Abstract
We used genome fragment enrichment and bioinformatics to identify several microbial DNA sequences with high potential for use as markers in PCR assays for detection of human fecal contamination in water. Following competitive solution-phase hybridization of total DNA from human and pig fecal samples, 351 plasmid clones were sequenced and were determined to define 289 different genomic DNA regions. These putative human-specific fecal bacterial DNA sequences were then analyzed by dot blot hybridization, which confirmed that 98% were present in the source human fecal microbial community and absent from the original pig fecal DNA extract. Comparative sequence analyses of these sequences suggested that a large number (43.5%) were predicted to encode bacterial secreted or surface-associated proteins. Deoxyoligonucleotide primers capable of annealing to a subset of 26 of the candidate sequences predicted to encode factors involved in interactions with host cells were then used in the PCR and did not amplify markers in DNA from any additional pig fecal specimens. These 26 PCR assays exhibited a range of specificity in tests with 11 other animal sources, with more than half amplifying markers only in specimens from dogs or cats. Four assays were more specific, detecting markers only in specimens from humans, including those from 18 different human populations examined. We then demonstrated the potential utility of these assays by using them to detect human fecal contamination in several impacted watersheds.
Waterborne diseases are a significant public health issue, and many originate from contact with water contaminated with human fecal material (2, 10, 19). An estimated 850 billion gallons of untreated human wastewater and storm water are discharged into U.S. surface waters each year (27). Because such combine sewer overflow runoff contains raw sewage capable of carrying numerous human pathogens (e.g., Shigella sonnei, noroviruses, and Cryptosporidium) (8), solids, debris, and toxic pollutants (i.e., antibiotics, hormones, and caffeine, as well as steroids, metals, and synthetic organic compounds) (27), it is an important public health concern. There is also now strong evidence that enterococci and other fecal indicator bacteria in recreational waters themselves contribute to gastrointestinal illness (6, 25), as well as eye, ear, nose, skin, respiratory, and other infections (12, 18, 26). Ensuring public water quality therefore requires that we develop improved methods to more accurately identify human fecal pollution.
The lack of accurate methods for identifying sources of fecal pollution has stimulated the recent development of a number of microbial source tracking (MST) methods. In general terms, these MST methods can be divided into culture-based and culture-independent techniques (23). Most culture-based methods for identification of human sources depend on matching panels of environmental bacterial isolates with known human fecal indicator type strains. A major limitation of this approach is its requirement for the development of large collections of isolates from both water and human fecal samples. Thus, MST methods that do not require cultivation, such as the direct detection of bacterial 16S rRNA gene sequences using PCR, are becoming more widespread (5, 9). While these assays have now been used in field applications (5, 9, 20), a recent study demonstrates understandable cross-reactivity when highly conserved genomic regions are targeted (7). Since ribosomal genes are not directly involved in microbe-host interactions, it is possible that other bacterial genetic markers encoding factors related to host specificity might be better candidates for MST assays. Although a significant number of bacterial genes have been identified as relevant to host-microbe interactions in the human gut (14, 15), the challenge remains to identify which of these genes are from bacteria that are truly restricted to this specific niche. We hypothesize that direct comparisons of the genetic coding capacities of entire human fecal bacterial communities can identify such factors involved in host-microbe interactions and that these would be the best targets for PCR assays designed to identify sources of human fecal contamination (21, 22).
We recently developed a nucleic acid analysis method called genome fragment enrichment (GFE) to identify differences in the genomes of phylogenetically related bacterial species and to identify differences in total microbial community DNA obtained from different sources (21, 22). Here we describe the extension of this approach to address the highly significant problem of identification of human fecal contamination in water, and we report important differences encountered relative to other animal sources. A large number of candidate marker sequences are described, with a major characteristic being that almost half are predicted to encode bacterially secreted or cell surface factors located at the interface with host cells. Four new human-specific PCR assays developed by this approach and potentially useful in diagnosing human fecal pollution in environmental samples are described.
MATERIALS AND METHODS
Sample collection and DNA extraction.
