Abstract
An approach is proposed for in situ detection of short signature DNA sequences present in single copies per bacterial genome. The site is locally opened by peptide nucleic acids, and a circular oligonucleotide is assembled. The amplicon generated by rolling circle amplification is detected by hybridization with fluorescently labeled decorator probes.
Fluorescence in situ hybridization (FISH) is a convenient method for bacterial identification (23, 25, 27, 31, 34). Due to their high copy numbers (104 to 105), rRNAs have long been regarded as the most suitable targets for bacterial FISH (2, 3, 8). With the advent of the era of “global” genome sequencing and the parallel development of potent signal amplification techniques (7, 21, 25, 29), it has recently become possible to target other nucleic acid sites (4, 13, 22, 24, 32), including regions of bacterial chromosomal DNA (34, 36). However, these new approaches require long probes, very high probe concentrations, and long hybridization (35, 36). A shorter target sequence would increase assay specificity: shorter probes are less tolerant of base pair mismatches and also can result in better discrimination between otherwise very similar bacteria. Recently, another approach has been described for the in situ detection of low-copy-number genomic sequences by using short (35- to 39-nucleotide [nt]-long) target sequences, rolling-circle amplification (RCA), and FISH. However, this approach cannot be performed directly on slides and involves embedding the cells into membrane filters and heating them to denature genomic DNA (19, 20).
An isothermal amplification method reported in the literature, the multiple displacement amplification reaction, uses phi29 DNA polymerase and random primers to amplify genomic DNA (6, 18). It is applied for DNA microarray-based screening and direct detection of pathogen-specific DNA (1, 14, 28, 30, 33).
We describe here a new approach to using low-copy-number genomic sequences as targets for the detection of specific bacteria. The technique expands both the utility and the resolving power of whole-cell FISH for the detection of microbes and may be useful in food, environmental, clinical diagnostic, and national security-related applications.
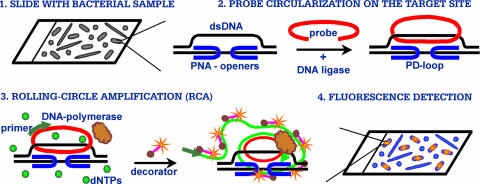
Our major tool is peptide nucleic acid (PNA) (Fig. 1). PNA openers have a unique ability to locally open double-stranded DNA (dsDNA) via binding to one of the two DNA strands, leaving the other strand accessible for hybridization with synthetic oligonucleotide probes. Such a complex between dsDNA, a pair of PNAs, and the oligonucleotide probe is called a PD-loop (5). The binding sites for each of the two PNA openers must be short (7- to 10-bp-long) homopurine-homopyrimidine tracts, and they are separated by an arbitrary sequence of nucleobases of up to 10 bp. We call such a site consisting of two PNA-binding sites and the linker sequence the PD-loop site. Statistically, such sites are expected to exist once per a few hundred base pairs of DNA sequence (see reference 9 and the supplemental material).
FIG. 1.
Major steps of a DNA-based assay for fluorescence in situ detection of short DNA sequences in a single bacterial cell. dNTPs, deoxynucleoside triphosphates.
After the opening of a PD-loop site with a pair of PNA openers, a circular probe is assembled by T4 DNA ligase by using a circularizable oligonucleotide with termini that are complementary to the displaced strand (11, 15, 16) (Fig. 1, step 2). This circular probe assembly is exceedingly sequence specific since only chosen sites are opened by PNA openers and the remaining DNA maintains its duplex form and is inaccessible for mismatch hybridization (10, 11, 16, 26).
Fluorescence labeling is achieved by an RCA reaction on the circular probe (17) in the presence of fluorescently labeled decorators. These decorators consist of linear oligonucleotides with fluorophores at their termini; their hybridization to single-stranded DNA produced by RCA yields the multiply fluorescently labeled product (Fig. 1, step 3). The fluorescent signal is readily detected by standard techniques using fluorescence microscopy (Fig. 1, step 4).
