Abstract
Manure samples were collected from 16 organic (ORG) and 9 low-input conventional (LIC) Dutch dairy farms during August and September 2004 to determine the prevalence of the STEC virulence genes stx1 (encoding Shiga toxin 1), stx2 (encoding Shiga toxin 2), and eaeA (encoding intimin), as well as the rfbE gene, which is specific for Escherichia coli O157. The rfbE gene was present at 52% of the farms. The prevalence of rfbE was higher at ORG farms (61%) than at LIC farms (36%), but this was not significant. Relatively more LIC farms were positive for all Shiga toxin-producing E. coli (STEC) virulence genes eaeA, stx1, and stx2, which form a potentially highly virulent combination. Species richness of Enterobacteriaceae, as determined by DGGE, was significantly lower in manure positive for rfbE. Survival of a green fluorescent protein-expressing E. coli O157:H7 strain was studied in the manure from all farms from which samples were obtained and was modeled by a biphasic decline model. The time needed to reach the detection limit was predominantly determined by the level of native coliforms and the pH (both negative relationships). Initial decline was faster for ORG manure but leveled off earlier, resulting in longer survival than in LIC manure. Although the nonlinear decline curve could theoretically be explained as the cumulative distribution of an underlying distribution of decline kinetics, it is proposed that the observed nonlinear biphasic pattern of the survival curve is the result of changing nutrient status of the manure over time (and thereby changing competition pressure), instead of the presence of subpopulations differing in the level of resistance.
Shiga-toxin producing Escherichia coli (STEC) is an important group of zoonotic human pathogens, with E. coli O157 being the best known and most studied serotype (10). STEC strains are generally carried asymptomatically by cattle and shed in their feces, which in turn may serve as a means of maintenance and spread of these pathogens among cattle herds (47). Since the reported increase in food-borne disease associated with the consumption of fresh vegetables (51, 54), the potential contamination of vegetable crops that are grown in fields enriched with manure has raised increasing concern (18, 29, 52). Understanding on-farm survival and spread of human pathogens is of utmost importance in preventing the spread of this organism to the environment, groundwater, food, and crops and subsequently back to cattle.
A considerable body of literature exists on the prevalence of STEC on cattle farms and statistically associated management factors which could be used for potential intervention strategies to reduce shedding of these pathogens (2, 53, 57). However, these studies are not unambiguous and have resulted in the identification of only a very few risk factors, like season and cattle age, which are ubiquitous. The direction of influence of the majority of the management practices like feeding regimen and housing conditions remains unclear and is under debate.
Dairy farm management practices (i.e., diet, housing conditions, and antibiotic use) influence both the abiotic and biotic characteristics of the digestive system of cattle, the manure produced by these animals, and the environment in which the pathogens are excreted. In turn, these abiotic and biotic factors will determine the susceptibility of cattle to STEC colonization, growth, and/or survival in the ruminant gut and subsequent survival in excreted manure. To date, little is known about the relationship between STEC prevalence in manure and the chemical and biological composition of this complex substrate.
E. coli O157:H7 is able to survive for extended periods of time in manure, but very little is known about the factors determining the fate of the pathogen in manure (19, 33, 60). Behavior of pathogens in broth culture as a function of different chemical and physical variables cannot be extrapolated to complex substrates like manure. Recently it was reported that the pH and fiber content of the manure were the main determinants of survival of E. coli O157:H7 in manure derived from cattle fed different diets, with the longest survival at lower pH and lower fiber content (19). However, there is still a need to study pathogen survival under a wider array of non-experimentally produced manure in order to identify the driving factors behind pathogen survival and decline in manure.
Besides the chemical composition, there are strong indications that the microbial community has an important influence on the survival capabilities of E. coli O157:H7 in environmental substrates. E. coli O157:H7 growth kinetics were influenced by the presence of a competitive microflora compared to pure culture (14), and E. coli O157:H7 survived significantly longer in manure-amended autoclaved soil than in manure-amended nonautoclaved soil (30). In addition, the decline rate of the soil-borne plant pathogenic bacterium Ralstonia solanacearum in soil was positively correlated with the diversity of microbes as estimated by denaturing gradient gel electrophoresis (DGGE) (N. A. S. Messiha, A. D. van Diepeningen, E. Franz, J. D. Janse, M. E. Schoeman-Weerdestijen, A. J. Termorsuizen, and A. H. C. van Bruggen, submitted for publication). However, the influence of the microbial community on the survival of E. coli O157:H7 in manure has never been investigated.
In order to conduct a risk analysis on pathogen spread in the environment and the (vegetable) food chain, models are needed which accurately describe pathogen survival in complex substrates of concern. Many bacterial inactivation curves do not seem to obey first-order kinetics but show initial shoulders and tails or upward or downward concavity. Survival of human pathogens in manure is also often nonlinear, but attempts to model it accordingly are scarce (3, 19).
The main objectives of this study were (i) to determine the natural prevalence of the E. coli O157-specific rfbE gene and STEC virulence genes stx1 (coding for Shiga toxin 1), stx2 (coding for Shiga toxin 2), and eaeA (coding for intimin) in manure from 16 organic (ORG) and 9 low-input conventional (LIC) dairy farms; (ii) to study the survival of an introduced E. coli O157:H7 strain in these manures; and (iii) to relate the prevalence of the STEC genes and the survival characteristics to chemical and biological characteristics of the manure.
MATERIALS AND METHODS
Farm sampling.
During August and September 2004, manure samples were collected from 9 LIC dairy farms in the province Friesland of The Netherlands and from 16 ORG dairy farms throughout The Netherlands. All the ORG farms were certified organic and were joined in the Bioveem Project, which aims at the development of solutions for organic dairy farming. The LIC farms were joined in the VEL and VANLA (VV) Project, which is a joint initiative of two environmental corporations with the aim of enhancing the knowledge concerning nutrient and manure management in order to develop solutions for sustainable dairy farming. Farms within the VV Project may not be considered as typical conventional dairy farmers; farmers within this project were chosen because of their willingness to participate and because of the existence of a database of management practices. From a herd of lactating cows at each farm, a number of fecal samples (depending on the herd size) were collected from the stable floor in plastic pots and were pooled according to the sampling protocol used by Bouwknegt et al. (7). None of the farms used antibiotics at the time of sampling. Pooled samples were transported to the laboratory in closed plastic pots and stored at 4°C, and microbiological analyses were started within 48 h after sampling.
Detection methods.
Each pooled sample was enriched by adding a portion of 20 g of manure from each pooled sample to 180 ml modified EC broth containing novobiocin (0.02 g liter−1) (Fluka Chemie GmbH, Switzerland). Enrichments were incubated overnight at 37°C on a rotary shaker (100 rpm). Total DNA was extracted from 300 μl of the enrichment culture with the Bio101 FastDNA1 spin kit for soil according to the manufacturer's specifications (Bio101, Carlsbad, CA), except that bead beating (three times for 90 s each with four 1-mm glass beads) was used instead of the FastPrep1 device.
For the detection of the E. coli O157-specific rfbE gene and STEC virulence genes stx1 (coding for Shiga toxin 1), stx2 (coding for Shiga toxin 2), and eae (coding for intimin), the DNA samples were subjected to two separate Taqman PCR assays. The first assay allowed the simultaneous detection of stx1, stx2, and eaeA. Primers for this assay were derived from Ibekwe et al. (28) and Sharma et al. (50). The second assay allowed the detection of the rfbE gene, which codes for the lipopolysaccharide O side chain of E. coli O157 (17). To reduce false-negative results, an internal amplification control (IAC) was used in the PCR assays mentioned above (32). Detection of this IAC was based on the coamplification of a green fluorescent protein (GFP) gene from the genetically modified Escherichia coli strain 99507GFP. Suboptimal amplification due to inhibitors is identified by a shift of the IAC cycle threshold (CT) value from a CT of 31.5 (optimal) to CT values up to 40. The PCR conditions were identical to the ones described by Klerks et al. (32). For each sample, the increase in fluorescence was measured during amplification with an ABI 7700 sequence detector (Applied Biosystems, Foster City, CA). Thresholds (the minimal fluorescence above which a sample is determined positive) were calculated for both the target and the IAC by taking the average of the fluorescence of the negative controls plus 4× the standard deviation of the fluorescence of these negative controls. For a positive sample, this gives a 99% probability of being positive.
Manure characterization. (i) Coliforms.
