Abstract
Xylella fastidiosa, a bacterium responsible for Pierce's disease in grapevines, possesses both type I and type IV pili at the same cell pole. Type IV pili facilitate twitching motility, and type I pili are involved in biofilm development. The adhesiveness of the bacteria and the roles of the two pili types in attachment to a glass substratum were evaluated using a microfluidic flow chamber in conjunction with pilus-defective mutants. The average adhesion force necessary to detach wild-type X. fastidiosa cells was 147 ± 11 pN. Mutant cells possessing only type I pili required a force of 204 ± 22 pN for removal, whereas cells possessing only type IV pili required 119 ± 8 pN to dislodge these cells. The experimental results demonstrate that microfluidic flow chambers are useful and convenient tools for assessing the drag forces necessary for detaching bacterial cells and that with specific pilus mutants, the role of the pilus type can be further assessed.
Xylella fastidiosa is a rod-shaped gram-negative bacterium with dimensions of 0.25 to 0.35 radius and 0.9 to 3.5 μm length (24). It is the causal agent of many economically important plant diseases, including citrus-variegated chlorosis, leaf scorches of oleander, coffee, and almond, and Pierce's disease of grapevines (8). In plant hosts, the bacteria are limited to colonizing the water-conducting conduits (xylem vessels), where they form polysaccharide-bacterial cell aggregates (biofilms) that block transpiration-induced water flow to the leaves, resulting in disease expression. Like many other gram-negative bacterial species, X. fastidiosa was recently shown to move on agar surfaces via “twitching motility” (5, 15), a process shown to be facilitated by hair-like type IV pili that extend, attach to a substratum, and then retract, pulling the cells forward (14, 16, 18). Meng et al. (15) demonstrated that X. fastidiosa exhibited twitching motility against the direction of flow of media in microfluidic chambers. In host plants, the bacteria were recovered upstream from the sites where they were inoculated into the plants. Such upstream recovery of the bacteria indicates that they similarly migrated by twitching against a flow of xylem sap to those locations. Because of the polar position of the pili on rod-shaped cells affixed to the substratum at the pilus end and the fluid dynamics of a stream pushing against the tethered cells coupled with the extension-retraction activities known for type IV pili (14, 16, 18), the cells would be expected to be propelled against a current, as has been reported for both X. fastidiosa and Pseudomonas aeruginosa (15). X. fastidiosa differs from other bacterial species in that, in addition to possessing type IV pili which are 1 to 6 μm in length for this bacterium, it also possesses shorter (0.4 to 1.0 μm in length) type I pili that are also positioned at the same pole of the cell (15).
Together, the two classes of pili function, in part, to attach X. fastidiosa cells to their environmental substrata as well as to each other. From the work of Meng et al. (15) and others (10, 23) it is apparent that biofilm development in X. fastidiosa is highly reliant upon the presence of type I pili, as was previously shown for Escherichia coli (17). While type IV pili may also have a role in biofilm formation, their primary activity appears to be associated with motility (14). Similarly, it is well established that type IV pili mediate twitching motility in other bacteria such as P. aeruginosa, Neisseria gonorrhoeae, and Myxococcus xanthus (14). Type I pili, best studied in Escherichia coli, are associated with attachment of cells to substrata and development of biofilms (17, 22). Adhesion to substrata by X. fastidiosa as well as by other bacterial species can also occur through the activity of afimbrial adhesins present on the cell surface (6, 9, 11, 19, 20); however, the pili of X. fastidiosa appear to often keep the bacterial cell surface from making intimate contact with the substratum, at least initially. Thus, contact with and adhesion of X. fastidiosa to substrata via pili are crucial for establishing spread and colonization. It is important, then, to better understand the role of both types of pili, together and separately, in contributing to the adherence of X. fastidiosa to various substrata.
Traditionally, bacterial adhesion has been measured in commercially available parallel-plate flow chambers (2, 19, 20). Using such chambers, Thomas et al. (19, 20) demonstrated that the adhesion force generated by type I pili (fimbrae) can increase 10-fold in the presence of an external shear force. Recently, laser tweezers and atomic force microscopy (AFM) were used to investigate forces generated by single and multiple pili (13, 21). Using these approaches, Sheetz and coworkers (13, 16), and Touhami et al. (21) determined that the forces generated by a single type IV pilus of N. gonorrhoeae and P. aeruginosa exceed 100 pN and 95 pN, respectively. The extension and retraction dynamics of type IV pili of P. aeruginosa was assessed by Skerker and Berg (18); in their experiments, they were able to monitor these activities by fluorescently labeling pili and capturing their movement microscopically. While both laser tweezer and AFM methods offer advantages in assessment of the adhesion force of individual bacteria or even a single pilus, these are difficult procedures and rather time consuming, making it difficult to obtain many data measurements. The parallel-plate flow chamber allows for examining many bacteria at the same time, although the relatively large size of the chamber does not allow for detailed microscopic examination of the dynamics of the bacteria subjected to a flow at single-cell resolution.