Our approach integrates the highly specific and sensitive recognition of target dsDNA sequences by using PNA openers and circularized probes with efficient signal amplification by RCA. We report here the results of proof-of-principle experiments performed with three bacterial species: Escherichia coli, Bacillus subtilis, and Streptococcus mutans.
Table 1 presents the PD-loop sites chosen for the three bacterial species along with corresponding PNA openers, circularizable probes, decorators, and primer sequences (see methods in the supplemental material). The chosen target sites (Table 1) correspond to various genomic regions and to various DNA strands: coding and noncoding regions and sense and antisense strands in coding regions. All these sites yielded similar signals (see below), which excluded the possibility that the signal was due to the targeting of mRNA transcripts. To additionally exclude RNA targeting by our probes, RNA was degraded by subjecting all samples to RNase A treatment (see methods in the supplemental material).
TABLE 1.
Signature sites, PNAs, circularizable oligonucleotides, decorator probes, and primer used in this work
| Bacterium | Signature sitea (description) | PNA(s)b | Decorator probe, ODNs, and primerc |
|---|---|---|---|
| E. coli | GGAGAGAGACTCAAAAGAAGG (major cold shock protein gene [csp] region; 1.050 Mb; antisense strand; mRNA for cspG) | PNA1, H-Lys2-CTCTCTCC-(eg1)3-JJTJTJTJ- Lys-NH2; PNA2, H-Lys2-TTTJTTJJ-(eg1)3- CCTTCTTT-Lys-NH2 | decR, 5′-Cy3-TCACGGAATGGTTACTTGCACAGC-biotin-3′; ODNcspG, 5′-p-tcaAAAGAAGG(tcacggaatggttacttgcCAGC)CAGCAGCC(TCACggaatggttacttgccagc)GGAGAGAGac-3′; ODNrpoN, 5′-p-gctGAAAGAAG(tcacggaatggttacttgcCAGC)CAGCAGCC(TCACggaatggttacttgccagc)GAAAGAAGatg-3′; ODNrnr, 5′-p-ccGGAAGAAG(tcacggaatggttacttgcCAGC)CAGCAGCC(TCACggaatggttacttgccagc)GAAAGAAGaagtg-3′ |
| GAAAGAAGATGTGCTGAAAGAAG (RNA polymerase sigma N factor gene rpoN; 3.343 Mb; sense strand) | PNA3, H-Lys2-JTTTJTTJ-(eg1)3-CTTCTTTC- Lys-NH2 | ||
| GAAAGAAGAAGTGCCGGAA GAAG (exoribonuclease R gene rnr; 4.405 Mb; sense strand) | PNA3 and PNA4, H-Lys2-JJTTJTTJ-(eg1)3- CTTCTTCC-Lys-NH2 | ||