Two subsamples of approximately 1.5 g of fresh manure from each pooled sample were collected in tubes containing 4.5 ml 0.1% peptone buffer. These bacterial suspensions were mixed by vortexing, sonicated in an ultrasonic cleaner (Bransonic 12; Branson Cleaning Equipment Co., Shelton, CT) for 30 s, and again vortexed. Samples were taken for further 10-fold serial dilutions. From two appropriate dilutions, 50 μl was plated in triplicate on MacConkey agar (Oxoid CM0007). The total numbers of fecal coliforms (facultative anaerobic gram-negative, rod-shaped, non-spore-forming bacteria present in the intestinal tract of warm-blooded animals that can ferment lactose to gas and acid within 48 h at 35°C) were counted after 24 h at 37°C.
(ii) Lactic acid bacteria.
An estimation of the total number of lactic acid bacteria was made by taking samples as described for coliforms and plating them on MRS agar (Oxoid CM0361) using the double-layer technique as indicated by the manufacturer. Numbers of CFU were counted after incubation for 2 days at 35°C.
(iii) Chemical characterization.
Dried samples (40°C) were ground and analyzed for total carbon by the Dumas method followed by detection by a Fisons type EA 1108 element analyzer (Milan, Italy) and for fiber content (58). Dissolved organic carbon (DOC) was measured by a carbon analyzer in a manure extract of 0.01 M CaCl2. Total dissolved nitrogen content (Nts) was measured in a segmented-flow analysis system (Skalar Analytical BV, The Netherlands). The dissolved organic nitrogen content (DON) was calculated by subtracting the amount of nitrogen present in ammonium and nitrate from the total dissolved nitrogen content. Nitrogen content was determined by the Kjeldahl method (9), and ammonium content was determined in a solution of trichloroacetic acid by an Autoanalyzer II (Technicon Instrument Corporation, Tarreytown, NY). The pH was measured in a watery suspension with an Inlab pH level 1 (WTW GmbH, Weilheim, Germany).
DGGE.
DNA was extracted from 300 mg (fresh weight) manure sample with the Bio101 Systems FastDNA spin kit for soil according to the manufacturer's specifications (Qbiogene, Inc., Carlsbad, CA), except that bead beating (three times for 90 s each) was used instead of the FastPrep1 instrument. The 16S rRNA genes of eubacteria and Enterobacteriaceae were amplified from manure DNA with the eubacterial primer pair U968-GC and L1401 (16) and the Enterobacteriaceae primer pair DG74f and RW01r (23), to which a GC clamp according to Felske et al. (16) was added. The eubacterial PCR was performed using a touchdown scheme (44) for 30 thermal cycles, and the enterobacterial PCR was performed using 3 min of denaturation at 95°C, after which 30 thermal cycles of 1 min at 95°C (denaturation), 1 min at 55°C (annealing), and 2 min at 72°C (extension) were performed, followed by a final extension step at 72°C for 10 min. The PCR products were examined by standard 1.2% (wt/vol) agarose-0.5× Tris-borate-EDTA (TBE) gel electrophoresis with ethidium bromide staining, to confirm product integrity and size. DGGE was performed using the DCode system (Bio-Rad Laboratories, Hercules, CA). We used 6% acrylamide gels (37.5:1 acrylamide-bisacrylamide) with a 45 to 60% denaturing gradient as defined by Muyzer et al. (38) to separate the generated amplicons (100% denaturant is 7 M urea and 40% formamide) and an 8% acrylamide stack without denaturing agents. The gels were poured from the top in the DCode template, prepared with Gelbond PAG film (Amersham Pharmacia Biotech AG, Uppsala, Sweden) to one side, using a gradient maker and a Heidolph Pumpdrive (Heidolph, Schwabach, Germany) set at 4 ml/min. Eubacterial and enterobacterial PCR products derived from DNA of each manure sample were loaded in adjacent slots.
Electrophoresis was performed in 0.5× Tris-acetate-EDTA buffer for 16 h at 100 V at a constant temperature of 60°C. Gels were stained with Bio-Rad silver stain (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's protocol, but using the protocol for gels >1 mm thick instead of 0.5 to 1 mm to compensate for the barrier formed by the Gelbond. After staining, the gels were preserved for at least 1 h in Cairn's preservation solution of 25% (vol/vol) ethanol and 10% (vol/vol) glycerol, covered by a permeable cellophane sheet (Amersham Pharmacia Biotech Ag, Uppsala, Sweden), and dried overnight at 60°C.
The gels were scanned using ScanSoft Omnipage pro. 14 at a resolution of 300 dots per inch. Scanned gels were analyzed with Phoretix 1D (NonLinear Dynamics Ltd., Newcastle upon Tyne, United Kingdom). Bands were selected manually. Data of different DGGE gels were standardized by referring to the DGGE marker. Diversity of bacteria was estimated in two ways: as species richness, S, and as the Shannon-Wiener index of bacterial diversity, H. S was defined as the number of DGGE-detected bands per soil type (56). The Shannon-Wiener diversity index was calculated as
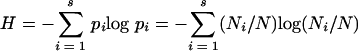 |
where Pi is the importance probability of the bands in a gel lane, S is the individual band, Ni is the band intensity for each band, and N is the sum of intensities of bands in a lane. DGGE analysis was done in duplicate where the replicas were on different gels. S and H were calculated as the mean of the two replicas.
Survival experiment. (i) Strain.
Escherichia coli O157:H7 B6-914 GFP-91 was kindly provided by Pina Fratamico (Eastern Regional Research Center, ARS, USDA, Wyndmoor, PA). This strain does not produce the Shiga-like toxins I and II (Stx1− Stx2−) and contains the pGFP cDNA vector (Clontech Laboratories, Inc., Palo Alto, CA) expressing GFP and ampicillin resistance. The survival characteristics of this strain were indistinguishable from those of the wild-type strain (20). Kudva et al. (33) reported no differences in survival in bovine manure and manure slurry between toxin-positive (Stx1+ Stx2+) and toxin-negative (Stx1− Stx2−) E. coli O157:H7 strains. In addition, survival in manure of E. coli O157:H7 that passed the intestinal tract of cattle was not different from the survival of the same strain directly inoculated into the manure (48). Bacteria were stored at −80°C and checked for viability prior to use by growing on Luria-Bertani medium supplemented with ampicillin (50 μg ml−1).
(ii) Inoculation and sampling of manure.
The pooled samples were mixed per farm, and the survival experiment was executed with two replicas per farm. Preparation of inoculum, inoculation of manure, and sampling over time were done as described in detail by Franz et al. (19). In brief, cells suspended in buffered peptone water (BPW) were added to 200 g manure with a final density of 1 × 107 CFU per g manure dry weight (gdw−1), taking the individual dry weights of the different manures into account. The manure and the inoculum were thoroughly mixed by kneading them in a plastic bag from the outside by hand. Subsequently, the inoculated manure was transferred to plastic pots (1 liter) which were closed (but with the ability of gas exchange) and incubated at 10°C in darkness. Moisture content remained constant (85% ± 3%) during the experiment. The inoculated pots were sampled over time using serial dilution series with BPW, sonication in a ultrasonic bath for 30 s (Branson 5200; 120 W; output power, 47 kHz), and plating on sorbitol MacConkey agar (SMAC; Oxoid CM813) supplemented with ampicillin (50 μg ml−1). Numbers of fluorescent CFU were determined using a dark-blue lamp (Philips PL-S 9W/08 Blacklight Blue; peak at 365 nm UV-A) after incubation for 18 to 20 h at 37°C.
Statistical analysis. (i) Prevalence.
Farm-level prevalence of E. coli O157 and STEC virulence genes was calculated as the ratio between the number of farms showing at least one positive pooled sample and the total number of farms. Chi-square analysis was used to test for differences in proportions of positive farms between ORG and LIC manures (PROC FREQ, SAS System for Windows version 8.02; SAS Institute Inc., Cary, NC). Based on the number of positive farms, the true prevalence, P, with associated uncertainty was modeled with a beta distribution: P = (s + 1)/(n − s + 1), where s is number of positives and n is the total number of farms or samples. True prevalence and 90% confidence interval (CI) were determined by running 1,000 iterations with Latin Hypercube sampling (59) with @Risk software (version 4.5.4; Palisade Corporation, Newfield, NY).
(ii) Fitting of survival data.
