Abstract
A specific quantitative real-time PCR (qPCR) method was developed for the quantification of hepatotoxin nodularin-producing Nodularia, one of the main bloom-forming cyanobacteria in the Baltic Sea. Specific PCR primers were designed for subunit F of the nodularin synthetase gene (ndaF), which encodes the NdaF subunit of the nodularin synthetase gene complex needed for nodularin production. The qPCR method was applied to water samples (a total of 120 samples) collected from the Baltic Sea in July 2004. As few as 30 ndaF gene copies ml−1 of seawater could be detected, and thus, the method was very sensitive. The ndaF gene copy numbers and nodularin concentrations were shown to correlate in the Baltic seawater, indicating the constant production of nodularin by Nodularia. This qPCR method for the ndaF gene can be used for detailed studies of Nodularia blooms and their formation. ndaF gene copies and nodularin were detected mostly in the surface water but also in deeper water layers (down to 30 m). Toxic Nodularia blooms are not only horizontally but also vertically widely distributed, and thus, the Baltic fauna is extensively exposed to nodularin.
Nodularia spumigena is a hepatotoxic cyanobacterium that occurs in brackish waters all around the world (e.g., Central Asia [5], North America [6], Australia [10], Southern Africa [34], and Europe [39]). Especially in the Baltic Sea (39), as well as in some Australian lakes (10) and estuaries, it forms toxic mass occurrences, posing health risks to humans (17), causing animal poisonings (see, e.g., references 4, 8, 26, and 43), harming fisheries (14), and interfering with recreational use of waters (1).
The cyanobacterial blooms in the Baltic Sea consist mainly of three nitrogen-fixing filamentous cyanobacterial species: N. spumigena, Aphanizomenon flos-aquae, and, in lesser amounts, Anabaena species (16, 41). N. spumigena is the only known toxin-producing, bloom-forming cyanobacterial species in the Baltic Sea (16, 39, 41). Nontoxic Nodularia species have also been found, but they occur mainly in coastal areas in benthic habitats (24).
The toxin produced by N. spumigena is a pentapeptide nodularin, cyclo-(d-MeAsp-l-arginine-Adda-d-glutamate-Mdhb) (33, 39). d-MeAsp is d-erythro-β-methylaspartic acid, Mdhb is 2-(methylamino)-2-dehydrobutyric acid, and Adda is (2S,3S,8S,9S)-3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid, an amino acid found only in cyanobacterial toxins (33, 39). Nodularin is unique to Nodularia: it has not been found in any other cyanobacterial genera (38). Nodularin is produced nonribosomally by the nodularin synthetase enzyme complex, which is encoded by the 48-kb nodularin synthetase genes ndaA to ndaI (27). The structure of nodularin is similar to that of the cyanobacterial hepatotoxin microcystin (31). Consequently, it has been suggested that nda genes have evolved from the microcystin synthetase (mcy) genes through the deletion of two nonribosomal peptide synthetase modules and a change in substrate specificity of one nonribosomal peptide synthetase module (27, 31).
To widen our knowledge of toxic Nodularia blooms and their initiation, sensitive methods are needed for the quantification of nodularin-producing Nodularia. There are no studies of the early stages of Nodularia blooms in the Baltic Sea, since there have been no methods that are sensitive or specific enough to quantify nodularin-producing Nodularia from environmental samples. Microscopy, for example, does not clearly separate nodularin-producing Nodularia isolates from non-nodularin-producing Nodularia isolates (24). Microscopy methods are also laborious and time-consuming. While it is possible to estimate the number of toxic Nodularia cells based on nodularin concentrations in the samples, nodularin concentrations in a Nodularia cell may vary, depending on environmental conditions and the growth phase of the cell (see, e.g., references 21, 22, and 32). Thus, nodularin concentrations may not accurately predict the number of nodularin-producing Nodularia cells. A sensitive, specific, and rapid method to detect and quantify the toxin producer N. spumigena itself is needed to enable accurate studies of N. spumigena blooms, e.g., bloom formation.
We developed a specific, rapid, and sensitive quantitative real-time PCR (qPCR) method targeting nodularin synthetase gene subunit F (ndaF). With this method, as few as 30 copies of ndaF containing genomes of potentially nodularin-producing Nodularia cells ml−1 of Baltic seawater were detected, which enables studies on the early stages of Nodularia blooms.