| B. subtilis | GAAAAGAAACCCTTCAGAGGA AG(serA region; 2.391 Mb; noncoding region)GGAAGAAGCGCACTAAAG AAAA (yxjA gene; 4.005 Mb; antisense strand) | PNA5, H-Lys3-JTTTTJTT-(eg1)3-TTCTTTTC- Lys-NH2; PNA6, H-Lys2-TJTJJTTJ-(eg1)3- CTTCCTCT-Lys-NH2PNA4 and PNA7, H-Lys2-TTTJTTTT-(eg1)3- TTTTCTTT-Lys-NH2 | decG, 5′-FITC-CCTCAATCGTCGTCGTGTACTAC-FITC-3′; ODNserA, 5′-p-ttcAGAGGAAGttatCAGCCAGCAGCCTCA(Cctcaatcgtcgtcgtgtactac)tattGAAAAGAAaccc-3′; ODNyxjA, 5′-p-cactAAAGAAAAagtCAGCCAGCAGCCTCA(Cctcaatcgtcgtcgtgtactac)taattGGAAGAAGcg-3′ |
| S. mutans | AAAGAAAAATATTTAAAGA GGAA (dnaK region; 0.085 Mb; sense strand) | PNA7 and PNA8, H-TTJTJJTT-(eg1)3- TTCCTCTT-Lys-NH2 | decG, 5′-FITC-CCTCAATCGTCGTCGTGTACTAC-FITC-3′; ODNdnaK, 5′-p-ttaAAGAGGAAttatCAGCCAGCAGCCTCA(Cctcaatcgtcgtcgtgtactac)tattAAAGAAAAatat-3′; ODNwapA, 5′-p-tttAAGAGGAAtattCAGCCAGCAGCCTCA(Cctcaatcgtcgtcgtgtactac)tattAAAAGAGGtat-3′; ODNhypP, 5′-p-ggtgAGAGGAAGttatCAGCCAGCAGCCTCA(Cctcaatcgtcgtcgtgtactac) tattGGAAGAAGttcg-3′; primer, 5′-GTGAGGCTGCTGGCTG-3′ |
| AAAAGAGGTATTTTAAGAGG AA (wapA region; 0.934 Mb; sense strand) | PNA9, H-Lys2-TTTTJTJJ-(eg1)3-CCTCTTTT- Lys-NH2; PNA8 | ||
| GGAAGAAGTTCGGGTGAGAG GAAG (hypothetical protein gene [hyp] region; 1.329 Mb; noncoding region) | PNA4 and PNA6 |
PNA binding sites are underlined.
eg1, Lys, and J denote the bis-PNA linker segment, the amino acid lysine, and the nucleobase pseudoisocytosine, respectively (12).
In circularizable probe sequences, letters both capitalized and underlined represent primer annealing sites, letters capitalized but not underlined represent sequences identical to those of PNA binding sites, and letters in parentheses represent sites for decorator hybridization. ODNs, circularizable oligodeoxynucleotides; FITC, fluorescein isothiocyanate.
We first validated our approach using E. coli and B. subtilis as model organisms. All steps were sequentially performed (Fig. 1) with cells fixed on polylysine slides (see methods in the supplemental material). Cells were counterstained by DAPI (4′,6′-diamidino-2-phenylindole), and slides were evaluated by fluorescence microscopy using standard FISH equipment and software. Figure 2 shows typical results of experiments with E. coli and B. subtilis. Both bacteria gave clear signals (red for E. coli and green for B. subtilis) when a set of site-specific genomic probes was applied (Fig. 2A and D). Figure 1 presents the data for only one site for each species (see the legend to Fig. 1). Very similar data (not shown) were obtained for three other sites (two for E. coli and one for B. subtilis).
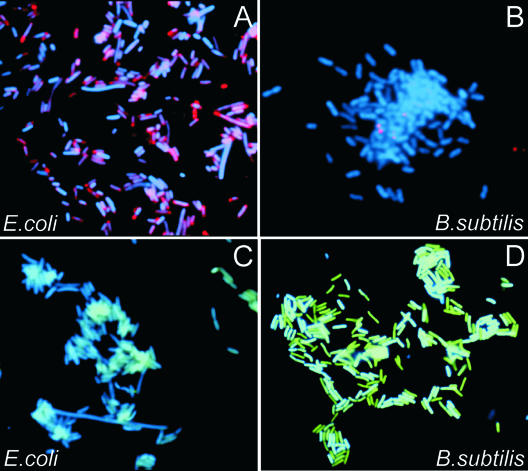
FIG. 2.