Plate counts of zero were replaced by 10 CFU gdw−1, which is the calculated detection limit of the dilution plating procedure. The microbial survival data of both replicas of each farm were averaged. The majority of the survival curves clearly showed shoulders, tails, and a biphasic pattern. Clearly, first-order kinetics were not appropriate. Therefore, the log-transformed survival data were fitted to a biphasic model as proposed by Geeraerd et al. (21) with the Geeraerd and Van Impe inactivation model-fitting tool (GInaFiT):
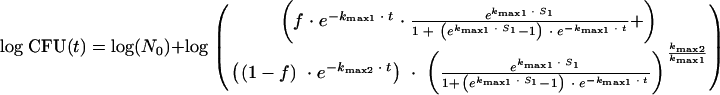 |
where t is time in days, N0 is the number of cells present at t0, f is the fraction of the initial population in a major less-resistant subpopulation, (1 − f) is the fraction of the initial population in a minor more-resistant subpopulation (at t0), kmax1 and kmax2 (day−1) are the specific inactivation rates of the two subpopulations, and sl is the initial shoulder length (time). In addition to the evaluation of the separate model parameters, the time necessary to reach the detection limit (ttd) was calculated with the biphasic model and subjected to the same statistical analysis described for the individual parameters. Model performance was evaluated by assessing the root mean square error (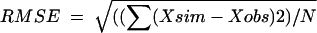 and the pseudo-R2 regression coefficient (R2 = 1 − SSresiduals/SStotal[corrected]). The standard error of estimated parameters and residuals were assessed visually for each fit and were checked for large values.
and the pseudo-R2 regression coefficient (R2 = 1 − SSresiduals/SStotal[corrected]). The standard error of estimated parameters and residuals were assessed visually for each fit and were checked for large values.
(iii) Data analysis of survival.
ORG versus LIC manure and PCR-positive versus PCR-negative manure were compared with respect to the estimated parameter values of the survival models and the chemical and biological variables by nonparametric Mann-Whitney tests since very few of the parameters and variables were normally distributed (SPSS version 12; SPSS, Inc., Chicago, IL). Nonparametric correlation tests (Spearman) were conducted to test for linear relationships between model parameters and measured variables (SPSS). Regression analysis was conducted in order to describe possible nonlinear relationships (SPSS). Multiple regression models that describe the expected values of the model parameters as functions of biotic and abiotic manure characteristics were constructed (stepwise method with a significance level of P = 0.15) (SAS System for Windows version 8.02; SAS Institute, Inc., Cary, NC). The following variables were considered: pH, dry matter content in percentage of total weight (DM), nitrate content (NO3), ammonium content (NH4), total nitrogen content (Ntotal), total DON, total dissolved organic carbon content (DOC), acid detergent fiber content (ADF), neutral detergent fiber content (NDF), number of lactobacilli, number of coliforms, species richness of Enterobacteriaceae as determined by DGGE (Sent), species diversity of Enterobacteriaceae (Hent), species richness of eubacteria (Seub), and species diversity of eubacteria (Heub).
RESULTS
Prevalence of E. coli O157 and STEC virulence genes.
The overall prevalence of the E. coli O157-specific rfbE gene on farms was 52% (90% CI = 0.36 to 0.67) (Table 1). ORG dairy farms (n = 16) had a prevalence of 61% (90% CI = 0.42 to 0.79), and LIC farms (n = 9) had a prevalence of 36% (90% CI, 0.15 to 0.61). Surprisingly, there was no single organic herd in which none of the four genes (rfbE, stx1, stx2, or eaeA) was detected, and only two LIC herds tested negative for all four genes. The presence of the various virulence genes and the combinations in which they occurred varied widely among the different farms, as well as among rfbE-positive herds. Variation was higher within the ORG herds. The majority of the toxin-positive farms (13/16 ORG and 7/9 LIC) tested positive for both toxin genes (8/13 ORG and 7/7 LIC); only 2 (ORG) farms tested positive for only stx1 and 3 (ORG) farms tested positive for only stx2. Nine herds were positive for all three virulence genes eaeA, stx1, and stx2 (5/16 ORG, 4/9 LIC), of which 6 herds (4/16 ORG, 2/9 LIC) were also positive for rfbE. With the exception of one farm, farms positive for eaeA (6 ORG, 4 LIC) were positive for both toxin genes. Seven farms (six ORG, one LIC) were positive for rfbE but not for eaeA. No statistical differences between ORG and LIC herds were observed with respect to prevalence of virulence genes and the number of virulence genes present per farm.
TABLE 1.
Prevalence of the E. coli O157-specific rfbE gene and STEC virulence genes stx1, stx2, and eaeA among 16 ORG and 9 LIC dairy herdsa
| Herd and parameter | Prevalence of gene (product)
|
|||
|---|---|---|---|---|
| rfbE | stx1 (Shiga toxin 1) | stx2 (Shiga toxin 2) | eaeA (intimin) | |
| ORG | + | |||
| + | + | |||
| + | + | + | + | |
| + | ||||
| + | + | + | + | |
| + | + | + | ||
| + | ||||
| + | + | + | ||
| + | ||||
| + | + | + | + | |
| + | + | |||
| + | + | + | + | |
| + | + | |||
| + | ||||
| + | + | |||
| + | + | |||
| Prevalence | 0.61 | 0.61 | 0.67 | 0.39 |
| 90% CI | 0.42-0.79 | 0.42-0.79 | 0.48-0.83 | 0.21-0.58 |
| Modusb | 3 | 1 | 3 | 1 |
| LIC | + | + | + | |
| + | + | |||
| + | + | + | + | |
| + | + | + | ||
| + | + | |||
| + | + | + | + | |
| + | + | + | ||
| Prevalence | 0.36 | 0.73 | 0.73 | 0.45 |
| 90% CI | 0.15-0.61 | 0.49-0.91 | 0.49-0.91 | 0.22-0.70 |
| Modusb | 3 | 1 | 2 | 2 |
| Total prevalence | 0.52 | 0.67 | 0.70 | 0.41 |
| 90% CI | 0.36-0.67 | 0.51-0.81 | 0.55-0.84 | 0.26-0.56 |
| Modusb | 3 | 1 | 2 | 1 |
+ indicates presence, as determined by Taqman PCR of the rfbE gene (E. coli O157) and STEC virulence genes stx1, stx2, and eaeA. Prevalence and associated 90% CIs are based on the binomial distribution, modeled in @Risk software.
Modus of the number of positive pooled samples (maximum is 3).
Chemical and biological characterization of the manure.
Several chemical and biological characteristics of the manure were found to be different between ORG manure and LIC manure (Table 2). Nitrate, total nitrogen, total soluble organic nitrogen, total soluble organic carbon, and number of lactobacilli were significantly higher in the low-input manure (P = 0.003, P = 0.009, P = 0.005, P = 0.011, P = 0.002, and P = 0.046, respectively). Fiber content (ADF and NDF) was higher in ORG manure (both P = 0.046).
TABLE 2.
Average values and standard deviations of chemical and biological characteristics of ORG and LIC manuresa
| Manure type | pH | Chemical characteristic (mg/kg)b
|
No. in functional group (log CFU)
|
Species richness/diversityc
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DM | NO3* | NH4 | Ntotal* | DON* | DOC* | ADF* | NDF* | Lactobacilli* | Coliforms | Sent | Hent | Seub | Heub | ||
| ORG (n = 16) | |||||||||||||||
| Mean | 6.7 | 11.4 | 7.6 | 238 | 897 | 661 | 7637 | 31.8 | 42.1 | 5.9 | 7.3 | 14 | 0.98 | 16 | 0.85 |
| SD | 0.5 | 1.8 | 1.1 | 104 | 194 | 177 | 2127 | 3.2 | 4.6 | 0.5 | 0.9 | 5 | 0.17 | 8 | 0.25 |
| LIC (n = 9) | |||||||||||||||
| Mean | 6.8 | 12.4 | 9.1 | 193 | 1145 | 918 | 9377 | 27.0 | 37.8 | 6.6 | 7.7 | 15 | 1.05 | 19 | 0.89 |
| SD | 0.4 | 1.5 | 0.3 | 94 | 218 | 227 | 1402 | 2.5 | 4.7 | 0.4 | 0.6 | 6 | 0.14 | 11 | 0.26 |
*, significant difference between ORG and LIC manure (P < 0.05).
DM, dry matter; NO3, nitrate; NO4, ammonium; Ntotal, total nitrogen; DON, dissolved organic nitrogen; DOC, dissolved organic carbon; ADF, acid detergent fiber; NDF, neutral detergent fiber.
Sent, species richness of Enterobacteriaceae; Hent, species diversity of Enterobacteriaceae; Seub, species richness of eubacteria; Heub, species diversity of eubacteria.
Risk factors for the presence of E. coli O157-specific rfbE and STEC virulence genes.
