Abstract
Genes of Sulfolobus metallicus that appeared to be upregulated in relation to growth on either ferrous iron or sulfur were identified using subtractive hybridization of cDNAs. The genes upregulated during growth on ferrous iron were found in a cluster, and most were predicted to encode membrane proteins. Quantitative reverse transcription-PCR of cDNA showed upregulation of most of these genes during growth on ferrous iron and pyrite compared to results during growth on sulfur. The highest expression levels observed included those for genes encoding proteins with similarities to cytochrome c oxidase subunits and a CbsA-like cytochrome. The genes identified here that may be involved in oxidation of ferrous iron by S. metallicus are termed fox genes. Of three available genomes of Sulfolobus species (S. tokodaii, S. acidocaldarius, and S. solfataricus), only that of S. tokodaii has a cluster of highly similar open reading frames, and only S. tokodaii of these three species was also able to oxidize ferrous iron. A gene encoding sulfur oxygenase-reductase was identified as the source of the dominant transcript in sulfur-grown cells of S. metallicus, with the predicted protein showing high identities to the previously described examples from S. tokodaii and species of Acidianus.
The efficient extraction of copper by the microbial degradation of the mineral sulfide chalcopyrite can be achieved by utilizing ferrous iron- and sulfur-oxidizing thermoacidophilic archaea at temperatures close to 80°C (16). A demonstration plant comprising six 1,268-m3 bioreactors with a design production capacity of 20,000 tons of copper per year has successfully utilized such cultures (4). The key reaction in the process is the microbial conversion of ferrous iron, by an as yet uncertain oxidation and electron transport system, to ferric iron, which is the mineral sulfide-oxidizing agent. In contrast, the oxidation of sulfur by the thermoacidophilic archaea, which contributes to the mineral dissolution and the maintenance of the acidity required to keep the ferric iron in solution, is known to involve sulfur oxygenase-reductase as a key enzyme and has been extensively studied using Acidianus species (8, 12, 26).
Spectroscopic studies with species of Sulfolobus and Metallosphaera have shown a differential expression of respiratory complexes dependent on the growth substrate. The current view of the aerobic respiratory chains of the most-studied species for which genomes are available (Sulfolobus acidocaldarius, Sulfolobus solfataricus, and Sulfolobus tokodaii) is that a SoxABCD complex (a heme aa3-CuB oxidase) and a SoxM (heme bb3-CuB) supercomplex serve as the terminal aerobic oxidases (13, 14, 19). Whereas SoxABCD is a quinol oxidase, SoxM in conjunction with the SoxH subunit, which has a CuA center binding motif, is thought to receive electrons from SoxE, sulfocyanin (13). The additional Rieske iron-sulfur proteins, SoxF and SoxL (22), as well as the CbsAB/SoxL2N complex (9), are assumed to be intermediate cytochrome bc1-like quinol oxidases. A further type of heme, aa3 quinol oxidase, was found in Acidianus ambivalens (20), where it apparently represents the entire respiratory system of the organism. The genomes of the sequenced Sulfolobus species possess open reading frames (ORFs) whose products have high similarities to the A. ambivalens subunit I protein of this oxidase, DoxB.
An absorption peak at 573 nm was found as a novel feature of cytochrome spectra of thermoacidophilic archaea (Sulfolobus metallicus, Metallosphaera sedula, and Acidianus brierleyi) grown on ferrous iron or pyrite (3, 6). The expression of soxN, soxL2, cbsA, soxM, and soxB in M. sedula grown on different substrates indicated cbsA to be the only gene out of these five that was highly expressed in pyrite-grown cells (10), and it was suggested that its gene product could be related to the 573-nm cytochrome absorption peak. However, since no iron-expressed genes for terminal oxidases were identified and expression of cbsA during growth on sulfur and yeast extract was also relatively high in comparison to that during growth on pyrite, it can be concluded that the genetic basis of iron oxidation in such organisms remains largely unknown.
The sequences, transcriptional organization, and energy substrate-dependent expression of some of the genes upregulated during growth of S. metallicus on ferrous iron are described in this paper, and the organism's sulfur oxygenase-reductase gene is identified. These genes were found using micro-representational-difference analysis (mRDA), a subtractive hybridization approach (5), starting with cDNAs from S. metallicus grown with the different substrates, ferrous iron and sulfur.
MATERIALS AND METHODS
Microorganisms and growth conditions.
