remove any extracellular enzymes and proteins. The remaining pellets were stored at 4°C in the dark until labeling.
(ii) Labeling protocol.
Labeling with antibodies was performed according to the manufacturer's protocol (MoBiTec, Germany), with some modifications (see above). The final volume of the antibody complexes was adjusted to 1 ml with C- and N-enriched medium, and the preparations were added directly to aliquots (2 × 105 spores) of purified pellets of germinated spores in Eppendorf cups and gently shaken. After incubation for 2 and 24 h, samples were analyzed by fluorescence microscopy and laser scanning confocal fluorescence microscopy.
Separation of GST proteins from germinated spores.
The technique used for separation of GST proteins consisted of the following steps: (i) germination of 5 × 107 M. hiemalis EH5 spores at 35°C for 2 days at 120 rpm, (ii) separation of the cells by centrifugation (15,000 × g, 5 min) to obtain pellets, (iii) purification of the pellets by three centrifugation and washing (PBS, pH 7.3) cycles, and (iv) homogenization of the pellets in chilled cell lysis buffer containing protease inhibitor cocktails (Roche, Germany) with precooled glass beads (acid washed; diameter, 105 μm; Sigma) in a vibratory cell disrupter (30 Hz; 3 min; Retsch, Germany). Supernatants containing proteomes were immediately collected by centrifugation (15,000 × g, 1 min) of the homogenate. The GST proteins were picked from the proteomes by using the following techniques: (i) magnetic GST affinity and elution protocol (Bio-Nobile, Finland), (ii) anti-GST Alexa Fluor conjugation (MoBiTec) using a standard immunoprecipitation protocol (Roche, Germany), and (iii) conjugation of anti-GST Alexa Fluor coupled to magnetic secondary anti-rabbit protein A antibody BioMag (QIAGEN, Germany) using a magnetic pick pen (Bio-Nobile, Finland). GST proteins were eluted from the antibodies (technique iii) using 50 mM glycine buffer with 0.1% IGEPAL (Sigma) and 0.15 M NaCl at pH 2.8; for longer storage the eluates were immediately neutralized with phosphate buffer (pH 7.3), dialyzed against ultrapure water using a MidEx kit (Serva, Germany), lyophilized, and stored at −20°C.
Electrophoresis and staining.
A vertical mini gel electrophoresis system (Anamed, Germany) with precast Tris-glycine gradient (4 to 20%) gels (10 by 10 cm; thickness, 1.5 mm; Anamed) was routinely used for separation of GST proteins from activated spores (see above). Fresh Tris-glycine-SDS running buffer was prepared from a ready-made stock solution (10×; Anamed). Mixtures of protein standards having molecular masses in the range from 2.5 to 220 kDa for SDS gel electrophoresis (Anamed) were used for calibration. GST protein samples were diluted 1:1 with SDS sample buffer (2×; Anamed) containing bromophenol blue. Electrophoretic separation of proteins was performed at 120 V (constant voltage). After separation, the gels were stained with Sypro-Ruby (Invitrogen, United States) and scanned (Typhoon 9400 fluorescence scanner; GE Healthcare Bio-Sciences, United Kingdom) by following the instructions of the manufacturer.
Statistical analysis.
The data were analyzed statistically (Student's t test, regression analysis) and fitted by using the SigmaPlot 8.0 for Windows software (SPSS Inc., United States). Nonparametric Mann-Whitney U tests were carried out with small sample numbers (52).
Strain deposition.
Strain EH5 has been deposited in the DSMZ (Braunschweig, Germany) as strain DSM 14200.
RESULTS
In order to determine the effects of sulfur stress on induction of GSTs in biofilms and M. hiemalis aquatic strain EH5 from cold sulfidic spring water, as well as on immunofluorescence localization and characterization of GSTs, especially during fungal spore germination, we investigated the chemical and physical properties of the springs (Table 2), as well as the occurrence of GSTs in biofilms and pure fungal cultures. We also examined GST induction by sulfurous substrates with M. hiemalis strain EH5 using various GST isoenzyme substrates. Prior to the use of polyclonal GST antibody, its specificity was verified.
TABLE 2.
Chemical and physical data for cold sulfidic spring watersa
| Parameter | Irnsing-H2S spring | Teugn spring |
|---|---|---|
| Spring discharge (liters/min)b | 200-400 | 150-200 |
| Temp (°C) | 9.7-10.4 | 12.4-12.7 |
| Electrical conductivity (μS/cm) | 646-669 | 665-697 |
| pH | 5.4-6.5 | 4.7-6.4 |
| Redox potential (mV) | −168 to −210 | −192 to −215 |
| Oxygen concn (mg/liter) | 2.1-2.2 | 0.7-1.2 |
| H2S concn (mg/liter) | <1 | NMd |
| Concn of cations (mg/liter) | ||
| Na+ | 9.3-11.4 | 50.7-54.7 |
| K+ | 1.1-1.5 | 6.5-6.9 |
| Mg2+ | 29.4-31.3 | 22.7-22.9 |
| Ca2+ | 73.2-85.4 | 72.7-74.2 |
| Mn2+ | NDe | 7.6-9.5 |
| Concn of cations and total metals (μg/liter) | ||
| Ba2+ | 13.5-17.3 | 47.0-58.2 |
| Total Co | 0-12.4 | 0-16.0 |
| Total Cu | 2.8-5.5 | 1.1-7.6 |
| Total Fe | 6.9-12.5 | 9.8-16.7 |
| Li+ | 5.9-7.3 | 41.8-49.5 |
| Sr2+ | 64.7-109.1 | 78.9-117.2 |
| Zn2+ | 378.3-427.9 | 265.2-288.4 |
| Concn of anions (mg/liter) | ||
| Total S2− | 0.4-0.6 | 1.1-1.3 |
| Cl− | 19.6-21.0 | 25.5-26.3 |
| NO3− | 7.1-12.5 | 0.2 |
| SO42− | 25.1-27.2 | 16.1-26.2 |
| HCO3− | 345.1-349.4 | 420.6-420.9 |
| Dissolved organic carbon concn (mg/liter) | 0.3-0.7 | 0.4-1.1 |
| Atrazine concn (mg/liter)c | 0.04 | ND |
| Desethylatrazine concn (mg/liter)c | 0.04 | ND |
The data for the Irnsing-H2S spring are compared to the data for the low-stress Teugn spring (lower levels of nitrate and zinc; atrazine and desethyltrazine not detectable in spring water) (September 2002). Some important contrasting chemical data are indicated by boldface type.