MATERIALS AND METHODS
Strains.
The cyanobacterial strains used in this study are listed in Table 1. All the strains are maintained in the culture collection of K. Sivonen, University of Helsinki. Nodularia strains were grown in Z8 medium (Z8) (28) without nitrogen and with 8.75 g liter−1 added NaCl and 3.75 g liter−1 MgSO4·7H2O. Anabaena, Aphanizomenon, and Nostoc strains were cultivated in Z8 without nitrogen, and Microcystis and Planktothrix strains were cultivated in the same medium with nitrogen. The strains were uniclonal or axenic (Table 1). They were grown in a 40-ml volume under conditions of continuous light (4 to 7 μE m−2 s−1) at room temperature (24 to 25°C).
TABLE 1.
Specificities of primers ndaF8452 and ndaF8640 for the ndaF gene in conventional PCR and qPCR tested with microcystin- or nodularin-producing and nonproducing cyanobacterial strains
| Strain(s) | Result
|
|||
|---|---|---|---|---|
|
ndaFb
|
16S rRNAc PCR | Nod or Md | ||
| PCR | qPCR | |||
| Anabaena sp. strains 82,a PH 57, and PH 189a | − | ND | + | NP |
| Anabaena sp. strains 186,a 299 B,a 315, 318, 202A1, 202A2/41, NIVA-CYA83/1, and PH256a | − | ND | + | M |
| Anabaena sp. strains 66A and 90 | − | UD | + | M |
| A. flos-aquae NIES 81 | − | UD | + | NP |
| Microcystis aeruginosa PCC 7806 | − | UD | + | M |
| Microcystis sp. strain GL280641 | − | ND | + | NP |
| Microcystis sp. strains 98,a GL260735, GL280646, IZANCYA 25, NIES 89, NIES 102, and PCC 7941 | − | ND | + | M |
| Microcystis sp. strain 205 | − | UD | + | M |
| N. harveyana BECID27,a Bo53,a and Hübel 1983/300 | − | UD | + | NP |
| Nodularia sp. strains BECID29,a BECID35,a BECID36,a HKVV, UP16a,a UTEX B 2092,a and UTEX B 2093a | − | ND | + | NP |
| Nodularia sp. strains 55/15,a AV3,a AV63,a GDR113,a GR8a,a GR8b, HEM, Hübel 1987/310,a Hübel 1988/306a/b,a NSPI-05, P38,a Teili,a and TR183a | + | ND | + | Nod |
| Nodularia sp. strains BY1, F81,a and PCC 7804 | + | D | + | Nod |
| Nodularia sphaerocarpa Fä19a and UP16f | − | UD | + | NP |
| N. spumigena AV1, Hübel 1987/311,a NSOR-12, and PCC 9350a | + | D | + | Nod |
| Nostoc sp. strain 152 | − | UD | + | M |
| Oscillatoria sancta PCC 7515 | − | ND | + | NP |
| Planktothrix sp. strains NIVA-CYA126, NIVA-CYA127, and NIVA-CYA128 | − | ND | + | M |
| Planktothrix sp. strains 49 and 97 | − | UD | + | M |
Nonaxenic strain.
ndaF PCR or qPCR, PCR product with primers ndaF8452 and ndaF8640; +, PCR product in conventional PCR; −, no PCR product in conventional PCR; D, CT value over detection limit in qPCR; UD, CT value under detection limit in qPCR; ND, no data.
16S rRNA, PCR product with primers targeting to the 16S rRNA gene (to ensure the quality of the strain DNA samples).
Nodularin or microcystin production based on the literature. M, microcystin; Nod, nodularin; NP, non-nodularin- or non-microcystin-producing isolates.
Baltic seawater samples.
Water samples were collected from the Baltic Sea Proper and the Gulf of Finland in July 2004 on the ship R/V Aranda of the Finnish Institute of Marine Research (Fig. 1). The water was taken from depths of 0, 3, 7, 18, and 30 m with a Rosette sampler (General Oceanics). Five hundred to 2,000 ml of water was filtered using three replicate 1-μm-pore-size polycarbonate filters (GE Osmonics Labstore). The filters were frozen in liquid nitrogen and stored at −70°C. The samples were used for DNA and nodularin extractions. Some of the samples used for nodularin extraction were stored at −20°C.