Sulfolobus metallicus DSM 6482T and Sulfolobus tokodaii DSM 16993T were grown at 65°C and 77°C, respectively, in a medium with MgSO4 · 7H2O (0.5 g liter−1), (NH4)2SO4 (0.4 g liter−1), K2HPO4 (0.2 g liter−1), FeSO4 · 7H2O (10 mg liter−1), and one substrate from pyrite (10 g liter−1), elemental sulfur (5 g liter−1), or ferrous iron (50 mM; FeSO4 · 7H2O, 13.9 g liter−1). The initial pH of the medium was adjusted with H2SO4 to pH 1.5, 2.0, or 2.5 for growth on ferrous iron, pyrite, or sulfur, respectively. There was a requirement for a reduced sulfur source for autotrophic growth with ferrous iron, as with most moderately thermophilic, iron-oxidizing bacteria (15), and this was met with addition of elemental sulfur (10 mg liter−1) or potassium tetrathionate (0.5 mM) to the medium. Cultures were shaken (140 rpm) and gassed with 1% (vol/vol) CO2 in air. Cells were harvested by centrifugation from mid to late exponential growth phases and washed by resuspension in water acidified to pH 1.7 with sulfuric acid and then in deionized water prior to storage at −80°C.
RNA extraction and DNase treatment.
Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. Contaminating DNA was removed with RNase-free DNase I treatment (New England Biolabs, Ipswich, MA), and the sample was considered to be DNA free when 0.5 to 1 μg of RNA used as a template in a PCR did not yield any products after 35 cycles.
mRDA.
Experiments were performed with tester cDNA from ferrous iron-grown cells and driver cDNA from sulfur-grown cells and with the reverse combination of these cDNAs. Double-stranded cDNAs were synthesized using the Universal RiboClone cDNA synthesis system (Promega, Madison, WI) with the times for first- and second-strand syntheses extended to 2 h and 4 h, respectively. Syntheses of amplicons and two rounds of subtractive hybridizations were carried out as described by Becker et al. (5). The difference products obtained after the second round of subtractive hybridization (DP2s) were shotgun cloned using the TOPO-TA cloning kit (Invitrogen). Clones were screened by colony PCR using vector primers M13f/r, and a number of inserts of the appropriate sizes were selected for sequencing.
DNA extraction and inverse PCR.
A series of treatments with neutral-pH sodium dodecyl sulfate (SDS), proteinase K, high salt, and hexadecyltrimethylammonium bromide (2) was applied to S. metallicus to extract DNA. RNA was removed by treatment with pancreatic RNase to allow an accurate spectrophotometric determination of the DNA concentration. The DNA was digested with a number of restriction enzymes and recircularized at a concentration of 5 ng μl−1. Inverse PCR (17) was conducted using these religated DNA samples as templates.
RT-PCR and real-time RT-PCR (quantitative RT-PCR [qRT-PCR]).
First-strand cDNA synthesis for reverse transcription-PCR (RT-PCR) was performed using the ImProm-II reverse transcription system (Promega) using random hexamers and 5 mM MgCl2. For RT-PCR, a cDNA amount corresponding to 50 ng RNA was used in a 25-μl reaction volume with 30 cycles using 2.5 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate, 0.2 μM of each primer (Table 1), and 0.02 U μl−1 of Platinum Taq DNA polymerase (Invitrogen). The cycles comprised 0.5 min at 94°C, annealing for 1 min at approximately 2°C below the lower primer melting temperature, and an extension of 1 min per 1,000-bp product length at 72°C. The program was preceded by a 2-min initial denaturation at 94°C and concluded with 5 min at 72°C. Primer combinations Orf1f and FoxDr2, FoxDf2 and FoxCr2, FoxCf2 and FoxBr, FoxBf and FoxGr2, FoxGf2 and FoxHr2, FoxHf2 and FoxIr, FoxIf and FoxJr, FoxJf and FoxEr2, FoxEf and FoxFr2, FoxEf and FoxAr2, and FoxAf2 and Orf2r were used to evaluate the transcriptional organization of the described ORFs.
TABLE 1.