One hundred six individual animal fecal samples were collected and stored as described previously (22). These specimens represented a total of 11 different animal species that likely impact watersheds in the United States. Sixteen wastewater primary effluent samples (Clearwater, FL; Largo, FL; Crystal Lake, IL; Port Huron, MI; Saginaw, MI; Morgan City, LA; St. Peter, MN; Vicksburg, MS; Oxford, AL; Las Vegas, NV; Little Falls, NY; Saranac Lake, NY; Mason, OH; Lowery, OH; Dry Creek, OH; and Fairfield, OH) were collected. In addition, six recreational water samples, two stabilization pond samples, a storm water runoff sample, and a treated wastewater effluent sample were collected from eight different locations in Southern Ohio and Northern Kentucky. One hundred milliliters of each water sample was filtered through 0.2-μm-pore-size Supor-200 filters (Whatman) and each filter placed in a sterile 1.5-ml microtube and stored at −80°C. All DNA extractions were performed using the FastDNA kit for soils (Q-Biogene; Carlsbad, CA) as discussed previously (22). DNA extract yields were determined with a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies; Wilmington, DE). A general Bacteroides-Prevotella 16S rRNA gene PCR assay was used to verify the presence of fecal bacterial DNA in each extract and to detect PCR inhibition (4).
GFE.
A single round of GFE was used to select potential human-specific fecal community genetic markers as described previously (22). Briefly, biotin-labeled sheared total fecal DNA from a single reference human specimen was first prehybridized with sheared DNA fragments from total fecal DNA from an individual reference pig specimen. This “blocked” biotin-labeled DNA was then hybridized to equilibrium in solution with additional DNA fragments from the original source (human total fecal DNA) that contained defined terminal sequence “tags” that had been added by primer extension as described previously (22). DNA hybrids were then isolated by streptavidin binding, and the captured tagged genomic fragments were amplified by PCR using a single primer complementary to the defined 5′ fragment tags (13). The required specificity of the final PCR using either the K9-PCR or F9-PCR tag complementary primer (11) was verified using reference sheared human and pig total fecal DNA as templates. Five GFE PCRs were performed in individual tubes and then mixed to reduce sample variability in amplifying complex nucleic acid pools. The same pig fecal sample used as the GFE “blocker” in a previous report (22) was selected for this study to allow a direct comparison of animal-specific PCR assay development success rates between studies and because human and pigs share similar anatomies, physiologies, and diets.
DNA sequencing.
Final GFE products from five identical parallel PCRs were pooled and incorporated into the pCR4-TOPO plasmid vector as described by the manufacturer (Invitrogen; Carlsbad, CA). Individual Escherichia coli clones were then cultured in 300 μl of Luria broth containing ampicillin (10 μg/ml) and screened for inserts by PCR using M13F and M13R primers. Prior to sequencing, PCR products were purified using the QiaQuick 96 PCR purification kit (QIAGEN; Valencia, CA). Sequencing was performed on both strands at the Cincinnati Children's Hospital Medical Center Genomics Core Facility (Cincinnati, OH) by the dye-terminator method, using an ABI PRISM 3730XL DNA analyzer (Applied Biosystems, Foster City, CA).
Dot blot hybridizations.
Dot blot hybridizations with cloned GFE sequences and pig fecal DNA (GFE blocker) probe were used to identify any “false-positive” plasmid clone inserts obtained by GFE that were not unique to the original human total fecal DNA source. Probe preparation, hybridization conditions, and detection were performed as described previously (22).
Data analysis.
DNA sequence reads were assembled using SeqMan II (DNAstar, Inc.; Madison, WI) and used to search the National Center for Biotechnology Information (NCBI) RefSeq database using BLASTx software (1) (http://www.ncbi.nlm.nih.gov/BLAST/); BLASTx hits with expectation values of ≤1E−03 were designated as homologous. Gene function attributes for DNA sequences were assigned based on annotations available in the Comprehensive Microbial Resource genome database (CMR) at The Institute for Genomic Research (http://www.tigr.org/tigr-scripts/CMR2/CMRGenomes.spl).
Selection of relevant markers for PCR assays.
Twenty-six DNA sequences were chosen for the development of PCR assays based on best BLASTx match annotated gene function attributes potentially relevant to interaction with hosts. Target DNA sequences selected were those where (i) the BLASTx best hit contained either a predicted transmembrane helix or a secretory signal region based on either SignalP (3) or TMHMM prediction (16) or where there was published experimental evidence of cell surface or extracellular activity; (ii) DNA sequences were annotated as encoding noncystolic protease or factors involved in capsular polysaccharide synthesis; or (iii) sequences were predicted to encode proteins with a function other than that associated with DNA metabolism, protein synthesis, antibiotic or multidrug resistance, or mobile element functions. PCR primers were designed using PrimerSelect (DNAstar, Inc.; Madison, WI) with default settings. Candidate primer sequences were aligned with homologous sequences (e-value of ≤e−03) from the NCBI BLASTx analysis using ClustalW (24) with default settings (MegAlign; DNAstar, Inc., Madison, WI). Primer sets that aligned to variable DNA regions among homologous sequences were selected for optimization, host specificity, and limit-of-detection assays.