The spring discharge varies depending on metereological parameters.
Data from pesticide analysis by Wasserwirtschaftsamt Schweinfurt (see Materials and Methods).
NM, not measured.
ND, not detected.
Chemical and physical data for springs and biofilms.
Although most chemical and physical parameters of the Irnsing-H2S and Teugn springs were similar, higher levels of zinc, nitrate, atrazine, and desethylatrazine were detected in the Irnsing-H2S spring than in the Teugn spring (Table 2). Enrichment of various metals, including heavy metals, occurred in the biofilms even when there were trace concentrations in the spring waters (Table 2), and the enrichment factors were as follows: in the Irnsing-H2S biofilm, 5.6 × 105 for Al, 5.6 × 102 for Co, 1.4 × 103 for Cr, 1.9 × 102 for Cu, 7.9 × 104 for Fe, 5.5 × 104 for Li, 7.9 × 104 for Mn, 4.2 × 103 for Ni, 5.4 × 103 for Pb, and 3.7 × 102 for Sr; and in the Teugn biofilm, 5.8 × 105 for Al, 3.1 × 102 for Co, 9.0 × 102 for Cr, 4.6 × 102 for Cu, 5.8 × 104 for Fe, 1.5 × 103 for Li, and 4.6 × 102 for Mn (15). Nickel, lead, and strontium were not detected in the Teugn biofilm.
Specificity of polyclonal anti-GST antibody.
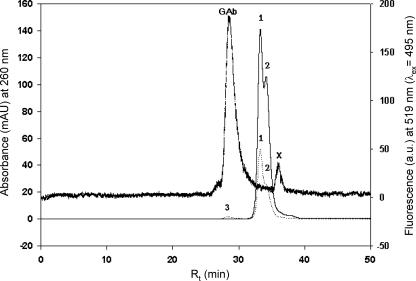
HPSEC separation of the Alexa Fluor-antibody-labeled, affinity-purified cytosolic GSTs (the absorption maximum was at an excitation wavelength of 495 nm for Alexa Fluor) of germinated EH5 spores revealed the presence of a fluorescence peak corresponding to Alexa Fluor-labeled conjugate and concomitant decreases in glutathione affinity-purified GST peaks 1 and 2 at ∼55.6 and ∼51.2 kDa, respectively (Fig. 1). The increase in the A260 of peak 3 for the GST-antibody complex was not proportional to the decrease in GST peaks 1 and 2, apparently due to a shift in the absorption maximum for the GST-antibody complex formed toward 495 nm for Alexa Fluor-labeled polyclonal anti-GST and due to a decrease in elution caused by nonspecific binding of GST-anti-GST complex in the gel matrix. The presence of an Alexa Fluor-specific fluorescence peak at 519 nm along with decreases in the GST peaks (A260) confirmed the presence of GSTs in cytosolic GST-active fractions.
FIG. 1.
HPSEC elution profiles of purified GST fractions from M. hiemalis strain EH5. The thin solid line indicates the profile for the cytosolic GST fraction (A260) with GST peaks 1 (major amount) and 2, the dotted line indicates the profile for the cytosolic GST fraction after treatment with a GST antibody (A260), and the thick dashed line shows the corresponding fluorescence signal (fluorescence at 519 nm) for the cytosolic GST fraction after treatment with the GST antibody. 1, native GST peak 1 (A260); 2, native GST peak 2 (A260); 3, GST antibody (A260); GAb, GST—anti-GST complex (fluorescence at 519 nm); mAU, milli absorbance units; a.u., arbitrary units; λex, excitation wavelength; x, low-molecular-weight impurity (Alexa Fluor).
Plots of the fluorescence signals at 519 nm (excitation wavelength, 495 nm) of Alexa Fluor-labeled anti-GST-GST complexes versus the amounts of protein determined in the HPSEC analysis of the cytosolic, soluble GST fractions from biofilms and pure cultures were linear (y = −7.264 + 81.208x; r2 = 1), demonstrating the specificity of the GST antibody technique employed for analysis of GSTs in biological samples.
Microbial community and GST activity.
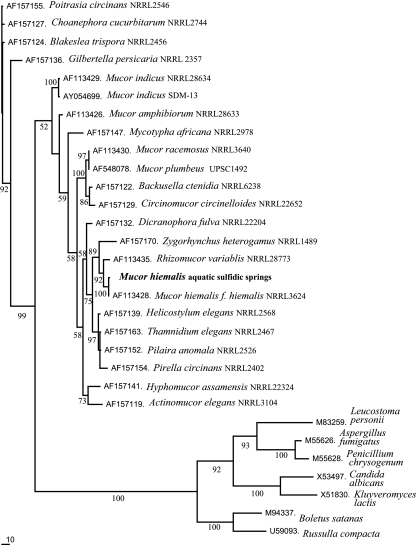
The biofilms from the Irnsing-H2S and Teugn springs contained diverse microbial communities, and the communities had similar compositions (approximately 15% Euryarchaeota, 5% α-Proteobacteria, 10% [Teugn] or 15% [Irnsing-H2S] β-Proteobacteria, 20% γ-Proteobacteria, 10% δ-Proteobacteria, 10% cytophagas and flavobacteria, 5% [Irnsing-H2S] or 15% [Teugn] high-G+C-content bacteria, 20% fungi) as detected by FISH. Pure cultures of the EH5 and EH7 fungi were isolated from the Irnsing-H2S and Teugn biofilms, respectively, and identified as aquatic M. hiemalis f. hiemalis (plus strand) on the basis of morphology and zygospore formation after mating with the minus strand of M. hiemalis f. hiemalis strain DSM 2656. Strain EH5 was also identified on the basis of the high level of similarity (98.6%) of its 18S rRNA gene sequence with that of M. hiemalis f. hiemalis (plus strand) (22). Based on 18S rRNA gene sequence phylogeny, the aquatic strain M. hiemalis EH5 is closely related to the terrestrial fungus M. hiemalis f. hiemalis DSM 2655. It also is phylogenetically related to some extent to Rhizomucor variablis, which clusters in a subset with Zygorhynchus heterogamus (Fig. 2). Some other aquatic M. hiemalis strains were found to occur in the microbial communities in biofilms from sulfidic spring waters from Bad Gögging, Marching, Sippenauer Moor, Schwandorf, and Künzing (Bavaria), as shown by cultivation, direct phase-contrast microscopy of biofilm samples, and FISH analysis (15).