Oligonucleotides used in this study
| Namea | Sequence (5′-3′) | Positionb |
|---|---|---|
| Oligonucleotides used in qRT-PCR | ||
| SM16Sf | ACGCTCTAAAAAGGCGTGGGAATA | |
| SM16Sr | TTGAGCTCGGGTCTTTAAGCAGTG | |
| Orf1f | CCGTTACCTAACGTCACATAACCGTACT | 34 |
| Orf1r | GCTGTAACTGAGACTTCCACCTCGTC | 185 |
| FoxDf | CAACGAATGCACCTACCGAACC | 1131 |
| FoxDr | GGGAAATGACATGGCGAGCATA | 1282 |
| FoxCf | GAGTGGAACCATCGGTGCTATCAG | 1862 |
| FoxCr | TTGCTGTATTCCAAGGAGGAGCTG | 2013 |
| FoxBf | TGGTCCCTTGTAGGAACCTAGAGCA | 4081 |
| FoxBr | TTTTTCGTGCTCGTGATTGTAGCTG | 4227 |
| FoxGf | AAGGTGGCCCATGAAGTACTCC | 5320 |
| FoxGr | CAAATAACTCCAGCACCGCAATG | 5471 |
| FoxHf | CCATGAGGAAGTGGAGGACTAAAGC | 7232 |
| FoxHr | GACGAGAACACTTTGACGTGACGA | 7379 |
| FoxIf | TTCCTCTATGCATATATAACGGGGTTG | 8300 |
| FoxIr | CATAGTTCACATGTGTCACAAAGGAAGA | 8446 |
| FoxJf | GGTTAACGAGATGAGTGGGAGCA | 9120 |
| FoxJr | AGGTTAACAATCCGAAGCCTGCTA | 9271 |
| FoxEf | GGGGTTGTGGAATTGAAGGACA | 9734 |
| FoxEr | AGACCAGCACCCCAAATGAGC | 9885 |
| FoxFf | ACATCCGTCTCCACAGCGATAATG | 11788 |
| FoxFr | CACGCTCACAAGAAAAGCGTATCC | 11937 |
| FoxAf | GGCAGAATTCCAGGCAGATTGATA | 13038 |
| FoxAr | TCCGAAGATGTTAATGCCAAGTCC | 13187 |
| Orf2f | CGTCACAAGCCCCACTTTGAG | 14818 |
| Orf2r | GCCGAACAAACGCGGTATCTC | 14965 |
| SORf | GGGAATGGAGAAGGAATTTGAGGA | |
| SORr | TGATGCCATGACTTTGCAGATAGC | |
| Additional oligonucleotides | ||
| FoxDr2 | CGGTGCACTGTCGATTTTAGCC | 952 |
| FoxDf2 | AAGGCTCTCCTTCCGCTTAGGTG | 1352 |
| FoxCr2 | CTTCATGTACCCCGAAAGGTTC | 2603 |
| FoxCf2 | CTCAGGACAAGCCATCCAGGAG | 3029 |
| FoxGr2 | TTCCAAGGATGTCTCCTATGAAGC | 5289 |
| FoxGf2 | AAAAGGTTCCCATGGCTTAACGAC | 5750 |
| FoxHr2 | TCCTCAATGTGACGGAAGGATCA | 6838 |
| FoxHf2 | CAAGGGTGACCTTAGGATAA | 7125 |
| FoxEr2 | GCTGGATTGAATATGATGTATACAAG | 10247 |
| FoxFf2 | ACAAGGTATCTATAAGGGCCAACG | 11035 |
| FoxAr2 | TGTAACGGATTCAGAAACGTGAAC | 12949 |
| FoxAf2 | AAGCTGAACCCACTCAACCAACTC | 13579 |
Oligonucleotides are named for the ORF in which they bind. Primers used in pairs have identical names and bind in forward (f) or reverse (r) orientation.
Position of the 5′ nucleotide on the DNA fragment containing the fox genes.
The program Primer3 (21) was used to design primers (Table 1) producing amplicons of 147 to 152 bp in length. These amplicons were used in qRT-PCR performed using 25-μl reaction volumes comprising 5 μl cDNA, 12.5 μl SYBR Green PCR master mix (Applied Biosystems, Foster City, CA), and 0.2 μM of each primer. The cDNA was diluted to appropriate concentrations before use, typically to values corresponding to 0.01 ng of RNA for 16S rRNA qRT-PCRs and 1 ng of RNA for mRNA qRT-PCRs. Standard curves were constructed for each primer pair based on serial 10-fold dilutions of genomic DNA from S. metallicus. The reactions were carried out in duplicate for cDNA obtained from three independently grown cultures for each data series. Arbitrary gene expression values were calculated based on the standard curves for each primer pair and corrected for differences in the template concentrations used. Each mRNA expression value was normalized against 16S rRNA gene expression.
2D blue native/SDS-PAGE analysis of S. metallicus membrane extracts.
Protein extracts were prepared from S. metallicus by suspension of the cells in ACA750 buffer (24), cell lysis by sonication, and removal of the cell debris by centrifugation at 15,000 × g for 10 min. Membranes were pelleted by centrifugation for 30 min at 100,000 × g, washed once in ACA750 buffer, and solubilized for 1 h at 4°C in ACA750 buffer containing 1% (wt/vol) n-dodecyl-β-d-maltoside. Solubilized membrane proteins were separated on a 5 to 15% (wt/vol) acrylamide blue native gel prior to second-dimension (2D) SDS-polyacrylamide gel electrophoresis (PAGE) (12% [wt/vol] acrylamide) following described protocols (24). Heme staining was performed using o-dianisidine (23).
Mass spectroscopy and bioinformatic analyses.
Sequences obtained from mRDA fragments were analyzed by using BLASTN and BLASTX searches (1) against GenBank (http://www.ncbi.nlm.nih.gov/BLAST/), and percent identity and similarity values of amino acid sequences from full ORFs were obtained using the Fasta3 algorithm (18) of the EBI database (http://www.ebi.ac.uk/fasta33/#). Theoretical molecular weights and isoelectric points were obtained using the ExPASy Compute pI/Mw tool (http://us.expasy.org/tools/pi_tool.html), and prediction of transmembrane helices used the TMHMM Server, v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Selected Coomassie blue-stained protein spots were excised from 2D gels. Tryptic-digest fragments were analyzed by liquid chromatography electrospray ionization tandem mass spectrometry with a Q-ToF instrument fitted with an in-line capillary LC system (Waters MS Technologies). Peptide mass fingerprints were analyzed using the Mascot v2.1 (MatrixScience) search engine aimed at the NCBInr database and the in-house database (consisting of UniProt sequences and the S. metallicus genome fragment sequenced in this work).