We report herein on the use of a microfluidic chamber designed to assess drag forces required to evaluate adhesiveness of X. fastidiosa to substrata (in this case, glass). Importantly, we were able to uniquely relate the adhesiveness of bacterial cells to the kind of pili (type I and/or type IV pili) present by comparing a wild-type (WT) strain which has both types of pili with mutants deficient with respect to genes expressing either type I or type IV pili.
MATERIALS AND METHODS
Bacterial isolate, mutants, and growth conditions.
The wild-type X. fastidiosa strain (ATCC 700964) used in this study was originally isolated from a grapevine grown in Temecula, CA, expressing symptoms of Pierce's disease (23). Mutants deficient in genes encoding type I and type IV pili were previously obtained by random mutagenesis using an EZ::TN Tn5 transposome system (6). The mutants used in this study were as follows: fimA null cells lacking short type I pili (identified previously as mutant 6E11) (15) and pilB null cells lacking longer type IV pili (identified previously as mutant 1A2) (15) (Fig. 1). Subsequent notation of these cells will be in the form “WT,” “fimA null,” and “pilB null.” The WT isolate and pilB null cells develop robust biofilms in culture, while fimA null cells are biofilm deficient. Also, the WT isolate and fimA null cells exhibit twitching motility, while pilB null cells are twitching deficient. Bacterial cultures were maintained at 28°C on PW agar (4) modified by omitting phenol red and by adding 3.5 g liter−1 bovine serum albumin (Invitrogen, Carlsbad, CA). PD2 broth (4) was used as the supporting medium when X. fastidiosa cells were introduced and studied in the microfluidic chambers. Stocks of X. fastidiosa cultures were stored in modified PW broth-7% dimethyl sulfoxide at −80°C.
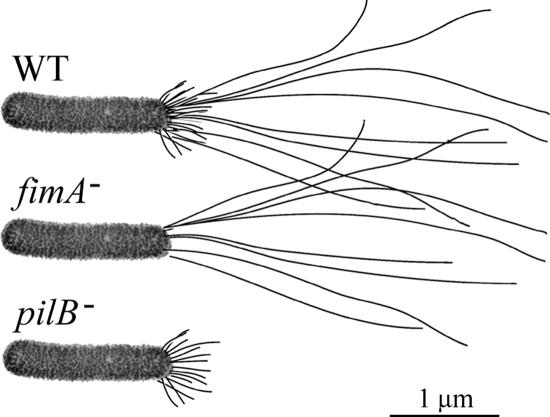
FIG. 1.
Illustration of Xylella fastidiosa strains with associated pili. The WT Temecula isolate possess both the shorter type I and longer type IV pili. fimA null cells (fimA−) (mutant 6E11) possess only type IV pili, and pilB null cells (pilB−) possess only type I pili.
Microfluidic chambers.
Microfluidic chamber fabrication involved photolithography and deep reactive-ion etching of a silicon wafer followed by replica molding of the wafer surface features with polydimethylsiloxane (PDMS) (Sylgard 184; Dow Corning, Midland, MI), a process similar to that previously described (15). In the present study, we used a newly designed dual-channel microfluidic device (Fig. 2). In brief, the chamber consisted of a molded PDMS body sandwiched between a cover glass and a supporting glass microscope slide. The interfacing surfaces were pretreated with an air plasma (Harrick Plasma, Ithaca, NY) prior to assembly to facilitate permanent covalent bonding (3). Each of the two parallel microchannels, separated by a 50-μm-wide PDMS partition, was 80 μm wide, 3.7 cm long, and 50 μm deep and had inlets and outlets for fluid flow as well as separate inlets for introduction of the bacteria. The primary difference between the design of the chamber used by Meng et al. (15) and the present chamber design is that the former had a single 100-μm-wide channel.
FIG. 2.
Schematic of the basic microfluidic chamber design. Microfluidic channels were 80 μm wide, 50 μm in depth, and 3.7 cm in length.
Microfluidic flow and bacterial cell attachment.