FIG. 2.
Phylogenetic tree based on 18S rRNA gene sequence data showing the position of aquatic M. hiemalis isolated from a sulfidic-sulfurous spring water biofilm. The topology of the phylogenetic tree was calculated by using the ClustalW and PAUP 4.0b software with 1,000 bootstrap trials and was visualized by using the TreeView software. Reference sequences were selected from the NCBI database to represent broad fungal diversity. Bar = 10% estimated difference in nucleotide sequences. Bootstrap values greater than 50% are indicated at the nodes.
The total GST (microsomal and cytosolic) activities in the Irnsing-H2S biofilm were statistically significantly (P ≤ 0.028, as determined by the Mann-Whitney test) greater than the total GST activities in the Teugn (low-stress spring) biofilm and the total GST activities of noninduced aquatic strain M. hiemalis EH5 (Irnsing-H2S) and EH7 (Teugn) and terrestrial M. hiemalis f. hiemalis strain DSM 2655 pure cultures for all of the compounds tested (Table 3). The microsomal GST activities of the Irnsing-H2S biofilm were always greater than the corresponding activities of the Teugn biofilm. The cytosolic (227,540 nmol min−1 mg−1) and microsomal (149,780 nmol min−1 mg−1) GST activities of the Irnsing-H2S biofilm were found to be relatively high when EPNP was used as a substrate. The cytosolic and microsomal GST activities with fluorodifen in biofilms were approximately 10 to 20% of the GST activities with EPNP. The lowest cytosolic GST activity (3,568 nmol min−1 mg−1) and the lowest microsomal GST activity (8,703 nmol min−1 mg−1) in the Irnsing-H2S biofilm were observed with the substrates IDNB and CDNB, respectively. Corresponding to the total in situ GST activities with EPNP in the Irnsing-H2S and Teugn biofilms, pure cultures of strains EH5 and EH7 from these biofilms exhibited similar variations in the total GST activities with EPNP. The cytosolic and the microsomal GST activities of EH5 and EH7 exceeded those of M. hiemalis f. hiemalis terrestrial strain DSM 2655, which was not adapted to the cold sulfidic spring water conditions (Table 3).
TABLE 3.
GST specific activities in cytosolic and microsomal protein fractions of biofilms and M. hiemalis strain EH5 from the cold sulfidic spring water of the Irnsing-H2S spring, of biofilms and M. hiemalis strain EH7 from the low-stress Teugn spring, and of the reference terrestrial strain M. hiemalis f. hiemalis DSM 2655a
| Sample | Protein fraction | GST sp act (nmol min−1 mg protein−1)
|
||||
|---|---|---|---|---|---|---|
| CDNB | IDNB | DCNB | EPNP | Fluorodifen | ||
| Biofilm, Irnsing-H2S | Cytosol | 4,331 | 3,568 | 5,664 | 227,540 | 27,532 |
| Microsomesb | 8,703 | 14,092 | 14,235 | 149,780 | 38,099 | |
| Totalb | 13,034 | 17,660 | 19,899 | 377,320 | 65,631 | |
| Biofilm, Teugn | Cytosolc | 7,883 | 5,645 | 6,809 | 32,640 | 8,777 |
| Microsomes | 135 | 234 | 160 | 1,579 | 2,154 | |
| Totalc | 8,018 | 5,879 | 6,969 | 34,219 | 10,931 | |
| Strain EH5, Irnsing-H2S | Cytosol | 235 | 422 | 280 | 15,542 | 1,011 |
| Microsomes | 255 | 589 | 150 | 14,673 | 1,646 | |
| Totald,e | 490 | 1,011 | 430 | 30,215 | 2,657 | |
| Strain EH7, Teugn | Cytosol | 491 | 733 | 255 | 7,385 | 1,192 |
| Microsomes | 516 | 737 | 307 | 15,911 | 2,693 | |
| Totald,e | 1,007 | 1,470 | 562 | 23,296 | 3,885 | |
| Strain DSM 2655 (terrestrial) | Cytosol | 153 | 160 | 138 | 4,701 | 316 |
| Microsomes | 188 | 171 | 46 | 6,303 | 635 | |
| Total | 341 | 331 | 184 | 11,004 | 951 | |
All the M. hiemalis strains investigated were plus-strand strains. The standard deviations for all the data were less than 10%. The statistical significance of the data was verified using the nonparametric Mann-Whitney test (52).
Probability of errors: P ≤ 0.028 for a comparison of the Irnsing and Teugn data.
Probability of errors: P ≤ 0.048 for a comparison of the Teugn and EH5 data.
Probability of errors: P ≤ 0.111 for a comparison of the EH5 and DSM 2655 data and P ≤ 0.075 for a comparison of the EH7 and DSM 2655 data.
No significant difference between the EH5 and EH7 data.