Primer optimization, host specificity, and limit of detection.
Optimal annealing temperatures were measured for each primer pair using thermal-gradient PCR as described previously (22). To assess specificity, each PCR assay was tested against DNA extracts (2 ng/reaction) from target (human) and nontarget animal fecal samples representing 106 individual specimens. Reference fecal samples represented 11 species of animals, including Bos taurus (cow), Gallus gallus (chicken), Anser sp. (Canadian goose), Canis familiaris (dog), Felis cattus (cat), Capra aegagrus (domestic goat), Sus scrofa (pig), Ovis aries (sheep), Equus caballus (horse), Odocoileus virginianus (whitetail deer), and Homo sapiens (human). The spatial robustness of each human-specific primer set was estimated by testing each PCR assay against a panel of wastewater samples (primary effluent and stabilization pond samples) representing 18 different human populations spanning 11 states. To test for nonspecific amplification of DNA from representative environmental microorganisms and explore the application of each human-specific primer set for water quality monitoring applications, PCR assays were performed using DNA extracted from recreational water (n = 6), storm water (n = 1), and treated wastewater effluent (n = 1) filtrates. The lower limit of detection for each primer set was estimated using serial dilutions of human total fecal DNA starting with a concentration of 10 ng/μl. All validation PCR assays were performed in duplicate. No-template, extraction blank, and water filtration blank PCR control assays were performed to test for the presence of extraneous DNA molecules introduced during laboratory experiments.
PCR assay controls.
Each sample tested yielded the expected PCR product when amplified with the Bacteroides-Prevotella 16S rRNA-specific primers 32F and 708R (4), indicating a lack of PCR inhibitors. To test for the presence of extraneous DNA molecules introduced during laboratory manipulations, no-template (n = 612), extraction blank (n = 35), and water filtration blank (n = 5) controls were included in PCR assays. In all cases the results were negative. Due to the requirement for the single-primer amplification step (13) amplifying DNA fragment pools via terminal tag sequences and the complex nature of metagenomic templates, we also tested the specificities of K9-PCR and F9-PCR primers. No amplification was seen when the K9-PCR primer was tested with sheared human metagenomic DNA, nor when the F9-PCR primer was tested against pig metagenomic DNA (data not shown).
RESULTS
Identification of unique genetic marker sequences for human fecal bacteria.
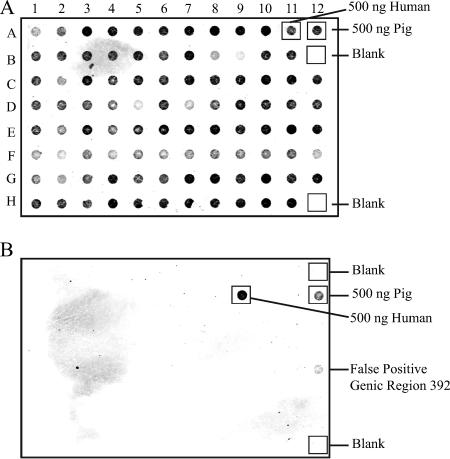
Three hundred fifty-one E. coli plasmid clones of DNA fragments averaging 323 base pairs in size were randomly selected for sequencing from a larger library obtained by a single round of GFE (21). Sequence analyses showed that this subset consisted of a total of 297 nonredundant sequences (i.e., not plasmid clone sibling; see Table S1 in the supplemental material). Dot blot hybridizations using the original pig total fecal DNA as a probe were then used to identify any “false-positive” GFE clones (Fig. 1). Analyses showed that only 6 of the 297 sequences were capable of hybridizing to the original pig total fecal DNA probe, demonstrating a very low false-positive rate (2%) among the nonredundant GFE clones.
FIG. 1.
Dot blot hybridization analysis of putative host-specific DNA fragments. PCR amplicons from all nonredundant clone sequences (93 shown) were transferred to nylon membranes and hybridized to a biotin-labeled human (A) or pig (B) fecal metagenomic DNA probe. Positive controls included 500 ng of human fecal metagenomic DNA (panel A, row A, column 11; panel B, row B, column 9) and 500 ng (panel A, row A, column 12; panel B, row B, column 12) of pig fecal metagenomic DNA. None of the “no DNA” controls (panel A, row B, column 12 and row H, column 12; panel B, row A, column 12 and row H, column 12) hybridized to the probe.
Annotation of nonredundant sequences obtained by GFE.