Four- to eight-day-old bacterial cultures on PW agar were used for the studies in which the cells were introduced into the microfluidic chambers (“adhesion force” experiments). The cells were gently scraped from the agar surface, suspended in PD2 broth, and dispersed by vortexing. The concentration of cell suspensions was assessed by microscopic observation and by determining absorbance at 600 nm. Microfluidic chambers and connecting tubing were flushed and filled with PD2 broth prior to introduction of bacterial cells. Flow of nutrient media (PD2 broth) through the microfluidic channels was controlled with a syringe pump (Pico Plus; Harvard Apparatus, Holliston, MA) fitted with two 5 ml Gastight syringes (Hamilton Company, Reno, NV), one for each channel. Introduction of bacterial cells was performed with 1 ml Gastight syringes. Once the cells attached to the glass surface of the microfluidic channels, medium flow was maintained at 0.1 to 1 μl min−1 (or at a shear stress of 0.4 dyn/cm2 to 4 dyn/cm2 at the substratum of the channel) for 60 to 90 min to stabilize the system and remove nonattached cells. During this time, images were captured every 30 s to confirm twitching motility activity of the introduced cells (for WT and fimA null cells only), indicating the presence of active type IV pili. Subsequently, the adhesiveness of the cells was assessed by sequentially increasing the flow of media by ramping up the syringe pump speed. Flow rates were increased from 2 to 220 μl min−1, which is equivalent to a shear stress of 8 to 878 dyn/cm2, with each maintained at a steady rate for 1 min.
All experiments were conducted at room temperature (ca. 22°C). For each set of experiments, the time between microfluidic chamber assembly and cell adhesion experiments, the tubing lengths, and the age of the cell cultures were kept as constant as possible. Each experiment was repeated three times using the side-by-side microfluidic chambers (Fig. 2); one of three different cell types, i.e., X. fastidiosa WT cells, fimA null cells, or pilB null cells, was placed in one of the paired chambers, and another of the cell types was placed in the other chamber. In addition, the experiment was repeated three times using the microfluidic chamber that was previously described by Meng et al. (15).
Microscopy and image analysis.
Microscopic observation of bacterial cell activities were assessed in the microfluidic chambers mounted on an inverted Olympus IMT-2 microscope using 40× phase-contrast optics. Time-lapse images were recorded with a SPOT-RT digital camera (Diagnostic Instruments, Inc., Sterling Heights, MI) controlled with MetaMorph Image software (Universal Imaging Corp., Downington, PA). Displacement of X. fastidiosa cells during the course of experimentation, viz., as the medium flow rate was increased, was recorded by capturing images every 5 s (= 13 images for each flow rate). The number of cells on each frame was scored using MetaMorph, and a number averaged from these 13 images was used as the number of cells attached at a specific flow rate. Cells were enumerated in a region comprising two-thirds of the channel width (avoiding boundary effects of channel side walls) and the full length of the channel captured in the image, viz., 53 μm by 280 μm.
Statistical analysis.
Experiments in which the drag force required to remove attached cells was determined (“adhesion force” experiments) were repeated six times. Highest and lowest adhesion force values for each bacterial strain treatment were discarded, providing trimmed means used for analysis. On the basis of assumption of equal population variances for each treatment, pair-wise trimmed t tests (25) were performed (P = 0.05).
Fluid dynamics and exerted forces on adherent cells. (i) Microfluidic channel design.
Relevant shear stress values are first determined by assessing the relation of shear stress S, volume flow rate Q, and pressure drop ΔP across the inlet and outlet of the channel by use of the Poseuille flow model (1). According to the analytical solution of a flow profile in a square channel, the shear stress at the bottom of the channel is given by S = 6 μQ/h2w, where μ is the viscosity of the water and h is the height and w the width of the channel. The pressure across the inlet and outlet of the channel is given by ΔP = 2SL/h, where L is the length of the channel. The analysis described above shows that a high shear stress can be achieved by decreasing the thickness of the channel h; however, h can not be arbitrarily small, as the pressure across the inlet and outlet of the channel will rise. Also, it is noted that in the Poiseuille flow model, a large aspect ratio (w/h) is assumed in which the flow velocity variation along the width w is ignored. This model provides an accuracy of ≤5% in comparison to the results from three-dimensional numerical simulations, given exact boundary conditions (12) for a large-aspect-ratio channel (w/h > 50). Note that the formula presented above is used only as the designing principle of this study and is an estimate of shear stresses at the bottom (surface) of the channel.
(ii) Computation of total drag force exerted on a bacterium.