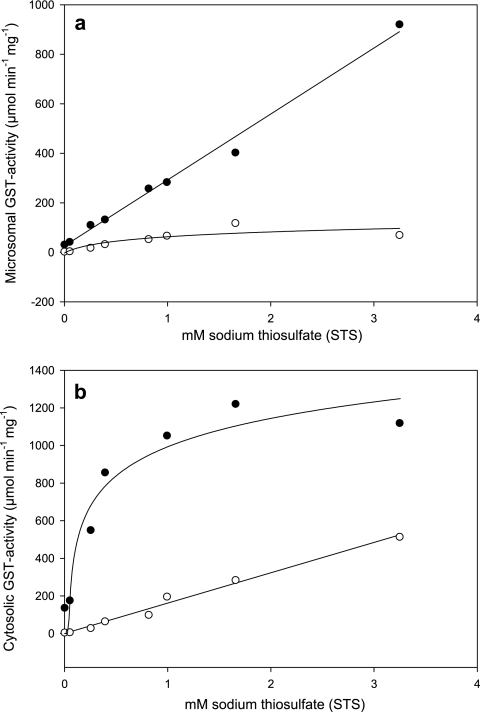
Effects of STS on GST activity.
STS at concentrations of ≥0.051 mM strongly induced statistically significant (P ≤ 0.05, as determined by Student's double t test) microsomal and cytosolic soluble GST activities in strain EH5 (Fig. 3a and b). The highest microsomal GST activity with CDNB (118.08 μmol min−1 mg protein−1), with 1.66 mM STS, was 44-fold greater than the activity of the control, and the activity decreased slightly as the STS concentration was increased to 3.25 mM. The response of the microsomal GST activity with CDNB in the presence of STS was found to be nonlinear with saturation-like effects at STS concentrations greater than 1 mM and could be described by a logarithmic three-parameter function (Fig. 3a) (r2 = 0.76). In contrast, activation of the microsomal GST activity with fluorodifen was strictly linear up to an STS concentration of 3.25 mM (Fig. 3a) (r2 = 0.99). At this STS concentration the microsomal GST activity with fluorodifen was 921 μmol min−1 mg protein−1, a 29-fold increase compared to the control. In contrast to the microsomal GST activity with CDNB (Fig. 3a), the soluble cytosolic GST activity with CDNB increased strictly linearly with addition of STS (Fig. 3b) (r2 = 0.99). With 3.25 mM STS, the cytosolic GST activity with CDNB was 94-fold greater (514.67 μmol min−1 mg protein−1) than the control activity. The cytosolic GST activity with fluorodifen that depended on the STS concentration was nonlinear, exhibited saturation-like effects around 1,000 μmol min−1 mg protein−1 at STS concentrations greater than 1 mM, and could be described by a logarithmic three-parameter function (r2 = 0.94) (Fig. 3b).
FIG. 3.
Induction of GST activity in M. hiemalis strain EH5 by STS. The microsomal (a) and soluble cytosolic (b) GST activity with CDNB (○) and GST activity with fluorodifen (•) of M. hiemalis strain EH5 (ca. 4-week-old cultures) induced by different STS concentrations are shown. The responses of cytosolic GST activity with CDNB and microsomal GST activity with fluorodifen to STS concentrations were found to be linear and could be described by the following regression equations: y = 0.6278 + 161.3227x (r2 = 0.99) and y = 26.0004 + 266.3944x (r2 = 0.99), respectively. The responses of microsomal GST activity with CDNB and cytosolic GST activity with fluorodifen to STS could be represented by the following nonlinear three-parameter logarithmic functions showing slow saturation effects at STS concentrations higher than ∼1 mM: y = 58.9046 + 31.0094[ln(x + 0.1364)] (r2 = 0.76) and y = 999.8441 + 213.1669[ln(x − 0.0306)] (r2 = 0.94), respectively. The standard deviation (n = 3) at each data point was usually less than 10%.
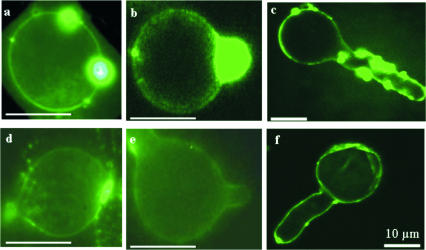
In situ imaging of GSTs after induction by STS.
GST production in germinating sporangiospores without activation by STS occurs at the plasma membrane and cell walls and especially in elongating hyphae of strain EH5. Figure 4 shows a comparison of the sequences of activation of GSTs by STS in sporangiospores during germination (Fig. 4a to c) and in a noninduced control (Fig. 4d to f). Large amounts of GST were formed during the pregermination phase and accumulated at a specific site. This site could have represented a germination site, as GST readily accumulated in the hyphal elongation zones (Fig. 4a and b). Laser scanning confocal fluorescence microscopy showed that GSTs were induced by STS in germinating sporangiospores of strain EH5 especially at the plasma membrane and cell walls (Fig. 4c). When the hyphae were elongating during induction by STS, GST was densely spread over the whole germinated spore and the hyphae themselves (Fig. 4c).
FIG. 4.
Localization of in situ GST induction by STS employing Alexa Fluor-labeled GST antibody at various growth stages of germinating sporangiospores of M. hiemalis strain EH5. Strong GST-specific green fluorescence using anti-GST Alexa Fluor was observed in sporangiospores grown in the presence of 0.82 mM STS (a to c), in contrast to the fluorescence without STS (d to f). Panels a, b, d, and e were obtained by conventional fluorescence microscopy (bars = 10 μm), whereas panels c and f were obtained by using laser scanning confocal fluorescence microscopy (bars = 10 μm). The GST antibody immunofluorescence staining in an elongated hypha activated by 0.82 mM STS (c) is stronger than the fluorescence staining in the control (f), analogous to the activation of GST enzyme activity by STS (Fig. 3).
SDS-PAGE.