Nucleotide sequence accession numbers.
The sequences of the S. metallicus DSM 6482T sulfur oxygenase-reductase gene, the genomic DNA region containing the fox gene cluster, and the partial aspartate oxidase gene have been deposited in the GenBank database with accession numbers EF040586, DQ813663, and EF040585, respectively.
RESULTS
mRDA with iron- and sulfur-grown cells.
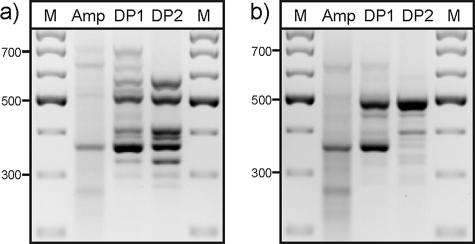
The electrophoretic band patterns of cDNA amplicons and the products obtained after one (difference product 1) or two (DP2) rounds of subtractive hybridizations using cDNA from ferrous iron-grown or sulfur-grown cells are shown in Fig. 1. In both cases, the second difference product was cloned.
FIG. 1.
Agarose gel electrophoresis of the cDNA amplicon (Amp) and the first (DP1) and second (DP2) difference products from the mRDA of ferrous iron- versus sulfur-grown S. metallicus. Tester cDNA was from (a) ferrous iron-grown cells and (b) sulfur-grown cells. M, 100-bp ladder.
With cDNA from sulfur-grown cells as the tester sample and from ferrous iron-grown cells as the driver sample, the DP2 profile showed one dominant fragment at 490 bp, which was cloned and sequenced. The deduced amino acid sequence showed high similarity to sequences of sulfur oxygenase-reductases. The complete gene sequence was obtained by inverse PCR and presumably coded for a 316-amino-acid protein with an apparent molecular weight of 36 kDa. The full amino acid sequence was similar to those of the sulfur oxygenase-reductases from S. tokodaii and Acidianus ambivalens (71% and 63% identities). A deduced amino acid sequence similar to that of l-aspartate oxidase of S. tokodaii (72% identity) was found from cloning and sequencing of the relatively minor fragment at 400 bp (Fig. 1b). There were also several weaker PCR products.
With cDNA from ferrous iron-grown cells as the tester sample and from sulfur-grown cells as the driver sample (Fig. 1a), 8 different sequences were found in 25 clones of the DP2 fragments. All deduced amino acid sequences showed highest percent identities to hypothetical proteins of S. tokodaii, and with one exception, these were to five hypothetical proteins, ST2587, ST2595, and ST2598 to ST2600, whose encoding genes are clustered in the S. tokodaii genome.
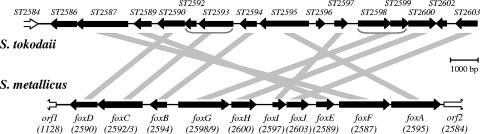
S. metallicus genes similar to genes ST2584 to ST2603 of S. tokodaii.
A 15-kb region of S. metallicus DNA was sequenced using a combination of inverse PCR and conventional PCR to bridge the gaps between sequences obtained from the mRDA DP2 products that were derived from the cDNA of ferrous iron-grown cells. Most of the identified ORFs were found to be very similar to ORFs in S. tokodaii (Table 2). ORFs ST2586, ST2596, and ST2602 within the ST2584-to-ST2603 ORF cluster in S. tokodaii had no counterparts in the S. metallicus sequence (Fig. 2). There are significant differences in the orders of the similar ORFs in the two organisms, and two of them in S. metallicus appear to be gene fusions, comprising the equivalent of ORFs ST2592 and ST2593 and ORFs ST2598 and ST2599 (Fig. 2). The genes in S. metallicus are here referred to as fox genes, for ferrous iron oxidation, in recognition of a presumed link to growth on ferrous iron. The amino acid sequences deduced from the S. metallicus ORFs indicate all proteins except FoxH would have at least one potential transmembrane region (Table 2). With the exception of FoxJ, with 35% and 37% identity to hypothetical proteins ST1860 of S. tokodaii and SSO2676 of S. solfataricus, respectively, only relatively low percent identities of the fox gene products to other than the S. tokodaii ORFs were detected. Nevertheless, these might have indicated possible functional assignments for FoxA, FoxB, and FoxC, as already noted by Kappler et al. with reference to their counterparts in S. tokodaii (10). FoxC has about 25% identity to cytochrome b558/566 subunits A (CbsA) from S. tokodaii (ST1664), S. acidocaldarius (Saci_1858), and S. solfataricus (SSO2801). FoxA and FoxB identities to cytochrome c oxidase subunits I and II were, for FoxA, 24% to DoxB of Acidianus ambivalens (P94117) and 26% to DoxB of S. solfataricus (SSO0044) and for FoxB, 30% to PoxB of Pyrobaculum oguniense (BAC56131) and 32% to SoxH of S. acidocaldarius (Q59825). Additionally, FoxG has a polyferredoxin domain (COG0348) and FoxH contains an N-terminal Fe-S binding domain (COG1146), as well as a C-terminal CBS domain (COG2905), which is frequently found in proteins involved in signal transduction processes.