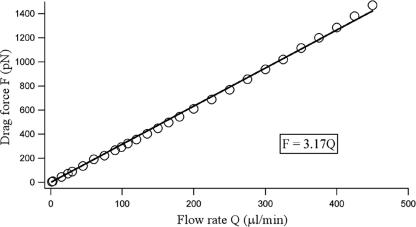
When a bacterium is attached to the surface of a flow chamber, the fluid flow exerts a shear stress as well as a pressure difference on the bacterium. The total force exerted on the bacterium by the flow is called the drag force (F) (contributed by both shear stress and pressure difference). To obtain a relationship between the drag force F and the volume fluid flow rate Q, we computed the drag force exerted on a bacterium by use of the exact channel geometry as employed in our experiments. A computation fluid dynamics software package, Fluent (Fluent Inc., Lebanon, NH), was employed for this purpose. Figure 3 describes the general setup for the numerical experiment, in which it was assumed that the bacterium had a capsule shape, a radius of 0.25 μm, and an end-to-end length of 1.90 μm and that it was located in the center and bottom of the channel. We chose the mesh size that resulted in a velocity field that was within 5% of its convergent value. In addition, we verified our code by solving a problem with a known analytical solution. The drag force of a rod in the center of the channel is F = 4πLU/[ln(L/R) − 0.5], where L is the length of the rod, R is the radius of the rod, and U is the velocity of the fluid flow along the center line of the bacterium (7). The total drag force exerted on the bacterium was then computed at various flow rates. The relationship between the drag force exerted on a bacterium in the center and bottom of the channel and the flow rate is shown in Fig. 4. Drag force was computed using F = 3.17Q, where Q is the flow rate. This relationship is computed for a channel width of 100 μm; for the experiments using channels with 80-μm widths, we estimated a 20% increase in drag force and thus used the relation F = 3.80Q. Two approximations were employed here: first, it was assumed that the cell body was prostrate and in contact with the substratum. Such close contact occurs as the bacteria are subjected to a strong unidirectional flow before they are moved or detached. Second, it was assumed that the drag force on a bacterium in the center two-thirds of the channel is the same as that on a bacterium in the median center of the channel. This assumption leads to a slight overestimate of our drag force; however, the uniformity with which the cells detach from the substratum in the center two-thirds of the channel justifies this assumption.
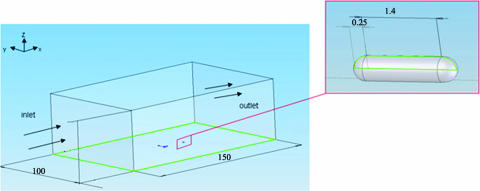
FIG. 3.
Schematic illustration of the experimental design used for numerical computation. The flow field was simulated using Fluent computation fluid dynamics software. The simulation box was 100 μm wide, 50 μm deep, and 150 μm long. The model bacterium attached to the channel surface was 0.5 μm in diameter and 1.90 μm in length.
FIG. 4.
Calibration curve generated by varying the flow rate in a test channel with total drag force exerted on the model bacterium (Fig. 3). Data points result from numerical calculations; the line represents a fit to a linear function.
RESULTS
Cell attachment.
Prior to and during introduction of X. fastidiosa cells into the microfluidic chambers, all conditions were maintained to be as constant as possible; nevertheless, differences were sometimes observed in the degree to which the cells attached to the surface in terms of both timing and number of cells. For example, cells with only short type I pili (pilB null cells) consistently attached as soon as they came into contact with the chamber surface. fimA null cells with solely longer type IV pili required a significantly longer time to attach to the surface. For this reason, it was necessary to reduce the flow rate of the medium to a minimum (0.1 μl min−1) for an hour to insure satisfactory attachment of enough cells to the chamber's surface. Therefore, for consistency we routinely left the system at a minimum flow rate for an hour for all strains following their introduction into the chambers. The strong attachment of pilB null cells is further demonstrated by the high number of cells (185; n = 6) attached to the chamber surface (within the field of view [53 μm by 280 μm]) immediately after cell seeding compared to the number of attached cells for fimA null cells (70; n = 6) and the wild-type X. fastidiosa isolate (65; n = 6). The type IV pilus-mediated twitching ability of the WT and fimA null cells was confirmed during the 1-h time period and before the medium flow was increased for the adhesion force experiments. In all instances WT and fimA null cells exhibited twitching motility in a significant number of cells.
Influence of flow on cell attachment.