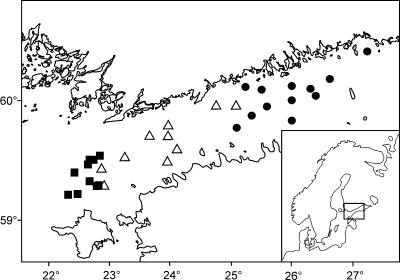
FIG. 1.
Sampling stations in the Baltic Sea Proper and in the Gulf of Finland. ▪, stations 1 to 32, with samples taken from 14 to 19 July 2004; ▵, stations 36 to 48, with samples taken from 20 to 23 July 2004; •, stations 49 to 64, with samples taken from 26 to 29 July 2004.
DNA extraction.
DNA was extracted from the cyanobacterial strains as follows. Cells were harvested for DNA extraction by filtering the cultures with 1- or 5-μm-pore-size polycarbonate filters (GE Osmonics Labstore). The cells were broken mechanically by bead beating with glass beads (425 to 600 and 710 to 1,180 μm; Sigma-Aldrich) with a FastPrep instrument (Savant Instruments) at a speed of 4.0 twice for 20 s. DNA was subsequently extracted using a DNeasy Plant Mini kit (QIAGEN). Concentrations of the DNA samples were determined spectrophotometrically at 260 nm with a BioPhotometer instrument (Eppendorf).
DNA was extracted from the Baltic seawater samples using the bead-beating and CTAB (N-cetyl-N,N,N-trimethyl-ammoniumbromide) method (15), with some modifications. The frozen sample filters were inserted into a tube containing glass beads (425 to 600 and 710 to 1,180 μm [1:1]; Sigma-Aldrich) and 0.8 ml cold lysis buffer (100 mM Tris-HCl [pH 8], 1.5% sodium dodecyl sulfate, 10 mM EDTA, 1% deoxycholate, 1% Igepal-CA630, 5 mM thiourea, 10 mM dithiothreitol). The cells were broken using a FastPrep instrument (Savant Instruments) for 30 s at a speed of 5.0, and the samples were placed on ice. The samples were centrifuged at 15,000 × g for 1 min, and the supernatant was carefully transferred into a new tube. Lysis buffer (0.5 ml) was added into the original tube containing the sample filter and the glass beads, and the FastPrep as well as the centrifuging steps were repeated. The supernatant was collected, combined with the supernatant collected earlier, and divided among three 2-ml tubes. Two hundred twenty-five microliters of 5 M NaCl and 170 μl of 10% CTAB (Merck KGaA) (in 0.7 M NaCl, warmed to +65°C) were added to each tube, and the samples were mixed and then incubated at +65°C for 20 min. An equal volume of chloroform was added, and the tubes were mixed and centrifuged at 10,000 × g for 10 min at +4°C. The upper phase was collected, and a 1/10 volume of 3 M Na acetate and a double volume of ice-cold 96% ethanol were added. The samples were then precipitated at −20°C and centrifuged at 15,000 × g for 15 min at +4°C, and the ethanol was discarded. The samples were subsequently washed with 70% ethanol, and the pellets were dried at +37°C and resuspended in 80 μl water per sample. To improve the quality and reduce the amount of PCR inhibitors, the extracted DNA samples were purified using a Geneclean Turbo kit (Q-Biogene) with two elution steps containing 30 μl of Tris-EDTA buffer (10 mM Tris [pH 8]-1 mM EDTA).
PCR.
PCR primers were designed to amplify the nodularin synthetase gene subunit F (ndaF) of Nodularia. The sequence of the forward primer ndaF8452 was 5′-GTG ATT GAA TTT CTT GGT CG-3′, and the sequence of the reverse primer ndaF8640 was 5′-GGA AAT TTC TAT GTC TGA CTC AG-3′. PCR was performed in 1× DyNAzyme II enzyme buffer (Finnzymes) with 0.35 μM of both primers, 65 μM of all four nucleotides (Finnzymes), 1 unit of DNA polymerase DyNAzyme II (Finnzymes), and 17.5 ng of template DNA in a reaction mixture volume of 20 μl. The following temperature protocol was used: 95°C for 3 min; 35 cycles of 94°C for 30 s, 61°C for 30 s, and 72°C for 20 s; and 72°C for 7 min. The PCR amplification was performed using a PTC-200 PCR instrument (MJ Research). The presence or absence of the ndaF amplification product was determined by 1.5% agarose gel electrophoresis.