TABLE 2.
Characteristics of deduced S. metallicus proteins and their sequence identities and similarities to corresponding predicted proteins of S. tokodaii
| Protein | No. of aaa | Mol wt (103) | pI | No. of TMb | % Sequence (aa) identity (similarity)c | Corresponding S. tokodaii ORF no. |
|---|---|---|---|---|---|---|
| FoxD | 330 | 36 | 8.9 | 11 | 38 (72) | ST2590 |
| FoxC | 543 | 59 | 6.3 | 1d | 64 (84)/55 (83)e | ST2592/ST2593 |
| FoxB | 197 | 22 | 5.4 | 1d | 77 (92) | ST2594 |
| FoxG | 613 | 69 | 9.2 | 11 | 64 (85)/58 (83)f | ST2598/ST2599 |
| FoxH | 309 | 34 | 8.6 | 0 | 41 (73) | ST2600 |
| FoxI | 154 | 17 | 5.0 | 2 | 37 (70) | ST2597 |
| FoxJ | 266 | 30 | 7.7 | 7 | 37 (74) | ST2603 |
| FoxE | 198 | 22 | 8.8 | 6 | 48 (81) | ST2589 |
| FoxF | 606 | 67 | 9.3 | 16 | 42 (74) | ST2587 |
| FoxA | 595 | 67 | 8.6 | 12 | 72 (92) | ST2595 |
aa, amino acids.
No. of transmembrane domains.
Comparison to corresponding predicted proteins of S. tokodaii.
One transmembrane domain plus two regions of increased hydrophobicity.
Amino acids 1 to 406 compared to ST2593 sequence/amino acids 428 to 543 compared to ST2592 sequence.
Amino acids 1 to 393 compared to ST2598 sequence/amino acids 394 to 613 compared to ST2599 sequence.
FIG. 2.
A comparison of ORFs in genome fragments of S. metallicus (ORFs of >300 bp) or S. tokodaii. S. tokodaii ORFs and corresponding fox genes are connected by diagonal lines, and the S. tokodaii gene numbers are given below the S. metallicus gene names.
Overall, the genomic region described here contains genes that code for membrane proteins involved in electron transport processes. These are present in S. tokodaii and S. metallicus but not in other representatives of the Sulfolobales for which sufficient genomic sequence is available for comparison. The partial ORFs 1 and 2 situated at the 5′ and 3′ ends of the obtained genomic sequence appear to represent the boundaries of the gene cluster in S. metallicus, because an ORF matching orf1 is not associated with the cluster in S. tokodaii and Orf2 shows similarity to AAA-type ATPases of the SpoVK family involved in cell division and protein folding.
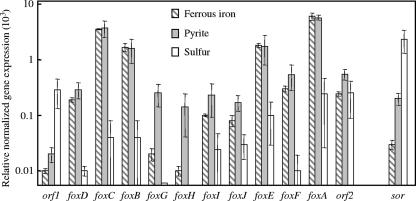
Expression of fox and sor genes under different growth conditions.
The fox genes that were relatively highly expressed in ferrous iron-grown versus sulfur-grown S. metallicus were also highly expressed in pyrite-grown cells, where ferrous iron was available from the mineral (Fig. 3). foxA, foxB, foxC, and foxE were most highly expressed, and foxA most of all. foxG and foxH most noticeably had higher expression levels with pyrite than when ferrous iron was added to the medium, although their final expression levels were quite low. orf2 did not appear to be differentially regulated by ferrous iron and sulfur, and orf1 had higher expression levels with sulfur as the substrate than with ferrous iron or pyrite, which is consistent with the assumption that these are not involved in the same functional task as the other genes. The highest expression level of the sor gene was observed during growth on sulfur (Fig. 3), with a level comparable to that of the fox genes that were highly expressed during growth on ferrous iron. The expression level was lower with pyrite than with sulfur but still higher than with ferrous iron.
FIG. 3.
Relative expression levels of ORFs of the sequenced S. metallicus genome fragment and of the sulfur oxygenase-reductase gene (sor) assessed by qRT-PCR and normalized against 16S rRNA gene expression in individual cultures. Mean values and standard deviations are from analyses of triplicate cultures grown autotrophically on ferrous iron, pyrite, or sulfur.