The 291 GFE clone insert sequences were then aligned and assembled into 289 unique genetic regions. This largely nonoverlapping assembly indicated that a far larger sampling would be needed to even begin to saturate the available DNA sequences that differ in the two reference specimens. BLASTx sequence similarity searches of these regions against the NCBI RefSeq database identified homologous sequences for 241 regions based on an expectation value cutoff of ≤1 × 10−3 (see Table S1 in the supplemental material). The BLASTx hit with the lowest expectation value for each region was designated the best BLASTx hit (see Table S1 in the supplemental material).
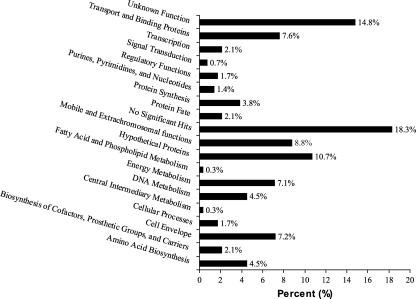
The top BLASTx hits for each region averaged only 57.6% sequence identity (ID) to the actual GFE plasmid clone sequences. Fifty-three of the 289 human fecal community DNA sequences (18.3%) (Fig. 2) showed no similarity to any previously reported gene sequences. All GFE sequence regions obtained were then assigned to 1 of the 18 functional groups described in the CMR based on annotations of best BLASTx sequence matches (Fig. 2) (see Table S1 in the supplemental material). The individual categories most frequently assigned were “unknown function” (14.8%) and “hypothetical proteins” (10.7%). Based on additional existing bioinformatic analyses of best-match genetic regions, 126 GFE sequences were annotated as likely to encode membrane-associated or putative extracellular proteins. Only two distinct DNA sequences were indicated by more than one nonredundant clone obtained by GFE. These sequences were similar to those encoding a hypothetical protein, DR1284 (28), isolated from Deinococcus radiodurans R1, and a previously uncharacterized DNA sequence of unknown function (see Table S1 in the supplemental material). Overall, DNA sequences obtained by GFE were therefore predominately representative of previously uncharacterized coding capacity of the bacteria that compose the human intestinal microbiota.
FIG. 2.
Functional group assignments for GFE nonredundant sequences. Functional groups are listed along the y axis, and the percentage of GFE sequences (total number = 290) for each group assignment is shown along the x axis.
Development of PCR assays for detection of human fecal DNA.
Twenty-six nonredundant GFE genetic regions (Table 1) were selected for development of human fecal community-specific PCR assays based on gene annotations reflecting potential roles in microbe-host interaction using the criteria described above. At optimal PCR annealing temperatures ranging from 56°C to 65°C (Table 1), all 26 assays amplified DNA fragments of the expected size from the original “target” GFE human fecal sample and produced negative results from the original “blocker” GFE pig fecal sample. To test for human fecal marker specificity, each putative primer set was tested with template DNA extracted from 106 different fecal samples representing 11 animal species (Table 2). None of the putative 26 human-specific detection assays amplified DNA from horse, pig, sheep, or goose composite fecal samples. Fourteen of these PCR assays did, however, amplify markers from either domestic dog or cat composite fecal samples (or both), potentially reflecting cohabitation with these species. Eight other PCR assays were also determined to lack necessary specificity in amplifying markers from two or more other animal species. This was a considerably larger amount of marker cross-amplification than in prior comparison of reference cattle (ruminant) and pig (omnivore) bacterial community DNAs, as discussed below.
TABLE 1.