Under no-flow or very-low-flow conditions, the orientation of many X. fastidiosa cells was observed to be more or less perpendicular to the substratum, with the cells being attached at the pilus-bearing pole. This cell orientation was readily noted in the form of spherical cellular shapes, since the rod-shaped cells were observed “on end.” As medium flow was increased, the cells, remaining attached by the one pole, gradually became oriented (leaning) with the direction of flow. In most instances, under increased flow conditions they appeared to be nearly prostrate with respect to the substratum. Upon further increases in flow rate, one of two distinct events was observed: (i) cells abruptly detached from the substratum and were washed downstream (and did not appear in subsequently captured images), or (ii) cells were dragged downstream on the glass surface and slid from the field of view or, sometimes, detached (Fig. 5). The former scenario was observed for most of the WT and fimA null cells, while the latter scenario always occurred for pilB null cells and sometimes for WT cells. Movies of both processes can be viewed in the supplemental material as well as at http://www.nysaes.cornell.edu/pp/faculty/hoch/movies/.
FIG. 5.
Image sequences depicting detachment of Xylella fastidiosa wild-type Temecula isolate cells (WT), fimA null cells (fimA−) (type IV pili only), and pilB null cells (pilB−) (type I pili only) from the microfluidic chamber surface. The results for three cells are demarcated (in a rectangle, an oval, and a circle) for each of the three cell types (with one cell type shown per column) in the frames in the first row to indicate the results seen at a flow rate of 2 μl min−1 (or at a shear stress of 8 dyn/cm2). At 90 μl min−1 (or at a shear stress of 360 dyn/cm2) one each of the WT and fimA null cells (oval) detached, and all three of the mutant pilB null cells were dragged downstream. At 220 μl min−1 (or at a shear stress of 880 dyn/cm2) few WT and no fimA null cells remained attached whereas significantly more pilB null cells remained attached, albeit displaced in position downstream. Flow direction is represented from left to right.
Drag forces necessary for cell detachment.
Xylella fastidiosa cells attached to the surface of the microfluidic chamber are exposed to drag forces that increase with increasing flow rates. The amount of force necessary for cell detachment, which we refer to as the “adhesion force of the bacterium,” is dependent on flow and the characteristics of the bacterium. In a typical experiment, as the flow of the medium was increased, the number of bacteria adherent to the substrate in the field of view (53 μm by 280 μm) was monitored. For example, the number of cells in the field of view decreased as the flow rate increased for the wild-type isolate and pilB and fimA null cells (Fig. 5). The rate at which the cells are detached from the substrate directly correlates to how strongly they adhere to the substrate. The number of cells in the center two-thirds of the channel (area of interest [AOI]) was counted for each captured frame, and the percentage of the cells remaining to be attached [(number of cells in the AOI/number of cells in the AOI at the lowest flow rate) × 100] was plotted as a function of flow rate (Fig. 6). For all cell types except pilB null cells, this was a straightforward process. For cells of pilB null cells which were dragged with the flow, most of which did not readily detach, it was necessary to confirm that the numbers present over time in the captured images represented an accurate count for MetaMorph analysis. This is because as the original cells were dragged out of the field of view, other cells upstream were dragged into view, with a potential for skewing of the data. Comparison of MetaMorph-derived cell counts with frame-by-frame visual counts confirmed the validity of the software approach. Total cell counts were nearly identical for the two approaches (data not shown); therefore, we used the MetaMorph software for all experiments.
FIG. 6.
Numbers of Xylella fastidiosa cells adherent to the microfluidic chamber surface as a function of the flow rate. Data represent the results observed for X. fastidiosa WT cells, fimA null cells (fimA−), and pilB null cells (pilB−).
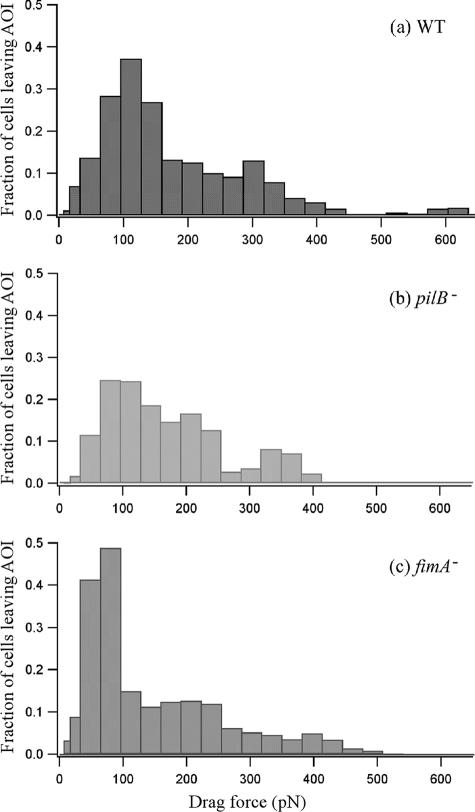
To assess the adhesion force of the bacteria for all three strains, we plotted the fraction of the cells [(number of cells at a given flow rate − number of cells at a lower consecutive flow rate)/number of cells at the lowest flow rate in the AOI] that detached from the substratum as a function of the drag force (Fig. 7). The data in Fig. 7 were generated from that in Fig. 6, and the flow rate values were converted to drag force values using the relationship shown in Fig. 4. The tendency of the cells with only the longer type IV pili (fimA null cells) to require lower drag forces for detachment from the substratum compared to the results seen with cells with only short type I pili (pilB null cells) is clearly apparent (Fig. 7). Using the data shown in Fig. 7, we then computed the average adhesion force for each cell type (integration of each curve in Fig. 7). Considering all six repetitions of the experiment, the average adhesion force observed for WT X. fastidiosa cells was 147 ± 11 pN; for pilB null cells, it was 204 ± 22 pN, and for fimA null cells it was 119 ± 8 pN (Fig. 8).