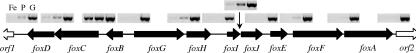
Transcriptional organization of the foxD-foxA locus.
Forward and reverse primers situated in adjacent ORFs and designed to give PCR products of approximately 1 kb were used in RT-PCR experiments to investigate transcriptional units present within the fox gene cluster (Fig. 4). The only strong RT-PCR products formed with cDNA from ferrous iron- and pyrite-grown cells were obtained with a primer pair connecting foxB and foxC. Primer pairs connecting orf1 and foxD, foxD and foxC, foxG and foxH, foxI and foxJ, or foxE and foxF gave weak products on cDNA from ferrous iron-grown cells and slightly stronger products on cDNA from pyrite-grown cells.
FIG. 4.
Investigation of the transcriptional organization of S. metallicus ORFs using RT-PCR with primers situated in adjacent ORFs. Products produced from cDNAs of a ferrous iron-grown culture (Fe) or a pyrite-grown culture (P) or from genomic DNA (G) are shown.
Oxidation of ferrous iron by Sulfolobus tokodaii.
The presence of a gene cluster in S. tokodaii which was similar to that with genes responding to ferrous iron in iron-oxidizing S. metallicus suggested that S. tokodaii would also grow on ferrous iron, but this has not been noted in previous descriptions of the organism (25). S. tokodaii was found to oxidize ferrous iron during growth supplemented with yeast extract. This was indicated by successful serial subculturing with oxidation of 90% of 50 mM ferrous iron, while only 7% was oxidized in a sterile control at 78°C. Ferrous iron oxidation during autotrophic growth was also maintained through serial cultures, although growth and oxidation were much weaker than with S. metallicus (data not shown). Confirmation of S. tokodaii culture purity after five autotrophic serial cultures with ferrous iron as the substrate was shown by direct sequencing of the 16S rRNA gene PCR product obtained from the last culture.
Identification of FoxA in membranes of Sulfolobus metallicus.
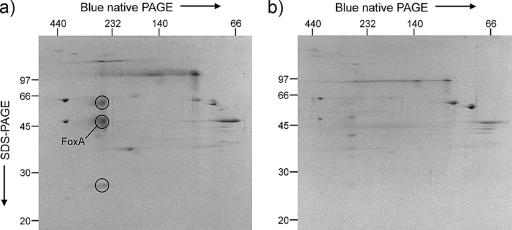
2D gel analyses of S. metallicus membrane proteins were performed to investigate differential protein expression with different substrates. Blue native PAGE of membrane proteins from pyrite-grown cells showed the presence of a complex with an apparent molecular weight of 270 kDa which was not seen in the membrane proteins from sulfur-grown cells. This complex separated into at least three subunits with apparent molecular sizes around 60, 50, and 27 kDa in SDS-PAGE (Fig. 5). The spot containing protein(s) of apparent relative molecular masses of about 50 kDa (Fig. 5) was analyzed by tandem mass spectrometry of tryptic digest fragments and yielded four different peptides (8 to 19 residues in length) with sequences identical to fragments of the deduced sequence of FoxA. Other individual peptides were identified in the sample, but no proteins in databases were indicated by more than one of these other peptides. A major protein spot appearing in the same relative position on a comparable gel of membrane proteins from ferrous iron-grown S. tokodaii was also analyzed (not shown). Five different peptides (9 to 21 residues) with sequences identical to those of fragments of the FoxA equivalent (ST2595) in S. tokodaii were found in database searches. One other protein, a hypothetical protein from the S. tokodaii genome, was indicated by two peptide sequences from this sample. The migration of these putative FoxA proteins in SDS-PAGE was not predicted from their indicated size of approximately 67 kDa (Table 2). The protein spot with an apparent relative mass of 60 kDa (circled in Fig. 5) showed heme-staining activity after SDS-PAGE.
FIG. 5.
Coomassie blue-stained 2D blue native/SDS-PAGE gels of membrane preparations of Sulfolobus metallicus cells grown on pyrite (a) and sulfur (b). The putative FoxA is indicated as one of three spots (circled) that represent a possible protein complex present in pyrite-grown cells but not sulfur-grown cells. The positions of molecular mass markers (in kDa) are shown on the borders of the gels.
DISCUSSION
The foxA, foxB, and foxC genes were highly induced during ferrous iron oxidation by S. metallicus: these genes probably code for membrane proteins involved in electron transport processes and possibly for SoxB/M-like, SoxH-like, and CbsA-like proteins, respectively. These might represent subunits of another type of aerobic terminal oxidase complex in the Sulfolobus-like thermoacidophilic archaea in addition to the SoxABCD proteins, the SoxM supercomplex, and the Dox-type oxidases. These three ORFs, together with foxE, were the most highly expressed within a gene cluster when S. metallicus was oxidizing ferrous iron and pyrite (where ferrous iron was available from the mineral), and FoxA was also detected as a dominant membrane protein in pyrite-grown cells but not in sulfur-grown cells. Some of the other genes in the identified cluster might be involved in different tasks, such as signal transduction or gene regulation. This assumes some proportionality between the levels of transcription and the final amounts of proteins synthesized.