The cytosolic and microsomal GSTs from germinated EH5 sporangiospores obtained by using the glutathione-affinity GST magnetic separation method (Bio-Nobile) and by using Alexa Fluor-labeled anti-GST antibody coupled to magnetic secondary antibody were eluted at pH 2.8, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and stained with Sypro-Ruby. Two GST bands at molecular masses of ∼27.8 kDa (major amount) and ∼25.6 kDa were found in the cytosolic GST fractions of EH5, whereas a single band at ∼27.8 kDa was found in the microsomal GST fraction of EH5 (data not shown). The glutathione-affinity GST magnetic separation method and analysis with Alexa Fluor-labeled GST antibody coupled to magnetic secondary antibody confirmed the presence of similar GST bands. Both GST bands corresponded to the rat liver GST subunits with molecular masses of ∼25.6 and ∼27.8 kDa. Major and minor GST bands detected by SDS-PAGE in cytosolic GST fractions of EH5 corresponded to the major and minor GST peaks around a retention time of 33 min measured by HPSEC (Fig. 1). Use of Alexa Fluor-labeled GST antibody bound to magnetic secondary antibody was found to be an interesting alternative for separating GSTs from bulk protein extracts, which could be used in addition to the glutathione-affinity magnetic bead separation technique for isolation and purification of diverse GST isoenzymes in a single step.
DISCUSSION
The continuous flow (150 to 400 liters min−1) of sulfidic spring waters means that there is a significant potential for accumulation of substances dissolved in spring water in biofilms even when the concentrations in the spring outflow are low (Table 2). Indeed, various metals accumulated in the biofilms with high enrichment factors compared to the low concentrations in spring water (see Results) (15). The higher concentrations of zinc, nitrate, atrazine, and desethylatrazine in Irnsing-H2S spring water and the greater accumulation of various heavy metals (Co, Cr, Mn, Ni, and Pb) (15) in the Irnsing-H2S spring biofilm than in the Teugn spring biofilm may be considered initial stress factors (9, 18, 25, 28) and thus may synergistically stimulate various GST isoenzyme activities in the cytosolic or microsomal GST fractions of sulfidic spring water biofilms. It can be argued that the biosynthesis of soluble cytosolic GSTs may be induced more quickly than the biosynthesis of microsomal GSTs so that the organisms can respond rapidly to stress situations. Thus, the microsomal GSTs in biofilms may record stress history for a longer period than the cytosolic GSTs. Using the specific polyclonal Alexa Fluor-labeled GST antibody (Fig. 1) and classical GST activity analysis method, we localized and detected GST-like proteins in situ in biofilms, especially in fungi and bacteria (data not shown), corresponding to the detection of GST activities. For the first time, we isolated aquatic strains EH5 and EH7 of M. hiemalis from sulfidic springs. By using RT-PCR with rRNA (Table 1), sequencing, and 18S rRNA gene phylogenetic analysis, we showed that the aquatic M. hiemalis strains are related to the terrestrial organism M. hiemalis f. hiemalis (Fig. 2). When the GST activities of the aquatic M. hiemalis strains and M. hiemalis terrestrial strain DSM 2655 were determined, the results showed that the activities of aquatic strains EH5 and EH7 were similar but that the activities of both of these strains were different from the activities of M. hiemalis terrestrial strain DSM 2655 (Table 3). The measured GST activities in biofilms (Table 3) and the measured GST activities of the isolated fungal strains (Fig. 1 and 3) supported the in situ observations for GSTs. Corresponding to the greater stress in the Irnsing-H2S spring than in the Teugn spring (Table 2) (see Results), the following sequence for total (cytosolic and microsomal) GST activities with all the xenobiotic substrates tested with statistical significance was found: Irnsing-H2S biofilm > Teugn biofilm > M. hiemalis aquatic strains EH5 and EH7 > M. hiemalis terrestrial strain DSM 2655 (Table 3). In support of our conclusion (see above), the microsomal GST activities of the Irnsing-H2S biofilm were found to be always statistically significantly higher (P ≤ 0.004) than those of the control Teugn biofilm. In contrast, only the soluble cytosolic GST activities with EPNP and fluoridifen of the Irnsing-H2S biofilm were higher than those of the Teugn biofilm, which may have been due to the short exposure of the biofilms to the stress conditions. Compared to the high levels of total GST activity with EPNP (Irnsing-H2S, ∼377,320 nmol min−1 mg protein−1; Teugn, 34,219 nmol min−1 mg protein−1) in the biofilms, the corresponding levels of total GST activity with fluorodifen were approximately three- to sixfold lower, indicating that the cyclic ether bond (epoxide) in EPNP (11) is a better target than the noncyclic ether bond in fluorodifen. The low levels of cytosolic and microsomal GST activities with CDNB, DCNB, and IDNB in biofilms indicated that the expression of the genes encoding these GST isoenzymes in biofilms and M. hiemalis strains was weak. Thus, the very high levels of GST activity that occur naturally in plants, algae, and microorganisms (40, 41, 50) were found in our study mainly in the aquatic biofilms (cytosolic fraction, ∼227,540 nmol min−1 mg−1; microsomal fraction, 149,780 nmol min−1 mg−1) and fungi (EH5 cytosolic fraction, 15,542 nmol min−1 mg−1; EH5 microsomal fraction, 14,673 nmol min−1 mg−1) from cold sulfidic spring waters using EPNP as a substrate. In contrast, the highest level of GST activity in the plant kingdom reported so far was found in Callitriche pallustris and was only 3,390 nmol min−1 mg protein−1 (recalculated) when fluorodifen was used as a substrate (41). The microsomal GST activity in the white rot fungus Phanerochaete chrysopsorium with CDNB was found to be 120 nmol min−1 mg protein−1 (recalculated) (40), but the maximal recorded GST activity (10,000 nmol min−1 mg protein−1) in other microorganisms was found previously in Escherichia coli K-12 strain JM105 using CDNB as a substrate (50). Levels of GST activity up to 4,800 nmol min−1 mg−1 were detected in some other bacteria using CDNB as a substrate (50).