Putative human-specific PCR assay primers, optimal annealing temperatures, and target DNA annotation
| PCR assay no. | Primer set | Sequences (5′ → 3′) | Amplicon length (bp) | Annealing temp (°C) | Target DNA annotation |
|---|---|---|---|---|---|
| 1 | hum5Fc | CCGGCGGTGGCTTTGACTA | 116 | 56 | Putative tricorn-like protease |
| hum5Rc | TCCTCCTTGTGCACTTACCATACC | ||||
| 2 | hum12Fb | GCAGGAAGGCAAATGGTT | 133 | 63 | Periplasmic beta-glucosidase precursor |
| hum12Rb | AGCAGATGCACTCAGGGCGATGCTC | ||||
| 3 | hum13Fa | TCATCATCCTCAAGGCGAACAAT | 115 | 58 | Serine protease precursor |
| hum13Ra | GGGAACATACCGGTGATAAACAAC | ||||
| 4 | hum14Fb | CATCTCCGGACTTGCCATTACTT | 127 | 57 | Putative membrane protein |
| hum14Rb | TTCCGCTCCTTTTATATCTTTCTG | ||||
| 5 | hum24Fb | CATCGGTGGTTCCCTTCAGTC | 141 | 61 | Putative outer membrane protein |
| hum24Rb | TCTTGGGATGGGTTTTTGGTAGTA | ||||
| 6 | hum33Fb | CGGATGTATCGGCAGGTA | 121 | 59 | Putative outer membrane protein |
| hum33Rb | CCGTTTCATAGTTCCAAGCATTAG | ||||
| 7 | hum39Fc | GCCATGAGAAGTTTGCAGAGATAG | 100 | 64 | Putative outer membrane protein |
| hum39Rc | TTGGGAGAAATGGAAAATACC | ||||
| 8 | hum63Fc | CATGAGGAATACTGCCCACTGAAT | 206 | 62 | Putative exported hydrolase |
| hum63Rc | TCCCAATGAACCACGAGACG | ||||
| 9 | hum64Fa | ACCGGTACCTGTTCGTTTGTGT | 228 | 61 | Xanthan lyase |
| hum64Ra | CATTGGCGGTGAAGTTTGTATCT | ||||
| 10 | hum70Fb | CATCACCGTGCAGCAGTATTAGG | 189 | 59 | Putative oligopeptide transporter |
| hum70Rb | AAGTTCGGGTGACATTTCGCTGAT | ||||
| 13 | hum86Fb | TAATGGAAGGATAGAATAAATAGT | 99 | 55 | Putative outer membrane protein |
| hum86Rb | AAAGGACAAAGCCAAAGCATA | ||||
| 14 | hum135Fc | TGGGCATTTACTTCATCC | 101 | 56 | Putative outer membrane protein |
| hum135Rc | GAGCATTTCCCGACAGA | ||||
| 15 | hum137Fa | ACCGGTCCGCTTTATGTGATT | 163 | 58 | Putative outer membrane protein |
| hum137Ra | AACGACCGCCTTTAGTAGTGACC | ||||
| 16 | hum144Fa | TCTCTGCATGGCTGACA | 158 | 64 | Putative membrane efflux protein |
| hum144Ra | CGCTTTGGCTATATTGGGAGGTA | ||||
| 17 | hum153Fa | TGCGTGGTACTAAATCTATCAT | 97 | 60 | Putative outer membrane protein |
| hum153Ra | AACTCTGTACCTCCTTCATTTGT | ||||
| 18 | hum162Fb | CAACGTAAACTCTCGGGTGATAA | 106 | 60 | Putative TonB-dependent receptor |
| hum162Rb | CGGTGCCAGCGGTAAGTTT | ||||
| 19 | hum163Fa | CGTCAGGTTTGTTTCGGTATTG | 165 | 60 | Hypothetical protein BF3236 |
| hum163Ra | AAGGTGAAGGTCTGGCTGATGTAA | ||||
| 20 | hum184Fa | TTGCCGCCAGATTCATAAAAA | 130 | 65 | Putative transmembrane spore maturation- |
| hum184Ra | AAGATAGGCGAGAAAGGGGGAGTC | like protein | |||
| 21 | hum172Fa | GTTACGGTACGCAGAAGAAGGTGA | 142 | 62 | Putative outer membrane protein |
| hum172Ra | CCCGACGAGGTAGTGACATT | ||||
| 22 | hum181Fb | GTAATTCGCGTTCTTCCTCACAT | 110 | 61 | Putative RNA polymerase extracytoplasmic function- |
| hum181Rb | ACCTGCAAACCGTACAAGAAAAA | type sigma factor | |||
| 24 | hum218Fb | TCAATTTTACCACGCCAGAA | 184 | 55 | Putative outer membrane protein |
| hum218Rb | CTACGCAAGATGAATATGAAGGTG | ||||
| 26 | hum243Fb | TTCCGGCATCTGTTCTACTATCTC | 97 | 64 | TonB |
| hum243Rb | CAATCAGGCTGTGGAAATCAAA | ||||
| 27 | hum245Fc | GCGGATGTCGAGCAGGAAAGTC | 98 | 60 | Glycosyltransferase |
| hum245Rc | GCTACCGGGGAGAAACCAAAGAAC | ||||
| 28 | hum327Fa | CGCATGGGCCGGATTTACG | 124 | 62 | Polysaccharide biosynthesis protein |
| hum327Ra | CACCGCAGCCAACAGCACATAG | ||||
| 29 | hum330Fb | CATCGCCCTTATCTTGGTT | 94 | 63 | Putative biopolymer transport protein |
| hum330Rb | TGGCGTATTAGCAGGTTCA | ||||
| 30 | hum336Fa | CCAACGGCGTAACTTCTTCA | 162 | 62 | Outer membrane efflux protein precursor |
| hum336Ra | ATTACCGGATTACAAACCTTATG |
TABLE 2.