The species richness of the Enterobacteriaceae (Sent) was significantly lower in those manures that were positive for E. coli O157 (Sent = 12 ± 4) than in the manures that tested negative for E. coli O157 (Sent = 18 ± 7) (P = 0.009). Similarly, although not significant, the presence of eae, stx1, and stx2 was associated with a lower species richness of eubacteria and Enterobacteriaceae as well as lower Shannon indexes. The species richness (expressed as the number of DGGE bands) was positively correlated with the microbial diversity (expressed as the Shannon index) for both eubacteria (r = 0.91, P = 0.000) and the Enterobacteriaceae (r = 0.79, P = 0.000). With respect to the chemical composition of the manure, total dissolved organic carbon and total nitrogen content were higher in manure positive for stx2 (P = 0.048 and P = 0.032).
Survival of GFP-expressing E. coli O157:H7. (i) Model performance.
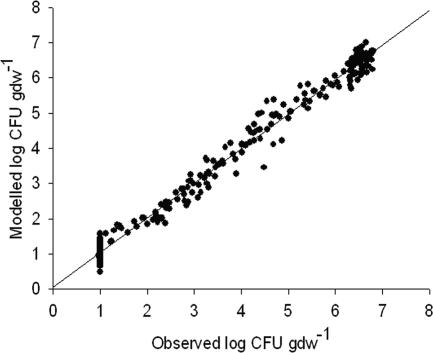
All decline curves showed nonlinearity, with the presence of shoulders and/or tailing. Clearly, fitting a model based on first-order decline kinetics was not appropriate. The biphasic model was found to be flexible enough to fit all survival curves: fitting to the biphasic model resulted in good fits with an average RMSE of 0.34 ± 0.11 and average R2 of 0.97 ± 0.01. In addition, observed and fitted values were highly correlated (r = 0.99, P < 0.00) (Fig. 1). The initial decline rate (kmax1) was positively correlated to the second-phase decline rate (kmax2) (r = 0.78, P = 0.000). The time necessary to reach the detection limit (ttd), was strongly correlated to sl (r = 0.67, P = 0.001), kmax2 (r = 0.79, P = 0.000), and, to a lesser extent, kmax1 (r = 0.49, P = 0.017).
FIG. 1.
Correlation between E. coli O157:H7 densities in manure (log CFU gdw−1), resulting from fitting survival data to the biphasic Geeraerd model, and observed densities (n = 253, r = 0.99). The solid line represents the isocline where fitted values are equal to observed values.
(ii) Survival in ORG versus LIC manure.
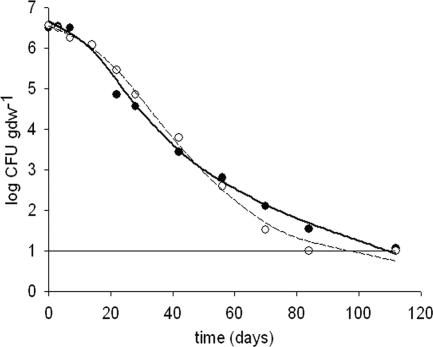
The decline of the introduced E. coli O157:H7 population was preceded by an initial shoulder of approximately 11 days which did not differ between ORG and LIC manures (Table 3). The initial decline rate was significantly higher for ORG manure (P = 0.022). The fitted survival curves of E. coli O157:H7 in ORG and LIC manures crossed after approximately 55 days (Fig. 2), caused by an earlier onset of tailing on ORG manure. This coincides with a lower kmax2 and a significant larger fraction of cells (1 − f) showing a reduced decline rate (kmax2) relative to the fraction showing the initial decline rate, kmax1 (Table 3). The overall survival (ttd) was shorter for LIC manure (96 ± 11 days) than for ORG manure (109 ± 14 days) (P = 0.026) (Table 3 and Fig. 2).
TABLE 3.
Mean values and standard deviations of the estimated parameters of the biphasic Geeraerd model and the Augustin model for survival of GFP-expressing E. coli O157:H7 in ORG and LIC manuresa
| Value for manure | kmax1* (day−1) | kmax2 (day−1) | f* (N1/N) | sl (days) | N0 (log CFU) | ttd* (days) | R2 | RMSE (log CFU) |
|---|---|---|---|---|---|---|---|---|
| ORG (n = 16) | ||||||||
| Mean | 0.32 | 0.06 | 0.999940 | 11.3 | 6.7 | 109 | 0.98 | 0.29 |
| SD | 0.02 | 0.02 | 1.1 × 10−4 | 9.1 | 0.2 | 11 | 0.01 | 0.10 |
| LIC (n = 9) | ||||||||
| Mean | 0.23 | 0.03 | 0.999994 | 10.2 | 6.6 | 94 | 0.96 | 0.41 |
| SD | 0.07 | 0.01 | 3.41 × 10−6 | 8.3 | 0.2 | 14 | 0.02 | 0.08 |
*, significantly different between ORG and LIC (P < 0.05).
FIG. 2.
Average course of decline in ORG (solid line, solid circles) and LIC manure (dashed line, open circles) as fitted by the biphasic Geeraerd model. Standard deviations are omitted for clarity (average of 0.61 for ORG and 0.52 for LIC). The horizontal line represents the detection limit.
(iii) Relationships between survival parameters and chemical manure characteristics.
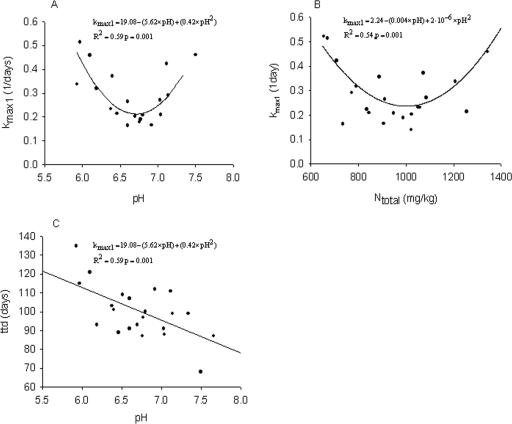
The initial decline rate (kmax1) was related to the pH of the manure in the form of an optimum curve and is best described with a quadratic relationship (Fig. 3). By taking the derivative of this function, a pH optimum of 6.71 was determined. By conducting linear regression separately on the data points left and right of the optimum (with a fixed intercept), it was observed that kmax1 decreased by −0.34 · pH, with increasing pH at values below the optimum, and increased by 0.40 · pH with increasing pH at values above the optimum. Thus, the effect of pH on the decline rate was stronger at pH values above the optimum of 6.7. Both sl and ttd were negatively correlated with the pH of the manure (r = −0.63, P = 0.001, and r = −0.60, P = 0.002, respectively) (Table 4 and Fig. 3). A similar optimum curve in the form of a quadratic relationship was observed between kmax1 and the total nitrogen content (R2 = 0.54, P = 0.001, optimum at 1,000 mg N/kg) (Fig. 3) and kmax2 and the total nitrogen content (R2 = 0.42 P = 0.006, optimum at 952 mg N/kg). A negative correlation was observed between kmax1 and the nitrate content (r = 0.48, P = 0.029) (Table 4). A linear trend (P < 0.1) was observed between sl and the ammonium (NH4) content.
FIG. 3.
Observed points (solid circles) and relationships (solid lines) between chemical manure characteristics and parameters of the biphasic model. (A) Initial decline rate, kmax1, and pH. (B) kmax1 and total nitrogen content, Ntotal. (C) ttd and pH.
TABLE 4.
Correlations between model parameters and chemical/biological manure characteristics
| Manure characteristica | Model parameterb
|
|||
|---|---|---|---|---|
| kmax1 | kmax2 | sl | ttd | |
| pH | −0.63** | −0.59** | ||
| NO3 | −0.48* | |||
| Coliforms | −0.52* | −0.42* | ||
| Sent | −0.58** | −0.42** | ||
NO3, nitrate; Coliforms, total number of coliforms; Sent, species richness of Enterobacteriaceae as determined by DGGE.
kmax1, decline rate of major sensitive subpopulation; kmax2, decline rate of minor more resistant subpopulation; *, correlation is significant at the 0.05 level (two-tailed test); **, correlation is significant at the 0.01 level (two-tailed test).
(iv) Relationships between survival parameters and microbial community in manure.