Strong expression of GST activity with EPNP indicated that particularly θ class GST isoenzymes (11) occurred in biofilms and EH5 from the Irnsing-H2S spring. The relatively low GST activities with CDNB, DCNB, and IDNB accompanied by the high GST activities with EPNP in EH5 suggested that a θ class GST was the dominant stress-responsive GST isoenzyme in this strain and this biofilm. Previously, a stress response protein in rats was characterized as a mammalian member of the θ class GSTs (27). By comparison with GST activities with EPNP we concluded that the eukaryote EH5 was probably not the major producer of the θ class GST isoenzyme in the microbial community of the cold sulfidic spring water biofilm. This conclusion is apparently valid for the cytosolic GST activities with EPNP in biofilms as bacterial GSTs are mostly cytosolic (50). As the cytosolic GST activity with EPNP in Irnsing-H2S biofilms was about 50% higher than the corresponding microsomal activity, the possibility that there was physiological induction of the cytosolic GST activity with EPNP and/or relocalization of microsomal GST into the soluble cytosolic fraction of the microbial community under stress conditions (27, 35) cannot be eliminated. The significance of the higher expression of a θ class GST in the Irnsing-H2S biofilm than in the Teugn biofilm seemed to be related to cellular defense and/or adaptation to altered cellular redox conditions (27) due to heavy metal-mediated oxidative stress in biofilms. Based on the GST activities with different xenobiotic substrates, various other GST isoenzymes may be expected to be present in biofilms and EH5 from the Irnsing-H2S spring. The GST activities with CDNB detected may have included GST isoenzyme π (16). Previously, GST isoenzmye π was shown to be correlated with tumorgenicity and oxidative stress in mammals (49). The GST activity with DCNB indicated that a μ class GST isoenzyme was present (29). The high GST activities with fluorodifen in biofilms and EH5 may indicate either that other GST isoenzymes were present or that there was cross-reactivity of the θ class GST with an electrophilic carbon in the ether bond of fluorodifen (11).
Due to the instability of sulfides under acidic or neutral conditions, toxic H2S is readily produced in sulfidic springs, and this compound can be recognized by its characteristic intense smell. In order to determine the effects of sulfurous compounds on microorganisms from sulfidic springs, preliminary experiments were conducted with sodium sulfide in fungal medium (pH 8.3), but they showed that the inhibition of protein synthesis in EH5 by increasing sulfide concentrations was accompanied by stimulation of GST activity (data not shown). Therefore, the relatively nontoxic compound STS was chosen to simulate sulfur stress in microorganisms from sulfidic spring waters.
The linear and nonlinear kinetics of GST induction by STS indicate that apparently differential induction of GSTs took place in microsomal and cytosolic cellular fractions differently for various GST isoenzyme classes represented by various xenobiotic model substrates (Fig. 3). The strictly linear increase in the GST activity with fluorodifen in the microsomal fractions indicates that there were nonsaturating conditions for GST isoenzyme classes targeting ether bonds. Similarly, the strictly linear increase in the GST activity with CDNB in the cytosolic fractions indicates that there were nonsaturating conditions for GST isoenzyme classes targeting universal GST substrates (11). In contrast, the kinetics of GST induction for GST activity with fluorodifen in the cytosolic fractions were nonlinear, and at saturation of this activity at STS concentrations greater than 1 mM the universal GST activity with CDNB targeting all class of reactive substrates was induced linearly. This reciprocal behavior suggests that after a certain threshold value for GST activity saturation with fluorodifen, other GST isoenzymes are activated linearly in the cytosol to protect cells from damage due to accumulation of reactive substances under stress conditions. The observed strong stimulation of microsomal and cytosolic GST activities with CDNB and fluorodifen in EH5 by STS is consistent with the activation of microsomal and cytosolic GST activities in biofilms (Table 3) by various stress factors (see above), sulfhydryl reagents (37), sulforaphane (12, 29), and diallyl sulfide (34). However, the low toxicity of STS, the high sensitivity of induction of GST by STS (Fig. 3), and the antidote function of STS open new horizons in medicine for recommending use of this substance as a universal GST stimulator for antioxidative and anticancer prophylactic treatment (E. Hoque, S. Pflugmacher, and M. Wolf, 25 September 2003, German patent application DE 102 08 119 A1.) and thus perhaps for greater longevity. It should be noted, however, that not only the sulfurous substrates described here but also other nonsulfur substances (e.g., butylated hydroxyanisole [39]) may stimulate GST activity. Both biotic and abiotic stimuli in the microbial communities of biofilms in cold sulfidic spring waters may induce GSTs. Thus, GST activity may be induced nonspecifically by general stress factors. During stress, microorganisms are likely to spend energy for biotransformation in order to retain their physiological balance; otherwise delays in growth, development, and mobility might occur and might lead to a reduction in physiological fitness (17).
As fungal spore germination involves activation of various genes (e.g., the RasA gene) and protein synthesis (e.g., adenylate cyclase, cAMP-dependent protein kinase, RasA GTPase, and trehalase [13]), germinated sporangiospores were used for detection of GST activation. Microscopic imaging of fluorescently labeled GST in germinating sporangiospores following induction by STS revealed in situ microsomal and cytosolic GST localization. Using anti-GST antibody, we observed preferred formation of GST in the hyphal initiation and elongation zones, especially during germination of EH5 sporangiospores.
SDS-PAGE, HPSEC analysis of glutathione-affinity-purified GST, and in situ GST localization by immunofluorescence in imaging in EH5 suggested that (i) there are two main θ class homodimeric GSTs (∼27.8 kDa [major amount] and ∼25.6 kDa), one of which (∼25.6 kDa) does not occur in microsomes, and (ii) GST is needed in the fast-growing regions of EH5, apparently to protect the apical cells from physical and chemical stress during penetration and growth. The presence of GST at cell division sites in the fast-growing regions of EH5 is consistent with the report that GST accumulates in plants during cell division (33). We suggest that GST activities may vary depending on the species, strain, cellular fractions, substrate, and/or developmental state.