FIG. 7.
Fraction of the cells detached from the microfluidic chamber surface as a function of the drag force. The fraction of cells is defined as follows: fraction = [(number of cells at a given flow rate − number of cells at a lower consecutive flow rate)/number of cells at the lowest flow rate in the middle two-thirds of the channel]. The plot is derived from the data presented in Fig. 6. The drag force was calculated using the relation F = 3.80Q for channel width 80 μm and F = 3.17Q for channel width 100 μm, where Q is the flow rate. fimA−, fimA null cells; pilB−, pilB null cells.
FIG. 8.
Adhesion force of Xylella fastidiosa WT strain cells and pilus-defective mutants (fimA−, fimA null cells; pilB−, pilB null cells). The different letters on the bars correspond to statistical significant differences according to pair-wise trimmed t test results (P = 0.05). Error bars, standard deviations.
DISCUSSION
In this study we have measured the force of X. fastidiosa type I and type IV pilus adhesion to a glass substratum by use of a microfluidic flow chamber. To our knowledge, this is the first time that shear forces generated by flow through a microfluidic device have been used to assess the degree to which two distinct pilus types, separately and in combination, influence adhesion of bacteria to a substratum. Recently, Lu et al. (12) demonstrated that microfluidic flow chambers could be used to measure the adhesion force of mammalian cells. Microfluidic chamber devices such as that reported herein offer a range of advantages over macroscale flow chambers for studying cell-substratum interactions in that they (i) provide a larger dynamic range of shear forces; (ii) provide a platform that is readily integrated with microscopy; (iii) can be assembled to mimic nano- and microscale features of plants; (iv) allow for good microscopic imaging of small cells such as bacteria; and (v) consume lower volumes of reagents and cell culture. As shown in the present study, adhesion forces can be determined for a population of bacterial cells with different cell surface features; alternatively, larger devices are more amendable to incorporations of other techniques such as AFM and laser tweezers, often used to examine one cell at a time.
Over the course of the experiments using different mutants of X. fastidiosa, clear behavioral differences that were dependent on the pilus type present were observed. Furthermore, the results for some of the mutants were far more consistent and reproducible with respect to the way in which they interacted with the substratum. For example, the results obtained with fimA null cells (with type IV pili only) were the most reproducible, with adhesion force values between 101 and 202 pN (Δ = 101 pN). On the other hand, the results observed with pilB null cells (type I pili only) were the most variable, with adhesion force values between 114 and 418 pN (Δ = 304 pN). WT results ranged between 44 and 247 pN (Δ = 203 pN). There were also clear differences in how these different mutants and the wild-type cells interacted with the substratum. Wild-type and fimA null cells, for the most part, abruptly detached from the substratum when the shear force exceeded their ability to remain attached. In a few instances the wild-type cells “slid” for short distances on the substratum with the direction of flow before abruptly detaching. pilB null cells, however, were exceptionally consistent in exhibiting a sliding behavior along the substratum and rarely became detached even at higher shear forces. This observation led us to conclude that type I pili are primarily responsible for the “anchorage” of the cells to the surface in a fast-flowing environment; this anchorage may be a strategy for survival in the sap flow of plants. This is supported by other studies indicating that this mutant develops robust and abundant biofilms whereas fimA null cells produce essentially no biofilm and only loose aggregates of cells in liquid media (5, 10). Why do the wild-type cells with both types of pili adhere less strongly than those with only short type I pili (Fig. 5)? We hypothesize that the presence of the longer type IV pili somehow physically obstructs maximum contact of short pili with the substratum. Also, the type IV pili are dynamic—constantly extending and retracting, keeping the cell in motion. This also possibly keeps wild-type cells from achieving maximum contact.