Summary of putative human-specific PCR assay specificity testsa
| Source composite | No. of samples | Amplification of target DNA
|
Total no. of PCR assays | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Human-specific PCR assays 7, 19, 22, 30 | Human + 1 animal; PCR assays:
|
Human + 2 animals; PCR assay(s):
|
Human + 3 animals; PCR assay(s):
|
Human + 4 animals; PCR assays 1, 6, 27 | ||||||
| 11, 17, 18 | 16, 20 | 2, 3, 4, 9, 15, 21, 24, 26, 28 | 29 | 8, 13, 14 | 5 | |||||
| Horse | 5 | − | − | − | − | − | − | − | − | 0 |
| Pig | 10 | − | − | − | − | − | − | − | − | 0 |
| Chicken | 12 | − | − | − | − | − | + | + | + | 7 |
| Goat | 10 | − | − | − | − | + | − | + | + | 5 |
| Sheep | 10 | − | − | − | − | − | − | − | − | 0 |
| Cow | 12 | − | − | − | − | + | − | − | − | 1 |
| House cat | 10 | − | − | + | + | − | + | − | + | 17 |
| Domestic dog | 10 | − | + | − | + | − | + | − | + | 18 |
| Goose | 5 | − | − | − | − | − | − | − | − | 0 |
| Whitetail deer | 6 | − | − | − | − | − | − | + | − | 1 |
| Human | 16 | + | + | + | + | + | + | + | + | 26 |
PCR assays 1 through 26 (see Table 1) are grouped based on number of detectable animal source composites ranging from human-specific (most specific) to human plus four additional composites (least specific). A minus sign indicates that a respective PCR assay did not amplify target DNA, and a plus sign denotes amplification of target DNA for a particular source composite. “No. of samples” depicts number of individual fecal samples in source composite.
The remaining four PCR assays (designated assay 7, 19, 22, or 30) amplified target DNA only from human fecal specimens (Table 2), indicating we had nonetheless achieved our primary goal of identifying candidate human-specific bacterial markers. The first marker (assay 7) targets a 326-bp enriched DNA fragment predicted to encode a putative outer membrane protein based on a best BLASTx hit (2.00E−21) for this sequence showing 59% sequence ID to a Bacteroides thetaiotaomicron protein (locus BT0483) (29). The second maker (assay 19), targets a 278-bp enriched fragment annotated as encoding a B. fragilis hypothetical protein potentially involved in remodeling of bacterial surface polysaccharides and lipopolysaccharides (locus BF3236) (17); 88% ID; 6.00E−43]. Assay 22 amplifies a 220-bp DNA marker predicted to encode a putative RNA polymerase extracytoplasmic function sigma factor (B. thetaiotaomicron locus BT0326 [29]; 59% ID; 1.00E−14), with a potential function in transducing signals from outside the bacterial cell to the transcription apparatus. Finally, assay 30 targets a 428-bp enriched DNA fragment annotated as encoding an outer membrane efflux protein precursor (B. thetaiotaomicron locus BT2795 [29]; 86% ID; 2.00E−41) whose mature product functions in substrate binding and transport. Interestingly, all of these assays target genes within the uncharacterized fecal human bacterial metagenomes with alleles that show maximal yet limited similarity to those of B. thetaiotaomicron. Assays 7, 19, and 30 consistently detected as little as 1 pg of purified human fecal DNA under optimal conditions, while assay 22 consistently amplified 10-pg quantities (data not shown).
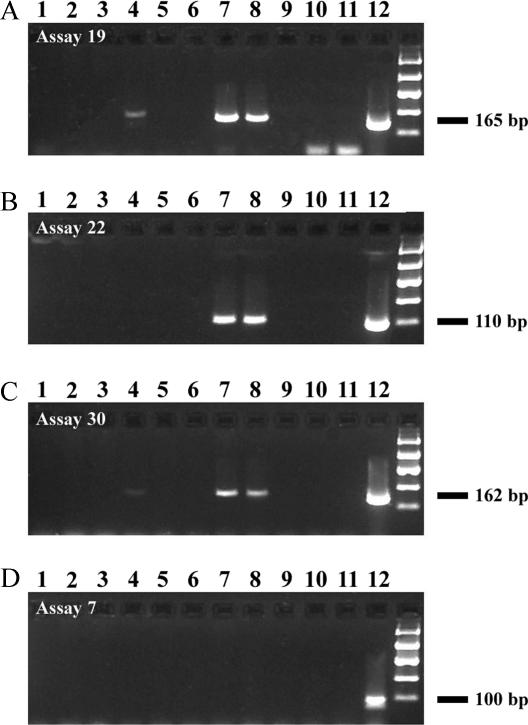
A series of initial studies was then performed to explore the future utility of these PCR assays for environmental monitoring of human fecal pollution. DNA extracts isolated from various wastewater facilities were used as a template to estimate the spatial robustness of each human-specific PCR assay. Three human fecal matter-specific PCR assays (assays 19, 22, and 30) yielded target products for all of the wastewater samples, while assay 7 showed the lowest spatial distribution, with amplification from only 61% of the samples.