Taken together, our observations demonstrated clearly that there was microbial cytosolic and microsomal GST activation by various stressors, especially the sulfurous compounds thiosulfate and sulfide. For the first time, the immunofluorescence GST visualization technique used in our study enabled verification of GST activities induced by thiosulfate directly in fungal cells. We propose that GST is a useful biomarker of ecological stress in the aquatic microbial environment. Differential cellular GST expression depending on spore and hyphal development was demonstrated in strain EH5. Due to the high sensitivity and the ease of handling of EH5, this organism may be a convenient eukaryotic model strain for rapid screening and interaction studies of GST effectors (Hoque et al., German patent application DE 102 08 119 A1.).
Acknowledgments
We thank Ernst Mannweiler, Michael Ziegler, and Günter Teichmann for technical assistance as well as Thomas Fröhlich and Rob Chapman for advice and critical reading of the manuscript.
Funding from the Franken Brunnen-Stiftung (Germany) to J. Fritscher is gratefully acknowledged.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipmann. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein data base search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arjmand, M., and H. Sandermann, Jr. 1985. Mineralization of chloroaniline/lignin conjugates and of free chloroanilines by the white-rot fungus Phanerochaete chrysosporium. J. Agric. Food Chem. 33:1055-1060. [Google Scholar]
- 4.Aßmus, B. 1996. In situ-Detektion von Bakterien aus Boden- und Gewässerhabitaten mit spezifischen Markierungen sowie optischen und zytometrischen Methoden. Ph.D. thesis. Faculty of Biology, Ludwig-Maximilians University, Munich, Germany.
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bucciarelli, T., P. Sacchetta, A. Pennelli, L. Cornelio, R. Romagnoli, S. Melino, R. Petruzelli, and C. Di Ilio. 1999. Characterisation of toad glutathione transferase. Biochim. Biophys. Acta 1431:189-198. [DOI] [PubMed] [Google Scholar]
- 7.Burggraf, S., T. Mayer, R. Amann, S. Schadhauser, C. R. Woese, and K. O. Stetter. 1994. Identifying members of the domain Archaea with rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 60:3112-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, B., W. Gimbel, T. Görblich, and T. Werner. 1993. HPLC-purification of PCR-products for direct sequencing. Trends Genet. 9:406. [DOI] [PubMed] [Google Scholar]
- 9.Choi, J. H., W. Lou, and A. Vancura. 1998. A novel membrane-bound glutathione S-transferase functions in the stationary phase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:29915-29922. [DOI] [PubMed] [Google Scholar]
- 10.Diesperger, H., and H. Sandermann, Jr. 1979. Soluble and microsomal glutathione S-transferase activities in pea seedlings (Pisum sativum L.). Planta 146:643-648. [DOI] [PubMed] [Google Scholar]
- 11.Dowd, C. A., C. M. Buckley, and D. Sheehan. 1997. Glutathione S-transferase from the white-rot fungus, Phanerochaete chrysosporium. Biochem. J. 324:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahey, J. W., and P. Talalay. 1999. Antioxidant functions of sulforaphane: a potent inducer of phase II detoxification enzymes. Food Chem. Toxicol. 37:973-979. [DOI] [PubMed] [Google Scholar]
- 13.Fillinger, S., M.-K. Chaveroche, K. Shimizu, N. Keller, and C. de'Enfert. 2002. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001-1016. [DOI] [PubMed] [Google Scholar]
- 14.Fjellstedt, T. A., R. H. Allen, B. K. Duncan, and W. B. Jacoby. 1973. Enzymatic conjugation of epoxides with glutathione. J. Biol. Chem. 248:3702-3707. [PubMed] [Google Scholar]
- 15.Fritscher, J. 2004. Untersuchungen über Sulfid-Schwefelquellen in Bayern. Beiträge zum ökologischen Monitoring und zur Entwicklung von biotechnologischen Methoden für die Grundwasserreinigung. Ph.D. thesis. Faculty of Natural Science, Friedrich-Alexander-Universität, Erlangen-Nürnberg, Germany.
- 16.George, S. G. 1994. Enzymology and molecular biology of phase II xenobiotic-conjugating enzymes in fish, p. 37-85. In D. C. Malins and G. K. Ostrander (ed.), Aquatic toxicology: molecular, biochemical and cellular perspective. Lewis, Searcy, AK.
- 17.Greulich, K., E. Hoque, and S. Pflugmacher. 2002. Uptake, metabolism, and effects on detoxification enzymes of isoproturon in spawn and tadpoles of amphibians. Ecotoxicol. Environ. Saf. 52:256-266. [DOI] [PubMed] [Google Scholar]
- 18.Gronwald, J. W., and K. L. Plaisance. 1998. Isolation and characterization of glutathione S-transferase isoenzymes from sorghum. Plant Physiol. 117:877-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habig, W., M. J. Pabst, and W. B. Jacoby. 1974. Glutathione S-transferase: the first step in mercapturic acid formation. J. Biol. Chem. 249:1730-1739. [PubMed] [Google Scholar]
- 20.Heinrichs, G., E. Hoque, M. Wolf, and W. Stichler. 2000. Hydrogeologische und biologische Besonderheiten der Schwefelquelle von Irnsing bei Neustadt a.d. Donau. Geol. Bl. Nordost-Bayern Angrenz. Geb. 50:1-16. [Google Scholar]
- 21.Hoque, E. 1995. High-performance size-exclusion chromatographic characterization of water-soluble polymeric substances produced by Phanerochaete chrysosporium from free and wheat cell wall bound 3,4-dichloroaniline. J. Chromatogr. A 708:273-281. [Google Scholar]
- 22.Hoque, E. June 2003. Verfahren zum Abbau von Xenobiotika durch Pilzarten mit Monooxygenase-/Dioxygenase-Aktivität in Gegenwart von Pilzen mit Glutathion-S-Transferase-Aktivität. German patent DE 101 25 365 C2.