Images of bacterial cells observed by using a fluorescence microscope in experiments performed according to the scheme presented in Fig. 1. The fluorescence signals were acquired separately using three filter sets (DAPI for DNA and Cy3 or fluorescein for the labeled RCA product). Each image is a superposition of two separate images, with DAPI and Cy3 or DAPI and fluorescein signals pseudocolored in blue and red or blue and green, respectively. (A) E. coli cells to which the probes corresponding to the 21-nt target site in the E. coli cold shock protein gene (csp) region, PNA1, PNA2, ODNcspG, and decR, were applied (Table 1). Virtually all cells displayed very bright spots. No such spots were observed in numerous negative control experiments in which any of the steps of the protocol given in Fig. 1 were omitted (see the supplemental material). (B) The same procedure as that described in the legend to panel A was carried out with a combination of all probes specific to E. coli (PNA1, PNA2, PNA3, PNA4, ODNcspG, ODNrpoN, ODNrnr, and decR) (Table 1) applied to B. subtilis cells. No signal was detected. (C) A combination of probes specific to B. subtilis (PNA4, PNA5, PNA6, PNA7, ODNserA, ODNyxjA, and decG) was applied to E. coli cells. No signal was detected. (D) B. subtilis cells to which the probes corresponding to the 23-nt target site in the B. subtilis phosphoglycerate dehydrogenase gene (serA) region (PNA5, PNA6, ODNserA, and decG) were applied.
No signal was observed in various negative control experiments, which are described in the supplemental material. To confirm the specificity of detection and to exclude the possibility of nonspecific probe binding to cell structures, a mixture of all E. coli-specific probes was applied to B. subtilis cells and vice versa. No cross signals were observed (Fig. 2B and C). The data shown in Fig. 2 demonstrate that our approach can specifically target single-copy, short DNA sequences in bacteria and can generate a detection signal readily seen by fluorescence microscopy.
We then performed our protocol with a mixture of E. coli and B. subtilis cells, combining the corresponding probes (Table 1). Nearly all cells on mixed slides displayed a red or green signal only (Fig. 3A). Although on these slides E. coli and B. subtilis cells can appear morphologically similar, the results of the control experiments (Fig. 2) support the premise that red fluorescence is specific to E. coli and green fluorescence is specific to B. subtilis.
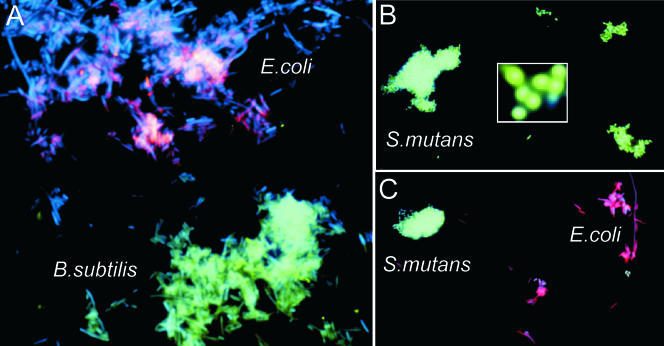
FIG. 3.
(A) Images of bacterial cells observed by using a fluorescence microscope in experiments performed with a mixture of E. coli and B. subtilis cells to which a combination of probes specific to E. coli and B. subtilis, PNA3, PNA4, PNA7, ODNrnr, ODNyxjA, decR, and decG, were applied. E. coli and B. subtilis bacteria can be distinguished from each other in the mixture of both types of cells. (B) S. mutans cells to which the probes corresponding to the 22-nt target site in the S. mutans wall-associated protein gene (wapA), PNA8, PNA9, ODNwapA, and decG, were applied. Virtually all cells are colored green. No such spots were observed in various negative control experiments in which any of the steps were omitted (data not shown). The insert shows an enlarged image of several S. mutans cells. (C) Images of bacterial cells observed by using a fluorescence microscope in experiments performed with E. coli and S. mutans cells with the combination of probes specific to E. coli (rpoN and rnr) and S. mutans (hypP): PNA3, PNA4, PNA6, ODNrpoN, ODNrnr, ODNhypP, decR, and decG. The fluorescent signals were acquired separately using three filter sets with DAPI, Cy3, and fluorescein.