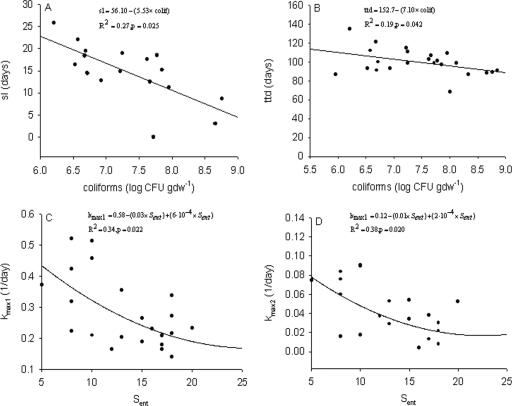
The initial decline rate, kmax1, was significantly lower in those manures which were positive for stx1 (P = 0.023), and the fraction of introduced cells which were characterized by a relatively slower decline rate, kmax2, was significantly lower in those manures positive for stx2 (P = 0.04). Both sl and ttd were negatively correlated with the number of coliforms (Table 4 and Fig. 4). Both of the decline rates kmax1 and kmax2 were negatively correlated with the species richness of the Enterobacteriaceae (Sent), which was defined as the number of detected DGGE bands (Table 5). However, quadratic fits explained more variation (Fig. 4).
FIG. 4.
Observed points (solid circles) and statistical relationships (solid lines) between biological manure characteristics and parameters of the biphasic model. (A)Length of the initial shoulder, sl, and the number of coliforms. (B) ttd and the number of coliforms. (C) Initial decline rate, kmax1, and species richness of Enterobacteriaceae Sent. (D) Second-phase decline rate, kmax2, and Sent.
TABLE 5.
Multiple regression equations with the parameters of the biphasic survival model as dependent variables and the biological and chemical characteristics of the manure as independent variables
| Dependent variable | Regression equationa | Model
|
|
|---|---|---|---|
| Significance | R2 | ||
| kmax1 | 0.53 (intercept, P < 0.001) − 0.02 × Sent (R2part = 0.33, P = 0.016) + 0.006 × NDF (R2part = 0.13, P = 0.073) − 0.11 × stx1 (R2part = 0.12, P = 0.104) − 0.003 × pH2 (R2part = 0.10, P = 0.073) | P = 0.006 | 0.69 |
| kmax2 | 0.23 (intercept, P = <0.001) − 0.02 × coliforms (R2part = 0.35, P = 0.010) − 0.003 × Sent (R2part = 0.20, P = 0.022) − 9.55 × 10−4 × ADF (R2part = 0.07, P = 0.139) | P = 0.003 | 0.61 |
| sl | 71.58 (intercept, P < 0.001) − 7.86 × coliforms (R2part = 0.70, P < 0.001) | P < 0.001 | 0.70 |
| ttd | 190.48-11.70 × coliforms (R2part = 0.51, P = 0.001) − 0.14 × pH (R2part = 0.09, P = 0.063) | P = 0.007 | 0.60 |
Sent, species richness of Enterobacteriaceae as determined by DGGE; NDF, neutral detergent fiber content (cellulose, hemicellulose, and lignin content as percentage of organic matter per unit dry weight); stx1, natural presence of the gene coding for Shiga toxin 1 as detected with Taqman PCR; coliforms, total number of coliform bacteria (log CFU/g dry weight); ADF, acid detergent fiber content (cellulose and lignin content as percentage of organic matter per unit dry weight).
The goodness-of-fit statistic R2 and RMSE of the biphasic model, which can be seen as a measure of the predictability of the model, were negatively correlated with the number of coliforms (r = −0.56, P = 0.005, and r = 0.52, P = 0.01) present in the manure. This means a better predictability of the decline of E. coli O157:H7 in manure with the biphasic Geeraerd model when fewer coliforms and copiotrophic bacteria are present. Indeed, as mentioned earlier, the number of coliforms was lower for organic manure and the predictability of decline was better for organic manure.
(v) Multiple regression-explaining parameters.
Multiple regression equations were constructed to relate the parameters of the survival models to chemical and biological characteristics of the manure. This resulted in significant models for kmax1, kmax2, sl, and ttd with a relatively high predictive value: i.e., high R2 (Table 5). The initial decline rate, kmax1, was best explained by the species richness of the Enterobacteriaceae (negative relationship). The remaining variation could be explained by the fiber content (positive relationship), the pH (quadratic relationship), and the presence of the gene coding for Shiga toxin 1 (negative relationship). Decline rate kmax2 could be explained by the total number of coliforms (negative relationship), the species richness of the Enterobacteriaceae (negative relationship), and the fiber content (positive relationship). Interestingly, kmax1 increased with increasing fiber content, while kmax2 decreased with increasing fiber content. The length of the initial shoulder of the decline curve was determined solely by the total number of coliforms present in the manure (negative relationship). The overall rate of decline, measured as the time to the detection limit, ttd, was also best explained by the number of coliforms (negative relationship) in the manure and the pH (negative relationship) (Table 5). In turn, the levels of coliforms were predominantly determined by the pH of the manure, with increasing numbers of coliforms with increasing pH (R2 = 0.36, P = 0.011). With respect to the model, variation in ttd was best explained by the variation in sl (partial R2 = 0.51).
(vi) Extremes and farming styles.
The four manures that best supported E. coli O157:H7 (i.e., four highest ttd values [all ORG]) were derived from farms with exclusively Frisian Holstein cows, while two of the four farms from which the manure supported the worst survival of E. coli O157:H7 (two ORG and two LIC) harbored another breed (both ORG) next to Frisian Holsteins. In addition, the four manures that best supported survival of E. coli O157:H7 had a significantly higher number of naturally present STEC virulence genes (P = 0.03), higher levels of NO3, higher levels of dissolved organic carbon (DOC) (P = 0.01), and lower levels of coliforms P = 0.001).
DISCUSSION
Prevalence of the E. coli O157-specific rfbE gene.
The survey conducted in this study showed that the rfbE gene, which is characteristic of E. coli O157, was present at 52% of 25 Dutch dairy farms from which samples were obtained in August and September 2004. This is relatively high compared to the prevalence at Dutch dairy farms in a previous study, namely 7.2% out of 678 farms (46). The same sampling strategy was used, but in the present study the E. coli O157-specific rfbE gene was detected by real-time PCR instead of the detection of E. coli O157 cells by immunomagnetic separation, which could lead to different results. In addition, the earlier reported prevalence of 7.2% was an average over 4 years including all seasons, while the present study included only the summer of 2004. The first study determining the prevalence of E. coli O157 on Dutch dairy farms was, just as the present study, a point estimate in time (fall 1996) and reported a prevalence of 70% (n = 10) (24). It is generally known that E. coli O157 prevalence shows seasonality, with the highest prevalences in the warmer months (57), and Schouten et al. showed peaks in prevalence occurring regularly: up to 36% in September 2000 (46). Monthly prevalence over 1999 and 2000 was positively correlated with mean and maximum temperatures of the corresponding months (r = 0.60, P = 0.002, and r = 0.53, P = 0.008) (see reference 46 and Koninklijk Nederlands Meteorologisch Instituut [http://www.knmi.nl]). August and September 2004 were characterized by high average temperatures compared to the yearlong average (18.8 versus 17.2°C and 15.2 versus 14.2°C) and by high maximum temperatures (32.5 and 28°C), which make the 52% prevalence on farms found in the present study plausible.
In other countries, reported E. coli O157 prevalences among dairy farms ranged from 7.1 to 80% (average of 39 ± 27.2; 20 studies), with generally a higher prevalence in the United States (50.4 ± 24.9; 10 studies) compared to Europe (31.8 ± 27.8, 10 studies). It must be stressed that direct comparison of prevalences across studies is confounded by the use of different sampling and detection methods, environmental conditions, and geographical locations.
Although prevalence of rfbE was higher among certified ORG farms (61%), this was not statistically different from the LIC farms (LIC) (36%). This trend is identical to a recently published report, where E. coli O157 was found in 4 out of 8 ORG dairy farms (50%) and in 3 out of 18 conventional dairy farms (16.7%) in the United States (11). In Switzerland, E. coli O157:H7 showed a herd prevalence of 25% at ORG farms and 17% at integrated (conventional) farms (both n = 60), but this was also not significantly different, and no difference in the risk to carry this pathogen was found between both farm types (34).
Risk factors for the presence of rfbE.