To demonstrate the potential of each human-specific PCR assay for environmental monitoring, each primer set was tested with DNA isolated from recreational water, storm water, and treated wastewater effluent samples. Both the Heiserman Stream sample (Milford, OH), taken from an area situated 100 m downstream of a treated wastewater discharge pipe, and the treated wastewater sample (Arrowhead, OH) tested positive in PCR assays 19, 22, and 30. A river sample (9-Mile Creek, OH) taken from an area approximately 1,000 m downstream of a treated wastewater discharge pipe also tested positive in assays 19 and 30. All assays yielded no detectable PCR product for the remaining storm water and river samples, as shown in Fig. 3.
FIG. 3.
Gel electrophoresis of PCR products from reactions with human-specific PCR assays 19, 22, 30, and 7 (A, B, C, and D, respectively). Each PCR assay was tested against DNA extracts from recreational water, storm water, and treated wastewater samples. Sources: Miami Trails, OH, column 1 (lane 1); Lower East Fork I and II, OH (lanes 2 and 6, respectively); O'Bannon, OH, (lane 3); 9-Mile, OH (lane 4); Middle East Fork, OH (lane 5); Heiserman, OH (lane 7); Arrowhead, OH (lane 8); extraction blank (lane 9); no template controls (lanes 10-11); and human fecal DNA (lane 12). PCR products in lanes 10 and 11 from panel A are primer dimers.
DISCUSSION
Several hundred candidate marker sequences were obtained, and four new PCR assays were developed using competitive solution-phase hybridization to identify fecal microbial community DNAs uniquely present in human sources. In extending our studies to the important question of human source contamination, we used GFE with total DNA extracted from only single reference human and pig fecal specimens. Although we did not know if these specimens would be representative of these two types of fecal microbial communities, our prior metagenomic analyses of cow and pig fecal samples (22) suggested that such a simplified approach could work. Enrichment for the desired marker sequences with regard to the two original sources was as anticipated (98% of DNA sequences); however, only 4 of the 26 putative human fecal community-specific PCR assays designed and optimized amplified markers only from human fecal community DNA (Table 2). In contrast, our prior examination of total DNA from individual reference cow and pig fecal samples yielded candidate markers where all GFE-derived DNA sequences selected for PCR assay development could be used for cattle fecal community-specific PCR assays (22). This lower success rate for development of human fecal community PCR assays suggests that genetic variation between the two types of reference fecal microbial communities selected for GFE, which likely reflect differences in at least anatomy, physiology, and diet, had an impact on the efficiency of the GFE approach for finding host-specific DNA sequences for the development of MST assays. Further studies of this type would therefore likely benefit from comparing different host microbial communities and perhaps combinations of competitor DNA pools that might obtain markers more efficiently with a higher degree of specificity.
Comparative sequence analysis of the GFE plasmid clone sequences obtained suggests that much of the genetic capacity of the reference human fecal microbial community not present in a pig specimen resided in previously uncharacterized microbial genes. Although many more GFE fragments would need to be sequenced to generate a complete assessment of the genetic differences between these two microbial communities, we were, perhaps surprisingly, able to obtain desired human bacterium-specific DNA marker sequences by analysis of only a relatively small number of plasmid clones and only two source specimens. Classification of a limited number of GFE clone sequences into functional groups indicated an abundance of genes that potentially encode bacterial membrane-associated or extracellular proteins. Among these, a striking 85% of 126 falling into the category of “surface-associated” factors (see Table S1 in the supplemental material) were predicted by SignalP (3) to encode secreted proteins. These findings suggest that a potential major difference between the reference human and pig fecal microbial communities is in their capacity for producing distinct secreted factors. We also previously observed this trend in the analyses of reference cow and pig fecal microbial communities (22). These findings are consistent with a hypothesis that highly specific bacterial markers may be found in genes related to host specificity, where surface and secreted factors are often involved in interactions with distinct types of host cells and tissues, in modifying the external bacterial cell surface, and in obtaining necessary nutrients from highly defined external environments.