We also investigated a mixture of bacillary and coccal bacteria. Figure 3C shows some resulting data for the mixture of E. coli and S. mutans, in which round cocci and rod-shaped bacilli show green and red signals, respectively.
In this study, every site arbitrarily chosen (eight sites total) from the genome database worked as a specific signature site for the corresponding bacterium. The data support the idea that the proposed approach could be used as a universal tool for specific bacterial detection.
We also verified that bacterial detection by our method is possible with clinical and environmental samples (see the supplemental material).
Supplementary Material
Acknowledgments
We thank Kevin Luk for technical help and useful comments and Nancy S. Miller for reading the manuscript and making many useful suggestions.
This work was supported by grants from the Coulter Foundation and from the National Institutes of Health (CA112418).
Footnotes
Published ahead of print on 9 February 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abulencia, C. B., D. L. Wyborski, J. A. Garcia, M. Podar, W. Chen, S. H. Chang, H. W. Chang, D. Watson, E. L. Brodie, T. C. Hazen, and M. Keller. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., F.-O. Glockner, and A. Neef. 1997. Modern methods in subsurface microbiology: in situ identification of microorganisms with nucleic acid probes. FEMS Microbiol. Rev. 20:191-200. [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakermans, C., and E. L. Madsen. 2002. Detection in coal tar waste-contaminated groundwater of mRNA transcripts related to naphthalene dioxygenase by fluorescent in situ hybridization with tyramide signal amplification. J. Microbiol. Methods 50:75-84. [DOI] [PubMed] [Google Scholar]
- 5.Bukanov, N. O., V. V. Demidov, P. E. Nielsen, and M. D. Frank-Kamenetskii. 1998. PD-loop: a complex of duplex DNA with an oligonucleotide. Proc. Natl. Acad. Sci. USA 95:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean, F. B., S. Hosono, L. Fang, X. Wu, A. F. Faruqi, P. Bray-Ward, Z. Sun, Q. Zong, Y. Du, J. Du, M. Driscoll, W. Song, S. F. Kingsmore, M. Egholm, and R. S. Lasken. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 99:5261-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 9.Demidov, V., N. Bukanov, and M. D. Frank-Kamenetskii. 1999. Duplex DNA capture, p. 266. In M. Egholm and P. E. Nielsen (ed.), Peptide nucleic acids: protocols and applications. Horizon Scientific, Wymondham, United Kingdom.
- 10.Demidov, V. V., and M. D. Frank-Kamenetskii. 2004. Two sides of the coin: affinity and specificity of nucleic acid interactions. Trends Biochem. Sci. 29:62-71. [DOI] [PubMed] [Google Scholar]
- 11.Demidov, V. V., H. Kuhn, I. V. Lavrentieva-Smolina, and M. D. Frank-Kamenetskii. 2001. Peptide nucleic acid-assisted topological labeling of duplex DNA. Methods 23:123-131. [DOI] [PubMed] [Google Scholar]
- 12.Egholm, M., L. Christensen, K. L. Dueholm, O. Buchardt, J. Coull, and P. E. Nielsen. 1995. Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucleic Acids Res. 23:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, D., R. I. Amann, and J. Zeyer. 1993. Detection of mRNA in Streptomyces cells by whole-cell hybridization with digoxigenin-labeled probes. Appl. Environ. Microbiol. 59:2753-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchison, C. A., H. O. Smith, C. Pfannkoch, and J. C. Venter. 2005. Cell-free cloning using phi29 DNA polymerase. Proc. Natl. Acad. Sci. USA 102:17332-17336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn, H., V. V. Demidov, and M. D. Frank-Kamenetskii. 1999. Topological links between duplex DNA and a circular DNA single strand. Angew. Chem. Int. Ed. Eng. 38:1446-1449. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn, H., V. V. Demidov, and M. D. Frank-Kamenetskii. 2000. An earring for the double helix: assembly of topological links comprising duplex DNA and a circular oligodeoxynucleotide. J. Biomol. Struct. Dyn. 11:221-225. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn, H., V. V. Demidov, and M. D. Frank-Kamenetskii. 2002. Rolling-circle amplification under topological constraints. Nucleic Acids Res. 30:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasken, R. S., and M. Egholm. 2003. Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol. 21:531-535. [DOI] [PubMed] [Google Scholar]