The value (108 pN) for adhesion of type IV pili to substrata seen with X. fastidiosa with fimA null cell characteristics is similar to those reported for other bacterial species as determined utilizing different approaches. Maier et al. (13) determined a “stall” force of 110 pN for type IV pili in N. gonorrhoeae in experiments performed using laser tweezers; recently, Touhami et al. (21) determined an adhesion force of 95 pN for type IV pili of P. aeruginosa in experiments performed using AFM. It is noted that both N. gonorrhoeae and P. aeruginosa lack type I pili; thus, only the values of the X. fastidiosa fimA null cells are applicable for comparison. Such closeness in values reflects upon the utility of the three methods of measurement and may also relate to the conserved structure of the pilus and function among diverse bacterial species. Our approach considered the bacterial cell and all appendages (pili) as a whole, while other researchers used a single pilus to measure the adhesion force (13, 21). We did not determine the number of type IV pili that attached individual cells to the substratum, in part because this value is not easily determined. The closeness of the adhesion force values for X. fastidiosa to the values observed in other studies (13, 21) may indicate that a single pilus, or at most a few pili, are involved in the attachment with the substratum at any given time; however, this hypothesis needs to be further investigated. One would expect that the adhesion force would increase with the number of pili attached to a substratum.
Factors other than type I and type IV pili have roles in cell attachment to substrata. Genome sequence data (23) and work by other researchers (6) show that the X. fastidiosa WT strain possesses an array of nonfimbrial adhesins present on the cell surface. These data are supported by our preliminary observations (unpublished results) showing that a double mutant lacking both types of pili still attaches to the substratum, albeit such mutants required longer durations and conditions of no flow or very slow to medium flow to achieve any significant degree of attachment.
We have presented the results of a comparative study in which the adhesion forces of type I and type IV pili were assessed using two mutant strains and a wild-type isolate of X. fastidiosa. The unique dual-pilus composition of the wild-type X. fastidiosa strain presented an opportunity to study the characteristics of the two types of pili comparatively in the same experimental setting. This study demonstrated that microfluidic chambers provide an accurate and easy system with which to measure adhesion force of bacterial cells. Insights gained during this study will lead to a better understanding of the roles played by type I and type IV pili in bacterial cell attachment to surfaces in nature.
Acknowledgments
This work was supported, in part, with funds from the National Science Foundation (CBET-0619626) and the New York State Office of Science, Technology and Academic Research (NYSTAR) to M.W. (in the form of a Center for Advanced Technology grant) and by grants from the Nanobiotechnology Center (NBTC), an STC Program of the National Science Foundation under agreement ECS-9876771, and from the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service administered through the University of California Pierce's Disease Research Grants Program to H.C.H. This work was also performed, in part, at the Cornell Nanofabrication Facility (a member of the National Nanofabrication Users Network), which is supported by the National Science Foundation under grant ECS-9731293, its users, Cornell University, and industrial affiliates.
We thank David Caughey for his assistance with numerical calculations of the drag force, Lauren Reese for her help with this project, and John Barnard for his help with the statistical analysis.
Footnotes
Published ahead of print on 9 February 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Batchelor, G. K. 1999. An introduction to fluid-dynamics, 2nd ed., p. 179-183. Cambridge University Press, Cambridge, United Kingdom.
- 2.Brown, D. C., and R. S. Larson. 2001. Improvements to parallel plate flow chambers to reduce reagent and cellular requirements. BMC Immunol. 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhury, M. K., and G. M. Whitesides. 1991. Direct measurement of interfacial interactions between semispherical lenses and flat sheets of poly(dimethylsiloxane) and their chemical derivatives. Langmuir 7:1013-1025. [Google Scholar]
- 4.Davis, M. J., W. J. French, and N. W. Schaad. 1981. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr. Microbiol. 6:309-314. [Google Scholar]
- 5.Galvani, C. D., Yaxin Li, T. J. Burr, and H. C. Hoch. 2007. Twitching motility among pathogenic Xylella fastidiosa isolates and the influence of bovine serum albumin on twitching-dependent colony fringe morphology. FEMS Microbiol. Lett. 268:202-208. [DOI] [PubMed] [Google Scholar]
- 6.Guilhabert, M. R., L. M. Hoffman, D. A. Mills, and B. C. Kirkpatrick. 2001. Transposon mutagenesis of Xylella fastidiosa by electroporation of Tn5 synaptic complexes. Mol. Plant-Microbe Interact. 14:701-706. [DOI] [PubMed] [Google Scholar]
- 7.Guyon, E., J. P. Hulin, L. Petit, and C. D. Mitescu. 2001. Physical hydrodynamics, 1st ed., p. 341-343. Springer, New York, NY.