In terms of the potential utility of the specific marker assays described for water quality monitoring, initial tests of the four human-specific PCR assays exhibited good spatial robustness across 11 states by consistently testing positive for almost all contaminated wastewater samples representing 18 different human populations. These preliminary experiments suggest that these PCR assays, particularly assays 19 and 30, may have a future utility in environmental monitoring and merit more extensive characterization. However, in order to realize the potential of these PCR assays for MST applications, several issues remain to be addressed. These include survival of target DNA molecules in the environment, relevance of each PCR assay to current culture-based fecal indicator methods used to monitor water quality, and establishing a link between the prevalence of genetic markers described and relevant public health risks. Both the broad distribution of these microbial genetic makers across human populations and the level of specificity established do encourage us to further explore the potential of the assays described for more accurately identifying human fecal contamination in our waters.
Acknowledgments
This research was funded in part by a New Start Award from the National Center for Computational Toxicology of the U.S. Environmental Protection Agency, Office of Research and Development.
We are grateful to Mark Meckes, Janet Blannon, Matt Morrison, Sam Myoda, and Don Stoeckel for providing fecal and wastewater samples.
Any opinions expressed in this paper are those of the author(s) and do not necessarily reflect the official positions and policies of the U.S. Environmental Protection Agency, and any mention of products or trade names does not constitute recommendation for use.
Footnotes
Published ahead of print on 5 January 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., F. Thomas, L. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balarajan, R., V. Soni Raleigh, P. Yuen, D. Wheeler, D. Machin, and R. Caftwright. 1991. Health risks associated with bathing in sea water. Br. Med. J. 303:1444-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SingalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding for 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabelli, V. J., A. P. Dufour, L. J. McCabe, and M. A. Levin. 1982. Swimming-associated gastroenteritis and water quality. Am. J. Epidemiol. 115:606-616. [DOI] [PubMed] [Google Scholar]
- 7.Carson, C. E., J. M. Christiansen, H. Yampara-Iquise, V. W. Benson, C. Baffaut, J. V. Davis, R. R. Broz, W. B. Kurtz, W. M. Rogers, and W. H. Fales. 2005. Specificity of Bacteroides thetaiotaomicron marker for human feces. Appl. Environ. Microbiol. 71:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC. 2002. Surveillance for waterborne disease outbreaks, United States 1999-2000. Morb. Mortal. Wkly. Rep. Surveill. Summ. 51:1-28. [PubMed] [Google Scholar]
- 9.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufour, A. P. 1984. Health effects criteria for fresh recreational waters. U.S. Environmental Protection Agency, Washington, DC.
- 11.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimes, D. J. 1991. Ecology of estuarine bacteria capable of causing human disease: a review. Estuaries 14:345-360. [Google Scholar]
- 13.Grothues, D., C. R. Cantor, and C. L. Smith. 1993. PCR amplification of megabase DNA with tagged random primers (T-PCR). Nucleic Acids Res. 21:1321-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooper, L. V., and J. I. Gordon. 2001. Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11:1R-10R. [DOI] [PubMed] [Google Scholar]
- 15.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 16.Krogh, A., B. Larsson, G. von Heijne, and L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 17.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruss, A. 1998. Review of epidemiological studies on health effects from exposure to recreational water. Int. J. Epidemiol. 27:1-9. [DOI] [PubMed] [Google Scholar]
- 19.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanks, O. C., C. Nietch, M. T. Simonich, M. Younger, D. Reynolds, and K. G. Field. 2006. A basin-wide analysis of the dynamics of fecal contamination and fecal source identification in Tillamook Bay, Oregon. Appl. Environ. Microbiol. 72:5537-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shanks, O. C., J. W. Santo Domingo, and J. E. Graham. 2006. Use of competitive DNA hybridization to identify differences in the genomes of bacteria. J. Microbiol. Methods 66:321-330. [DOI] [PubMed] [Google Scholar]
- 22.Shanks, O. C., J. W. Santo Domingo, R. Lamendella, C. A. Kelty, and J. E. Graham. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson, J. M., D. J. Reasoner, and J. W. Santo Domingo. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.U.S. Environmental Protection Agency. 1986. Ambient water quality criteria for bacteria—1986 EPA 440/5-84/002. Criteria and Standards Division, U.S. Environmental Protection Agency, Washington, DC.
- 26.U.S. Environmental Protection Agency. 1994. Combined sewer overflow (CSO) control policy 59. 40 C.F.R. Part 122. U.S. Environmental Protection Agency, Washington, DC.
- 27.U.S. Environmental Protection Agency. 2004. Report to Congress: impacts and control of CSOs and SSOs. EPA 833 R-04-001. U.S. Environmental protection Agency, Washington, DC.
- 28.White, O., J. A. Eisen, J. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]