The RT-PCR experiments investigating the transcriptional organization of the foxD-foxA locus are not entirely conclusive. foxA appears to form a monocistronic mRNA. Only foxB and foxC form a definite transcriptional unit, which may also include foxD. Additional bicistronic mRNAs may be formed by foxG and foxH, foxI and foxJ, and foxE and foxF. The possible transcriptional links between the ORFs would coincide with these genes having the same order in S. metallicus and S. tokodaii. However, the weak RT-PCR products observed here may also have been formed due to an incomplete termination of transcription. Since the expression level of foxC was approximately double that of foxB, it is possible that foxC also has an additional, independent promoter.
A sulfur oxygenase reductase was expected in S. metallicus based on its similar growth characteristics and phylogenetic relationship to the Acidianus species with which the enzyme has been studied. The high level of transcription of sor in sulfur-grown S. metallicus in contrast to the low level in iron-grown cells indicated a growth substrate-dependent transcriptional regulation, an observation also made by Kletzin (11) when RNA from aerobically and anaerobically grown cells of Acidianus ambivalens was compared.
The subtractive hybridization technique used in this study does not lead to an exhaustive sampling of all differentially regulated ORFs. The probability of targeting a differentially regulated gene depends on the formation of restriction fragments which have a suitable size for PCR amplification. Therefore, large ORFs and a group of adjacent ORFs which are all differentially regulated will have a higher likelihood of detection in mRDA than short and functionally isolated ORFs. For example, the ORFs foxE, foxC, and foxB were not detected directly by subtractive hybridization. Similarly, only the sor gene of the genes involved in the sulfur oxidation pathway was identified here. In addition, any constitutively expressed genes involved in iron or sulfur oxidation would not have been revealed by the subtractive hybridization method.
The 16S rRNA gene is often used for normalization of transcriptional data obtained by real-time PCR, but the data here should be regarded as semiquantitative and considered with caution, because it is not known if the 16S rRNA reference gene expression was at the same level during growth with the three substrates. However, the proteomic data showing differential FoxA expression supports the transcriptional trends described here. The gene for an antioxidant protein of the alkyl hydroperoxide reductase family which was shown to be a major cell protein induced in ferrous iron-oxidizing S. metallicus (7) was also found to be upregulated in iron- and pyrite-grown cells but not in sulfur-grown cells, using the same normalization procedure (data not shown).
A possible link between the CbsA protein of the cbsBA-soxL2N gene cluster in M. sedula and the previously noted cytochrome with an absorption peak at 573 nm, which was induced during ferrous iron oxidation, has been considered (10). However, the observed gene expression patterns suggest a significant involvement of the fox genes in iron oxidation, and one of their products may be a more likely candidate for this cytochrome. foxA and foxB homologues have been found in the type strain of M. sedula and in another iron-oxidizing Metallosphaera species, strain J1 (Bathe and Norris, unpublished data). However, additional genetic and protein biochemical work, including the investigation of the redox properties of the fox gene products, is required for elucidation of the potential roles of the individual genes and their products in ferrous iron-oxidizing, thermoacidophilic archaea.
The sequences and expression patterns of the genes described here can be used in development of functional gene probes for investigation of the abundance, diversity, and activity of iron- and sulfur-oxidizing archaea in natural environments and commercial biomining operations. Such probes should also be useful in the assessment of the relative activities of iron and sulfur oxidation in the degradation of different mineral sulfides where, for example, sulfur oxidation is considered to be more critical in efficient dissolution of the relatively refractory chalcopyrite than in dissolution of pyrite. The intermediate transcription level of sor in pyrite-grown cells together with the overall high levels of the foxD-foxA genes might indicate that the dominant metabolic activity on this substrate is iron oxidation but with a certain extent of simultaneous sulfur oxidation. However, the threshold substrate concentrations necessary for induction of the genes, as well as their time-dependent expression during growth on mineral sulfides, are unknown.
Acknowledgments
We appreciate expert technical assistance from Carly F. Brown and thank Susan E. Slade of the University of Warwick Proteomics Service for the mass spectroscopy analysis.
This work was supported by EU FP6 BioMinE project NMP2-CT-2005-500329 and by BHPBilliton PLC.
Footnotes
Published ahead of print on 23 February 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingson, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Short protocols in molecular biology. Wiley & Sons, New York, NY.