- 23.Hoque, E., and G. Remus. 1994. Native and atrazine-induced fluorescence of chloroplasts from palisade and spongy parenchyma of beech (Fagus sylvatica L.) leaves. Remote Sens. Environ. 47:77-86. [Google Scholar]
- 24.Hoque, E., and G. Remus. 1999. Natural UV-screening mechanisms of Norway spruce (Picea abies [L.] Karst.) needles. Photochem. Photobiol. 69:177-192. [DOI] [PubMed] [Google Scholar]
- 25.Jablonkai, I., and K. H. Hatzios. 1993. In vitro conjugation of chloracetanilide herbicides and atrazine with thiols and contribution of nonenzymatic conjugation to their glutathione-mediated metabolism in corn. J. Agric. Food Chem. 41:1736-1742. [Google Scholar]
- 26.Kirk, T. K., E. Schultz, W. J. Connors, L. F. Lorenz, and J. G. Zeikus. 1978. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch. Microbiol. 117:277-285. [Google Scholar]
- 27.Kodym, R., P. Calkins, and M. Story. 1999. The cloning and characterization of a new stress response protein. A mammalian member of a family of θ class glutathione S-transferase-like proteins. J. Biol. Chem. 274:5131-5137. [DOI] [PubMed] [Google Scholar]
- 28.Lamoureux, G. L., D. G. Rusness, P. Schröder, and H. Rennenberg. 1991. Diphenyl ether herbicide metabolism in a spruce cell suspension culture: the identification of two novel metabolites derived from a glutathione conjugate. Pestic. Biochem. Physiol. 39:291-301. [Google Scholar]
- 29.Lopez, M. F., W. F. Patton, W. B. Sawlivich, H. Erdjument-Bromage, P. Barry, K. Gmyrek, T. Hines, P. Tempst, and W. M. Skea. 1994. A glutathione S-transferase (GST) isoenzyme from broccoli with significant sequence homology to the mammalian theta-class of GSTs. Biochim. Biophys. Acta 1205:29-38. [DOI] [PubMed] [Google Scholar]
- 30.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 32.Manz, W., R. Amann, R. Szewszyk, U. Szewszyk, T. A. Stenström, P. Hutzler, and K. H. Schleifer. 1995. In situ identification of Legionellaceae using 16S rRNA-targeted oligonucleotide probes and confocal laser scanning microscopy. Microbiology 141:29-39. [DOI] [PubMed] [Google Scholar]
- 33.Marrs, K. A. 1996. The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:127-158. [DOI] [PubMed] [Google Scholar]
- 34.Maurya, A. K., and S. V. Singh. 1991. Differential induction of glutathione transferase isoenzymes of mice stomach by diallyl sulfide, a naturally occurring anticarcinogen. Cancer Lett. 57:121-129. [DOI] [PubMed] [Google Scholar]
- 35.Mehlen, P., and A.-P. Arrigo. 1994. The serum-induced phosphorylation of mammalian hsp27 correlates with changes in its intracellular localization and levels of oligomerization. Eur. J. Biochem. 221:327-334. [DOI] [PubMed] [Google Scholar]
- 36.Morgenstern, R. J., J. W. DePierre, and H. Jörnvall. 1985. Microsomal glutathione S-transferase. J. Biol. Chem. 260:13976-13983. [PubMed] [Google Scholar]
- 37.Morgenstern, R., J. W. DePierre, and L. Ernster. 1979. Activation of microsomal glutathione S-transferase activity by sulfhydryl reagents. Biochem. Biophys. Res. Commun. 87:657-663. [DOI] [PubMed] [Google Scholar]
- 38.Park, H.-J., K.-S. Lee, S.-H. Cho, and K.-H. Kong. 2001. Functional studies of cysteine residues in human glutathion-S-transferase P1-1 by site-directed mutagenesis. Bull. Korean Chem. Soc. 22:77-82. [Google Scholar]
- 39.Pearson, W. R., J. Reinhart, S. C. Sisk, K. S. Anderson, and P. N. Adler. 1988. Tissue-specific induction of murin glutathione transferase mRNAs by butylated hydroxyanisole. J. Biol. Chem. 263:13324-13332. [PubMed] [Google Scholar]
- 40.Pflugmacher, S., P. Schröder, and H. Sandermann, Jr. 2000. Taxonomic distribution of plant glutathione S-transferase acting on xenobiotics. Phytochemistry 54:267-273. [DOI] [PubMed] [Google Scholar]
- 41.Pflugmacher, S., and C. E. W. Steinberg. 1997. Activity of phase I and phase II detoxification enzymes in aquatic macrophytes. J. Appl. Bot. 71:144-146. [Google Scholar]
- 42.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K. H. Schleifer. 1994. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 43.Rudolph, C., G. Wanner, and R. Huber. 2001. Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology. Appl. Environ. Microbiol. 67:2336-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schüßler, A., D. Schwarzzott, and C. Walker. 2001. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105:1413-1421. [Google Scholar]
- 45.Seel, F. 1966. Grundlagen der Analytischen Chemie unter besonderer Berücksichtigung der Chemie in wässerigen Systemen. Verlag Chemie, Weinheim, Germany.
- 46.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systematics. John Wiley and Sons, Chichester, England.
- 47.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenberge, K., P. Stellamanns, G. Plenz, and H. Robenek. 1998. Nonradioactive in situ hybridisation for detection of hydrophobin mRNA in the phytopathogenic fungus Claviceps purpurea during infection of rye. Eur. J. Cell Biol. 75:265-272. [DOI] [PubMed] [Google Scholar]
- 49.Upham, B. L., and J. G. Wagner. 2001. Toxicant-induced oxidative stress in cancer. Toxicol. Sci. 64:1-3. [DOI] [PubMed] [Google Scholar]
- 50.Vuilleumier, S. 1997. Bacterial glutathione S-transferases: what are they good for? J. Bacteriol. 179:1431-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, M., R. Erhart, W. Manz, R. Amann, H. Lemmer, D. Wedi, and K. H. Schleifer. 1994. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 60:792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber, E. 1986. Grundriss der biologischen Statistik. VEB Gustav Fischer Verlag, Jena, Germany.
- 53.Weiss, J. 1991. Ionenchromatographie. VCH, Weinheim, Germany.