The species richness of Enterobacteriaceae was significantly lower in those manures which were positive for rfbE. Ecological theory argues that, at small spatial scales, diverse communities are more competitive and therefore more resistant to invasion than less diverse communities (15, 31). For example, the invasibility of wheat rhizosphere communities by Pseudomonas aeruginosa was inversely related to the level of diversity of microbes (37). It has been hypothesized that species richness and/or diversity will increase with increased resource heterogeneity (36) and that a community becomes more susceptible to invasion whenever there is an increase in the amount of unused (available) resources because the invader will encounter less intense competition from resident species (12). With respect to cattle and STEC, diet is a major factor in determining the abundance and nature of nutrient resources in the digestive tract and might therefore be an important factor in controlling the susceptibility to STEC invasion. A higher level of unused available nutrients and/or lower resource heterogeneity may result in a lower diversity of microbes and an increased susceptibility to pathogen invasion. Fewer bovine commensal E. coli serotypes were isolated from the feces of cattle on feedlot/grain diets compared with cattle on pasture/roughage-based diets (4), and some studies concluded that the shedding of E. coli O157:H7 is reduced when cattle are fed a low-energy hay-based diet compared to a high-energy grain-based diet (22, 45). We observed higher levels of total dissolved organic carbon and total nitrogen in manure positive for stx2, which might be an indication of a possible relationship between the nutrient status of the gut/manure and the presence of verotoxin-producing E. coli virulence factors.
Prevalence and risk factors for the presence of STEC virulence genes.
Overall, 80% of the farms tested stx positive. Very high prevalence of Shiga toxins was also demonstrated for Norway (100%; n = 50) and Ohio in the United States (70%; n = 50) (35). Quite a number of ORG farms (7/9) were positive for rfbE but negative for intimin. This could indicate the presence of less virulent O157 serotypes lacking intimin like those described for E. coli O157:H7, O157:H8, and O157:NM (17, 26). From epidemiological studies, it is known that clinical isolations of STEC from humans showed a significant association between the presence of eaeA and stx2 (5). Our results indicate that although there is a trend of increased prevalence of rfbE with ORG farms, more LIC farms tested positive for all verotoxin-producing E. coli virulence genes (both eaeA and stx2). It must be stressed that the detected virulence genes on a single farm do not necessarily originate from one cell or serotype since we did not isolate serotypes. However, given the fact that most STEC virulence markers are on mobile genetic elements, we may assume that the chance of the existence or future emergence of virulent serotypes by horizontal gene transfer is higher at the LIC farms.
Modeling survival of GFP-expressing E. coli O157:H7 in manure.
The survival curves obtained typically were nonlinear and showing considerable variation. The use of first-order kinetics and log-linear modeling of survival was clearly not appropriate. Survival of human pathogens in manure is also often nonlinear, but attempts to model it accordingly are scarce. Recently, the decline of E. coli O157:H7 in manure was described with a logistic model (19) and an exponential decay model (3). Unfortunately, these relatively simple models were not able to cover the range of survival curves obtained with the present study. The biphasic Geeraerd model was flexible to such an extent that all survival curves could be fitted to this model with good accuracy (significant and high R2). Biphasic inactivation curves, in which the initial rate of decline is followed by a slower second phase (tailing), have been observed regularly within food microbiology (1, 27, 49). Also in environmental substrates, pathogen die-off occurs often in two stages (41). For example, a two-stage die-off model was used to describe the decline of E. coli O157 in soil (40).
Manure characteristics affecting the survival of E. coli O157:H7.
The survival curves showed initial shoulders in 17 out of the 25 cases, with an average length of 11 days. No differences were observed in sl between ORG and LIC manures. The variation in sl and overall survival time, ttd, were best explained by the variation in the level of coliforms in the manure, with shorter initial shoulders and shorter overall survival with higher numbers of coliforms. Native coliforms may occupy the same ecological niche as E. coli O157:H7 (niche exclusion principle), and/or an increase in the level of coliforms may increase the level of competition for available nutrients. In turn, the number of coliforms was predominantly determined by the pH of the manure, with increasing numbers with increasing pH.
The pH of the manure also entered the regression model for kmax1. Decline rate kmax1 showed a quadratic relationship with pH, which makes sense, since organisms usually show curvilinear relationships with environmental variables. We observed an optimum pH of 6.7, which is close to the optimal pH of 6.9 for growth of E. coli O157:H7 in broth (39). Higher and lower pH levels, within a range of 5.9 to 7.2, resulted in increased decline rates, but the effect of higher pH was stronger than that of lower pH. The decline rate of E. coli O157:H7 increased linearly with the pH when survival was studied with manure from cows fed three different diets (19). However, the pH range in which a linear increase in decline rate was most evident ranged from 6.8 up to 7.7, which is the same range in which we observed the positive relationship between pH and decline rate in the present study. It is known that E. coli O157:H7 possesses several systems in order to survive exposure to low pH and can be considered quite acid tolerant (6). It therefore might have a selective advantage at low pH (13). In addition, organic nitrogen might be converted to ammonia at higher pH. It has been demonstrated that ammonia can be toxic to E. coli O157:H7 and can cause a significant reduction in numbers (25). A significant reduction of E. coli O157:H7 was also achieved by a combination of high concentrations of carbonate and ammonia, which require a high pH (42).
Explaining the nonlinear and biphasic nature of survival curve.
The vitalistic concept proposes that in a genetically homogeneous population, phenotypic variation in physiological responses exists such that resistance to a lethal agent is not uniform (27). Subsequently, the nonlinearity of the survival curve reflects the inactivation of a population of cells which is heterogeneous with respect to decline kinetics (43, 55). The biphasic model used in the present study assumes the existence of two subpopulations, each with its own inactivation characteristics. Theoretically, tailing occurs because the relative size of the more resistant subpopulation and the associated weight of kmax2 then increase, which results in a net slower decline and tailing. The earlier onset of tailing in ORG manure can therefore be explained from the significantly higher fraction of E. coli O157:H7 in the more resistant subpopulation.
Although the overall decline curve could be explained as the cumulative distribution of an underlying distribution of decline kinetics, the differences in decline kinetics between the different manures are difficult to explain from innate differences among E. coli O157:H7 cells within the starting culture. We propose that the nonlinear survival pattern is the result of a changing competition pressure over time due to a changing nutrient availability. Likewise, differences in decline kinetics between manures can be explained from differences in the substrate composition and differences in the changes over time. During the first days after inoculation, E. coli O157:H7 uses the easily available nutrients and maintains equilibrium between growth and death within the population. When the easily available nutrients become limited, this equilibrium is disturbed and the overall death rate of the population will exceed the overall growth rate, resulting in a decline phase. Additionally, the manure in the pots will become more anaerobic over time and E. coli O157:H7 has to switch from an aerobic to anaerobic metabolism. The levels of easily available nutrients are usually lower in manure with higher fiber content. Possibly, the higher fiber content and lower levels of total soluble organic carbon and nitrogen of ORG manure resulted in an increased initial nutrient stress for E. coli O157:H7 which resulted in a faster initial decline. The more abundant easily available nutrients in LIC manure became limited after 40 to 50 days, while the higher fiber content of ORG manure resulted in continued release of nutrients as a result of the decomposition of complex cellulose and lignin structures by cellulolytic organisms. As a result, the average survival curve for ORG manure leveled off and crossed the average curve for LIC manure after approximately 50 days. Indeed, according to the multiple regression analysis the initial decline rate was increased with higher fiber content while the decline rate of the second phase was decreased with higher fiber content. Earlier results showed a significantly faster overall decline of E. coli O157:H7 in manure from dairy cattle placed on diets differing in fiber content (19). However, the fiber content of the manures resulting from the high-fiber diet (pure straw) was significantly higher (ADF = 50% and NDF = 66%) than the fiber contents of ORG and LIC manures in the present study (Table 2). Probably, other limiting factors overrule the advantage of a slow but continuous nutrient gift from the fiber polymers at very high fiber contents. One such factor might be the pH, which was substantially higher with the high-fiber manure (pH 7.4) than with the ORG manure in the present study (pH 6.7; Table 2).
Additional factors explaining variation in survival.
In the present study, no effect of lactobacilli on the natural presence E. coli O157 rfbE gene and the survival of GFP-expressing E. coli O157:H7 was observed. Certain strains of Lactobacillus exert growth inhibition of E. coli O157 and/or bactericidal activities on E. coli O157:H7 (8). Supplementation of cattle with Lactobacillus acidophilus NP51 resulted in significant lower prevalence of E. coli O157 among experimental cattle herds (61). Probably, the inhibitory effect of lactobacilli is limited to certain strains and even varies with different E. coli O157 strains. This might be related to our finding that among the four manure types which were the least supportive of E. coli O157, two were derived from farms where in addition to Frisian Holstein cows another breed was present. It could be that some specific antagonistic bacteria were present in the manure from these more rare cattle breeds. Moreover, the lack of a relationship between survival and the number of lactobacilli might also have to do with the fact that the conditions in the manure were not anaerobic, while most lactobacilli favor anaerobic conditions when sugars are fermented to lactic acid.