- 3.Barr, D. W., W. J. Ingledew, and P. R. Norris. 1990. Respiratory chain components of iron-oxidizing, acidophilic bacteria. FEMS Microbiol. Lett. 70:85-90. [Google Scholar]
- 4.Batty, J. D., and G. V. Rorke. 2005. Development and commercial demonstration of the BioCOP™ thermophile process. Hydrometallurgy 83:83-89. [Google Scholar]
- 5.Becker, P., W. Hufnagle, G. Peters, and M. Herrmann. 2001. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl. Environ. Microbiol. 67:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blake, R. C., II, E. A. Shute, M. M. Greenwood, G. H. Spencer, and W. J. Ingledew. 1993. Enzymes of aerobic respiration on iron. FEMS Microbiol. Rev. 11:9-18. [DOI] [PubMed] [Google Scholar]
- 7.Burton, N. P., T. D. Williams, and P. R. Norris. 1995. A potential anti-oxidant protein in a ferrous iron-oxidizing Sulfolobus species. FEMS Microbiol. Lett. 134:91-95. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z. W., C. Y. Jiang, Q. She, S. J. Liu, and P. J. Zhou. 2005. Key role of cysteine residues in catalysis and subcellular localization of sulfur oxygenase-reductase of Acidianus tengchongensis. Appl. Environ. Microbiol. 71:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiller, A., T. Henninger, G. Schäfer, and C. L. Schmidt. 2003. New genes encoding subunits of a cytochrome bc1-analogous complex in the respiratory chain of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J. Bioenerg. Biomembr. 35:121-131. [DOI] [PubMed] [Google Scholar]
- 10.Kappler, U., L. I. Sly, and A. G. McEwan. 2005. Respiratory gene clusters of Metallosphaera sedula—differential expression and transcriptional organization. Microbiology 151:35-43. [DOI] [PubMed] [Google Scholar]
- 11.Kletzin, A. 1992. Molecular characterization of the sor gene, which encodes the sulfur oxygenase/reductase of the thermoacidophilic archaeum Desulfurolobus ambivalens. J. Bacteriol. 174:5854-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kletzin, A., T. Urich, F. Müller, T. M. Bandeiras, and C. M. Gomes. 2004. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36:77-91. [DOI] [PubMed] [Google Scholar]
- 13.Komorowski, L., and G. Schäfer. 2001. Sulfocyanin and subunit II, two copper proteins with novel features, provide new insight into the archaeal SoxM oxidase supercomplex. FEBS Lett. 487:351-355. [DOI] [PubMed] [Google Scholar]
- 14.Komorowski, L., W. Verheyen, and G. Schäfer. 2002. The archaeal respiratory supercomplex SoxM from S. acidocaldarius combines features of quinole and cytochrome c oxidases. Biol. Chem. 383:1791-1799. [DOI] [PubMed] [Google Scholar]
- 15.Norris, P. R., and D. W. Barr. 1985. Growth and iron oxidation by acidophilic moderate thermophiles. FEMS Microbiol. Lett. 28:221-224. [Google Scholar]
- 16.Norris, P. R., and J. P. Owen. 1993. Mineral sulphide oxidation by enrichment cultures of novel thermoacidophilic bacteria. FEMS Microbiol. Rev. 11:51-56. [Google Scholar]
- 17.Ochman, H., F. J. Ayala, and D. L. Hartl. 1993. Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol. 218:309-321. [DOI] [PubMed] [Google Scholar]
- 18.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 19.Pereira, M. M., T. M. Bandeiras, A. S. Fernandes, R. S. Lemos, A. M. Melo, and M. Teixeira. 2004. Respiratory chains from aerobic thermophilic prokaryotes. J. Bioenerg. Biomembr. 36:93-105. [DOI] [PubMed] [Google Scholar]
- 20.Purschke, W. G., C. L. Schmidt, A. Petersen, and G. Schäfer. 1997. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J. Bacteriol. 179:1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 22.Schmidt, C. L., S. Anemüller, and G. Schäfer. 1996. Two different respiratory Rieske proteins are expressed in the extreme thermoacidophilic crenarchaeon Sulfolobus acidocaldarius: cloning and sequencing of their genes. FEBS Lett. 388:43-46. [DOI] [PubMed] [Google Scholar]
- 23.Schulz, H., E. C. Pellicioli, and L. Thöny-Meyer. 2000. New insights into the role of CcmC, CcmD and CcmE in the haem delivery pathway during cytochrome c maturation by a complete mutational analysis of the conserved tryptophan-rich motif of CcmC. Mol. Microbiol. 37:1379-1388. [DOI] [PubMed] [Google Scholar]
- 24.Stenberg, F., P. Chovanec, S. L. Maslen, C. V. Robinson, L. L. Ilag, G. von Heijne, and D. O. Daley. 2005. Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280:34409-34419. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, T., T. Iwasaki, T. Uzawa, K. Hara, N. Nemoto, T. Kon, T. Ueki, A. Yamagishi, and T. Oshima. 2002. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 6:39-44. [DOI] [PubMed] [Google Scholar]
- 26.Urich, T., C. M. Gomes, A. Kletzin, and C. Frazao. 2006. X-ray structure of a self-compartmentalizing sulfur cycle metalloenzyme. Science 311:996-1000. [DOI] [PubMed] [Google Scholar]