- 8.Hopkins, D. L., and A. H. Purcell. 2002. Xylella fasidiosa: cause of Pierce's disease of grapevine and other emergent diseases. Plant Dis. 86:1056-1066. [DOI] [PubMed] [Google Scholar]
- 9.Lambais, M. R., M. H. Goldman, L. E. Camargo, and G. H. Goldman. 2000. A genomic approach to the understanding of Xylella fastidiosa pathogenicity. Curr. Opin. Microbiol. 3:459-462. [DOI] [PubMed] [Google Scholar]
- 10.Li, Y., G. Hao, C. D. Galvani, Y. Meng, L. De La Fuente, H. C. Hoch, and T. J. Burr. 2007. Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation, and cell-cell aggregation. Microbiology 153:719-726. [DOI] [PubMed] [Google Scholar]
- 11.Lindow, S. E. 2005. Effects of fimbrial (FimA, FimF) and afimbrial (XadA, HxfB) adhesins on the adhesion of Xylella fastidiosa to surfaces, p. 173-176. In proceedings of the 2005 Pierce's Disease Research Symposium. California Department of Food and Agriculture, Sacramento, CA.
- 12.Lu, H., L. Y. Koo, W. C. M. Wang, D. A. Lauffenburger, L. G. Griffith, and K. F. Jensen. 2004. Microfluidic shear devices for quantitative analysis of cell adhesion. Anal. Chem. 76:5257-5264. [DOI] [PubMed] [Google Scholar]
- 13.Maier, B., L. Potter, M. So, C. D. Long, H. S. Seifert, and M. P. Sheetz. 2002. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. USA 99:16012-16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 15.Meng, Y., Y. Li, C. D. Galvani, G. Hao, J. N. Turner, T. J. Burr, and H. C. Hoch. 2005. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 187:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 17.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type 1 pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 18.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas, W. E., L. M. Nilsson, M. Forero, E. V. Sokurenko, and V. Vogel. 2004. Shear-dependent ‘stick-and-roll’ adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol. 53:1545-1557. [DOI] [PubMed] [Google Scholar]
- 20.Thomas, W. E., E. Trintchina, M. Forero, V. Vogel, and E. V. Sokurenko. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109:913-923. [DOI] [PubMed] [Google Scholar]
- 21.Touhami, A., M. H. Jericho, J. M. Boyd, and T. J. Beveridge. 2006. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 188:370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Houdt, R., and C. W. Michiels. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156:626-633. [DOI] [PubMed] [Google Scholar]
- 23.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, L. E. Camargo, A. C. da Silva, D. H. Moon, M. A. Takita, E. G. Lemos, M. A. Machado, M. I. Ferro, F. R. da Silva, M. H. Goldman, G. H. Goldman, M. V. Lemos, H. El-Dorry, S. M. Tsai, H. Carrer, D. M. Carraro, R. C. de Oliveira, L. R. Nunes, W. J. Siqueira, L. L. Coutinho, E. T. Kimura, E. S. Ferro, R. Harakava, E. E. Kuramae, C. L. Marino, E. Giglioti, I. L. Abreu, L. M. Alves, A. M. do Amaral, G. S. Baia, S. R. Blanco, M. S. Brito, F. S. Cannavan, A. V. Celestino, A. F. da Cunha, R. C. Fenille, J. A. Ferro, E. F. Formighieri, L. T. Kishi, S. G. Leoni, A. R. Oliveira, V. E. Rosa, Jr., F. T. Sassaki, J. A. Sena, A. A. de Souza, D. Truffi, F. Tsukumo, G. M. Yanai, L. G. Zaros, E. L. Civerolo, A. J. Simpson, N. F. Almeida, Jr., J. C. Setubal, and J. P. Kitajima. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells, J. M., B. C. Raju, H. Y. Hung, W. G. Weisburg, L. Mandelco-Paul, and D. J. Brenner. 1987. Xylella fastidiosa gen. nov., sp. nov.: gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int. J. Syst. Bacteriol. 37:136-143. [Google Scholar]
- 25.Yuen, K. K., and W. J. Dixon. 1973. The approximate behavior and performance of the two-sample trimmed t. Biometrika 60:369-374. [Google Scholar]