Conclusions.
Although the prevalence of the E. coli O157-specific rfbE gene was higher at ORG farms than at LIC farms, relatively more LIC farms tested positive simultaneously for STEC virulence genes eaeA, stx1, and stx2. The species diversity of Enterobacteriaceae was higher in manure positive for rfbE, indicating the possible important role of microbial diversity in STEC epidemiology. The biphasic decline model was flexible enough to describe all 25 obtained survival curves with good fits. Initial shoulder length and overall survival were determined by and negatively related to the numbers of coliforms present in the manure and the pH. It is proposed that the nonlinear survival pattern is the result of a changing competition pressure over time due to changing nutrient availability, instead of the presence of subpopulations differing in the level of resistance.
Further research should focus on isolation and serotyping of STEC strains on ORG and conventional farms in order to investigate whether different populations of STEC are present on ORG farms compared to conventional farms. In addition, a detailed understanding of the interaction between pathogen, the nutrient status of the substrate and the native microbial community is needed in order to develop control strategies that reduce pathogen survival and thus spread to the environment and the food chain.
Acknowledgments
This research was supported by the Technology Foundation STW, Applied Science Division of NWO and the Technology Program of the Ministry of Economic Affairs, and by the Dutch National Product Board for Horticulture.
We thank Martin Northolt of the Louis Bolk Institute for obtaining samples from the organic farms, P. Fratamico for providing the gfp-modified E. coli O157:H7 strain, H. D. Halm for the chemical analyses, and A. V. Semenov for laboratory assistance and discussions.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Alvarez, I., R. Virto, J. Raso, and S. Condon. 2003. Comparing predicting models for the Escherichia coli inactivation by pulsed electric fields. Innovative Food Sci. Emerg. Technol. 4:195-202. [Google Scholar]
- 2.Bach, S. J., T. A. McAllister, D. M. Veira, V. P. J. Gannon, and R. A. Holley. 2002. Transmission and control of Escherichia coli O157:H7—a review. Can. J. Anim. Sci. 82:475-490. [Google Scholar]
- 3.Bach, S. J., K. Stanford, and T. A. McAllister. 2005. Survival of Escherichia coli O157:H7 in feces from corn- and barley-fed steers. FEMS Microbiol. Lett. 252:25-33. [DOI] [PubMed] [Google Scholar]
- 4.Bettelheim, K. A., A. Kuzevski, R. A. Gilbert, D. O. Krause, and C. S. McSweeney. 2005. The diversity of Escherichia coli serotypes and biotypes in cattle faeces. J. Appl. Microbiol. 98:699-709. [DOI] [PubMed] [Google Scholar]
- 5.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth, I. R. 2002. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 78:19-30. [DOI] [PubMed] [Google Scholar]
- 7.Bouwknegt, M., W. D. C. Dam-Deisz, J. M. Schouten, W. J. B. Wannet, W. van Pelt, G. Visser, and A. W. van de Giessen. 2003. Surveillance of zoonotic bacteria in farm animals in The Netherlands. Results from January 1998 until December 2000. Rijksinstitut voor Volksgezondheid en Milieu, Bilthoven, The Netherlands.
- 8.Brashears, M. M., D. Jaroni, and J. Trimble. 2003. Isolation, selection, and characterization of lactic acid bacteria for a competitive exclusion product to reduce shedding of Escherichia coli O157:H7 in cattle. J. Food Prot. 66:355-363. [DOI] [PubMed] [Google Scholar]
- 9.Bremnar, S. M. 1982. Nitrogen total, p. 595-624. In A. L. Page, R. H. Miller, and D. R. Keeney (ed.), Methods of soil analysis, part 2. Chemical and microbiological properties, 2nd ed. American Society of Agronomy, Soil Science Society of America, Madison, WI.
- 10.Caprioli, A., S. Morabito, H. Brugère, and E. Oswald. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289-311. [DOI] [PubMed] [Google Scholar]
- 11.Cho, S., J. B. Bender, F. Diez-Gonzalez, C. P. Fossler, C. W. Hedberg, J. B. Kaneene, P. L. Ruegg, L. D. Warnick, and S. J. Wells. 2006. Prevalence and characterization of Escherichia coli O157 isolates from Minnesota dairy farms and county fairs. J. Food Prot. 69:252-259. [DOI] [PubMed] [Google Scholar]
- 12.Davis, M. A., J. P. Grime, and K. Thompson. 2000. Fluctuating resources in plant communities: a general theory of invasibility. J. Ecol. 88:528-534. [Google Scholar]
- 13.Diez-Gonzalez, F., T. R. Callaway, M. G. Kizoulis, and J. B. Russell. 1998. Grain feeding and the dissemination of acid-resistant Escherichia coli from cattle. Science 281:1666-1668. [DOI] [PubMed] [Google Scholar]
- 14.Duffy, G., R. C. Whiting, and J. J. Sheridan. 1999. The effect of a competitive microflora, pH and temperature on the growth kinetics of Escherichia coli O157:H7. Food Microbiol. 16:299-307. [Google Scholar]
- 15.Dunstan, P. K., and C. R. Johnson. 2006. Linking richness, community variability, and invasion resistance with patch size. Ecology 87:2842-2850. [DOI] [PubMed] [Google Scholar]
- 16.Felske, A., B. Engelen, U. Nübel, and H. Backhaus. 1996. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl. Environ. Microbiol. 62:4162-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortin, N. Y., A. Mulchandani, and W. Chen. 2001. Use of real-time polymerase chain reaction and molecular beacons for the detection of Escherichia coli O157:H7. Anal. Biochem. 289:281-288. [DOI] [PubMed] [Google Scholar]
- 18.Franz, E. 2007. Quantification of infection of lettuce by GFP-expressing Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium. Food Microbiol. 24:106-112. (First published 24 March 2006; doi: 10.1016/j.fm.2006.03.002.) [DOI] [PubMed] [Google Scholar]
- 19.Franz, E., A. D. Van Diepeningen, O. J. De Vos, and A. H. C. van Bruggen. 2005. Effects of cattle feeding regimen and soil management type on the fate of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in manure, manure-amended soil, and lettuce. Appl. Environ. Microbiol. 71:6165-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fratamico, P. M., M. Y. Deng, T. P. Strobaugh, and S. A. Palumbo. 1997. Construction and characterization of Escherichia coli O157:H7 strains expressing firefly luciferase and green fluorescent protein and their use in survival studies. J. Food Prot. 60:1167-1173. [DOI] [PubMed] [Google Scholar]
- 21.Geeraerd, A. H., V. P. Valdramidis, and J. F. Van Impe. 2005. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int. J. Food Microbiol. 102:95-105. [DOI] [PubMed] [Google Scholar]
- 22.Hancock, D. D., T. E. Besser, C. Gill, J. B. Russell, and F. Diez-Gonzalez. 1999. Cattle, hay, and E. coli. Science 284:51-53. (Letter.) [DOI] [PubMed] [Google Scholar]
- 23.Handschur, M., G. Pinar, B. Gallist, W. Lubitz, and A. G. Haslberger. 2005. Culture free DGGE and cloning based monitoring of changes in bacterial communities of salad due to processing. Food Chem. Toxicol. 43:1595-1605. [DOI] [PubMed] [Google Scholar]
- 24.Heuvelink, A. E., F. L. A. M. van den Biggelaar, J. T. M. Zwartkruis-Nahuis, R. G. Herbes, R. Huyben, N. Nagelkerke, W. J. G. Melchers, L. A. H. Monnens, and E. de Boer. 1998. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 36:3480-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Himathongkham, S., S. Bahari, H. Riemann, and D. Cliver. 1999. Survival of Escherichia coli O157:H7 and Salmonella Typhimurium in cow manure and cow manure slurry. FEMS Microbiol. Lett. 178:251-257. [DOI] [PubMed] [Google Scholar]