- 19.Maruyama, F., T. Kenzaka, N. Yamaguchi, K. Tani, and M. Nasu. 2005. Visualization and enumeration of bacteria carrying a specific gene sequence by in situ rolling circle amplification. Appl. Environ. Microbiol. 71:7933-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maruyama, F., K. Tani, T. Kenzaka, N. Yamaguchi, and M. Nasu. 2006. Quantitative determination of free-DNA uptake in river bacteria at the single-cell level by in situ rolling-circle amplification. Appl. Environ. Microbiol. 72:6248-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris, R. M., M. S. Rappe, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 22.Moter, A., and U. B. Gobel. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41:85-112. [DOI] [PubMed] [Google Scholar]
- 23.Mothershed, E. A., and A. M. Whitney. 2006. Nucleic acid-based methods for the detection of bacterial pathogens: present and future considerations for the clinical laboratory. Clin. Chim. Acta 363:206-220. [DOI] [PubMed] [Google Scholar]
- 24.Pernthaler, A., and R. Amann. 2004. Simultaneous fluorescence in situ hybridization of mRNA and rRNA in environmental bacteria. Appl. Environ. Microbiol. 70:5426-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potaman, V. N. 2003. Applications of triple-stranded nucleic acid structures to DNA purification, detection and analysis. Exp. Rev. Mol. Diagn. 3:481-496. [DOI] [PubMed] [Google Scholar]
- 27.Procop, G. W. 2002. In situ hybridization for the detection of infectious agents. Clin. Microbiol. Newsl. 24:121-125. [Google Scholar]
- 28.Raghunathan, A., H. R. Ferguson, Jr., C. J. Bornarth, W. Song, M. Driscoll, and R. S. Lasken. 2005. Genomic DNA amplification from a single bacterium. Appl. Environ. Microbiol. 71:3342-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schönhuber, W., B. Fuchs, S. Juretschko, and R. Amann. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vora, G. J., C. E. Meador, D. A. Stenger, and J. D. Andreadis. 2004. Nucleic acid amplification strategies for DNA microarray-based pathogen detection. Appl. Environ. Microbiol. 70:3047-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, M., M. Horn, and H. Daims. 2003. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr. Opin. Microbiol. 6:302-309. [DOI] [PubMed] [Google Scholar]
- 32.Wagner, M., M. Schmid, S. Juretschko, T. Karl-Heinz, A. Bubert, W. Goebel, and K.-H. Schleifer. 1998. In situ detection of a virulence factor mRNA and 16S rRNA in Listeria monocytogenes. FEMS Microbiol. Lett. 160:159-168. [DOI] [PubMed] [Google Scholar]
- 33.Wu, L., X. Liu, C. W. Schadt, and J. Zhou. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwirglmaier, K. 2005. Fluorescence in situ hybridisation (FISH): the next generation. FEMS Microbiol. Lett. 246:151-158. [DOI] [PubMed] [Google Scholar]
- 35.Zwirglmaier, K., K. Fichtl, and W. Ludwig. 2005. In situ functional gene analysis: recognition of individual genes by fluorescence in situ hybridization. Methods Enzymol. 397:338-351. [DOI] [PubMed] [Google Scholar]
- 36.Zwirglmaier, K., W. Ludwig, and K.-H. Schleifer. 2004. Recognition of individual genes in a single bacterial cell by fluorescence in situ hybridization: RING-FISH. Mol. Microbiol. 51:89-96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.