- 26.Hornitzky, M. A., B. A. Vanselow, K. Walker, K. A. Bettelheim, B. Corney, P. Gill, G. Bailey, and S. P. Djordjevic. 2002. Virulence properties and serotypes of Shiga toxin-producing Escherichia coli from healthy Australian cattle. Appl. Environ. Microbiol. 68:6439-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humpheson, L., M. R. Adams, W. A. Anderson, and M. B. Cole. 1998. Biphasic thermal inactivation kinetics in Salmonella enteritidis PT4. Appl. Environ. Microbiol. 64:459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibekwe, A. M., P. M. Watt, C. M. Grieve, V. K. Sharma, and S. R. Lyons. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Islam, M., M. P. Doyle, S. C. Phatak, P. Millner, and X. Jiang. 2004. Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J. Food Prot. 67:1365-1370. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, X., J. Morgan, and M. P. Doyle. 2002. Fate of Escherichia coli O157:H7 in manure-amended soil. Appl. Environ. Microbiol. 68:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy, T. A., S. Naeem, K. M. Howe, J. M. H. Knops, D. Tilman, and P. Reich. 2002. Biodiversity as a barrier to ecological invasion. Nature 417:636-638. [DOI] [PubMed] [Google Scholar]
- 32.Klerks, M. M., C. Zijlstra, and A. H. C. van Bruggen. 2004. Comparison of real-time PCR methods for detection of Salmonella enterica and Escherichia coli O157:H7, and introduction of a general internal amplification control. J. Microbiol. Methods 59:337-349. [DOI] [PubMed] [Google Scholar]
- 33.Kudva, I. T., K. Blanch, and C. J. Hovde. 1998. Analysis of Escherichia coli O157:H7 survival in ovine or bovine manure and manure slurry. Appl. Environ. Microbiol. 64:3166-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhnert, P., C. R. Dubosson, M. Roesch, E. Homfeld, M. G. Doherr, and J. W. Blum. 2005. Prevalence and risk-factor analysis of Shiga toxigenic Escherichia coli in faecal samples of organically and conventionally farmed dairy cattle. Vet. Microbiol. 109:37-45. [DOI] [PubMed] [Google Scholar]
- 35.LeJeune, J. T., D. Hancock, Y. Wasteson, E. Skjerve, and A. M. Urdahl. 2006. Comparison of E. coli O157 and Shiga toxin-encoding genes (stx) prevalence between Ohio, USA and Norwegian dairy cattle. Int. J. Food Microbiol. 109:19-24. [DOI] [PubMed] [Google Scholar]
- 36.Lynch, J. M., A. Benedetti, H. Insam, M. P. Nuti, K. Smalla, V. Torsvik, and P. Nannipieri. 2004. Microbial diversity in soil: ecological theories, the contribution of molecular techniques and the impact of transgenic plants and transgenic microorganisms. Biol. Fertil. Soils 40:363-385. [Google Scholar]
- 37.Matos, A., L. Kerkhof, and J. L. Garland. 2005. Effects of microbial community diversity on the survival of Pseudomonas aeruginosa in the wheat rhizosphere. Microb. Ecol. 49:257-264. [DOI] [PubMed] [Google Scholar]
- 38.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nauta, J. N., and J. B. Dufrenne. 1999. Variability in growth characteristics of different E. coli O157:H7 isolates, and its implications for predictive microbiology. Quant. Microbiol. 1:137-155. [Google Scholar]
- 40.Ogden, I. D., D. R. Fenlon, A. J. A. Vinten, and D. Lewis. 2001. The fate of Escherichia coli O157 in soil and its potential to contaminate drinking water. Int. J. Food Microbiol. 66:111-117. [DOI] [PubMed] [Google Scholar]
- 41.Pachepsky, Y. A., A. M. Sadeghi, S. A. Bradford, D. R. Shelton, A. K. Guber, and T. Dao. 2006. Transport and fate of manure-borne pathogens: modeling perspective. Agric. Water Manag. 86:81-92. [Google Scholar]
- 42.Park, G. W., and F. Diez-Gonzalez. 2003. Utilization of carbonate and ammonia-based treatments to eliminate Escherichia coli O157:H7 and Salmonella Typhimurium DT104 from cattle manure. J. Appl. Microbiol. 94:675-685. [DOI] [PubMed] [Google Scholar]
- 43.Peleg, M. 2003. Microbial survival curves: interpretation, mathematical modeling, and utilization. Comments Theor. Biol. 8:357-387. [Google Scholar]
- 44.Rosado, A. S., G. F. Duarte, L. Seldin, and J. D. Van Elsas. 1998. Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl. Environ. Microbiol. 64:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rugbjerg, H., E. M. Nielsen, and J. S. Andersen. 2003. Risk factors associated with faecal shedding of verocytotoxin-producing Escherichia coli O157 in eight known-infected Danish dairy herds. Prev. Vet. Med. 58:101-113. [DOI] [PubMed] [Google Scholar]
- 46.Schouten, J. M., M. Bouwknegt, A. W. Van De Giessen, K. Frankena, M. C. M. De Jong, and E. A. M. Graat. 2004. Prevalence estimation and risk factors for Escherichia coli O157 on Dutch dairy farms. Prev. Vet. Med. 64:49-61. [DOI] [PubMed] [Google Scholar]
- 47.Schouten, J. M., E. A. M. Graat, K. Frankena, A. W. Van De Giessen, W. K. Van Der Zwaluw, and M. C. M. De Jong. 2005. A longitudinal study of Escherichia coli O157 in cattle of a Dutch dairy farm and in the farm environment. Vet. Microbiol. 107:193-204. [DOI] [PubMed] [Google Scholar]
- 48.Scott, L., P. McGee, J. J. Sheridan, B. Earley, and N. Leonard. 2006. A comparison of the survival in faeces and water of Escherichia coli O157:H7 grown under laboratory conditions or obtained from cattle faeces. J. Food Prot. 69:6-11. [DOI] [PubMed] [Google Scholar]
- 49.Shadbolt, C., T. Ross, and T. A. McMeekin. 2001. Differentiation of the effects of lethal pH and water activity: food safety implications. Lett. Appl. Microbiol. 32:99-102. [DOI] [PubMed] [Google Scholar]
- 50.Sharma, V. K., and E. A. Dean-Nystrom. 2003. Detection of enterohemorrhagic Escherichia coli O157:H7 by using a multiplex real-time PCR assay for genes encoding intimin and Shiga toxins. Vet. Microbiol. 93:247-260. [DOI] [PubMed] [Google Scholar]
- 51.Sivapalasingam, S., C. R. Friedman, L. Cohen, and R. V. Tauxe. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67:2342-2353. [DOI] [PubMed] [Google Scholar]
- 52.Solomon, E. B., S. Yaron, and K. R. Matthews. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevens, M. P., P. M. van Diemen, F. Dziva, P. W. Jones, and T. S. Wallis. 2002. Options for the control of enterohemorrhagic Escherichia coli in ruminants. Microbiology 148:3767-3778. [DOI] [PubMed] [Google Scholar]
- 54.Tauxe, R. V. 1997. Emerging foodborne diseases: an evolving public health challenge. Emerg. Infect. Dis. 3:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Boekel, M. A. J. S. 2002. On the use of the Weibull model to describe thermal inactivation of microbial vegetative cells. Int. J. Food Microbiol. 74:139-159. [DOI] [PubMed] [Google Scholar]
- 56.van Diepeningen, A. D., O. J. De Vos, G. W. Korthals, and A. H. C. van Bruggen. 2006. Effects of organic versus conventional management on chemical and biological parameters in agricultural soils. Appl. Soil Ecol. 31:120-135. [Google Scholar]
- 57.Vanselow, B. A., D. O. Krause, and C. S. McSweeney. 2005. The Shiga toxin-producing Escherichia coli, their ruminant hosts, and potential on-farm interventions: a review. Aust. J. Agric. Res. 56:219-244. [Google Scholar]
- 58.van Soest, P. J. 1963. Use of detergents in the analysis of fibrous feeds. A rapid method for determination of fiber and lignin. J. Assoc. Off. Agric. Chem. 46:829-835. [Google Scholar]
- 59.Vose, D. 2000. Risk analysis, a quantitative guide, 2nd ed. John Wiley and Sons, Ltd., Chichester, United Kingdom.
- 60.Wang, G., T. Zhao, and M. P. Doyle. 1996. Fate of enterohemorrhagic Escherichia coli O157:H7 in bovine feces. Appl. Environ. Microbiol. 62:2567-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Younts-Dahl, S. M., G. D. Osborn, M. L. Galyean, J. D. Rivera, G. H. Loneragan, and M. M. Brashears. 2005. Reduction of Escherichia coli O157 in finishing beef cattle by various doses of Lactobacillus acidophilus in direct-fed microbials. J. Food Prot. 68:6-10. [DOI] [PubMed] [Google Scholar]