To check the quality of the DNA samples, for example, for the presence of PCR inhibitors, the DNA was amplified in PCR with cyanobacterium-specific 16S rRNA gene primers CYA359F (29) and 23S30R (23). PCR was performed in a volume of 20 μl in 1× DyNAzyme II enzyme buffer (Finnzymes), 0.5 μM of both primers, 250 μM of all four nucleotides (Finnzymes), 0.4 U DNA polymerase DyNAzyme II (Finnzymes), 1 μg μl−1 bovine serum albumin (Promega), and 2 μl DNA. The temperature protocol was as follows: 94°C for 5 min; 10 cycles of 94°C for 45 s, 57°C for 45 s, and 68°C for 2 min; 25 cycles of 92°C for 45 s, 54°C for 45 s, and 68°C for 2 min; and 68°C for 7 min. The PCR amplification was performed using a PTC-200 PCR instrument (MJ Research). The presence or absence of the 16 S rRNA gene amplification product was determined by 1.5% agarose gel electrophoresis.
qPCR.
ndaF genes were quantified by qPCR using primers ndaF8452 and ndaF8640 and a LightCycler instrument with LightCycler software version 3.5 (Roche). qPCR was performed in a 10-μl volume with 3.5 mM MgCl2, 0.35 μM both primers, 1 μl enzyme-nucleotide-dye mix (LightCycler FastStart DNA Master SYBR green I; Roche), and 2 μl template DNA. The amplification program consisted of the following steps: (i) preheating at 95°C for 10 min, with a heating rate of 20°C s−1; (ii) quantification, including 45 cycles (95°C for 0 s, 63°C for 5 s, and 72°C for 8 s), fluorescence measurement at the end of each cycle at 72°C through channel F1 (530 nm), and a heating rate of 20°C s−1; and (iii) melting curve analysis, which included heating from 58°C to 95°C at rate of 0.1°C s−1 and fluorescence measurement continuously through channel F1 (530 nm).
A standard curve for qPCR was determined as a correlation between the ndaF gene copy numbers and the cycle threshold (CT). The DNAs of Nodularia spumigena strains AV1, NSOR-12, and PCC 9350 were chosen as standard samples, and 10-fold serial dilutions of these samples were analyzed by qPCR. The ndaF gene copy numbers of the standard samples were determined using the following formula: [DNA concentration (g μl−1) × Avogadro constant (6.022 × 1023 copies mol−1) × sample volume (μl)] × [genome molecular weight (g mol−1)]−1 = number of ndaF gene copies in a sample. The genome size of (3.34 ± 0.17) × 109 Da (or g mol−1) of non-nodularin-producing Nodularia sp. strain PCC 73104 analyzed previously by Herdman et al. (9) was used as a reference value for the Nodularia genome size. Based on the unfinished N. spumigena genome sequence (GenBank accession number AAVW00000000), there seems to be only one copy of the ndaF gene in the Nodularia genome.
The ndaF gene copy numbers of the Baltic seawater samples were determined by qPCR immediately after DNA extraction and purification. The DNA samples were analyzed, each in three replicates as 100-fold dilutions. Two microliters of diluted DNA was added to a 10-μl qPCR reaction mixture, and the qPCR was performed as described above. In each run, a standard curve was determined by analyzing a dilution series (101 to 106 ndaF gene copies per 10 μl of reaction mixture) of Nodularia spumigena AV1 standard DNA. For each measurement, a standard deviation of three replicates was determined.
Background DNA in qPCR.
In order to test if background DNA (DNA containing no ndaF gene sequences) has any effect on the reliability of the qPCR quantification, different DNA mixes were analyzed by qPCR. The mixes contained the DNA of one nodularin-producing (and ndaF gene-containing) strain, N. spumigena AV1, and one non-nodularin-producing (not containing the ndaF gene) strain, either Nodularia harveyana Hübel 1983/300 or A. flos-aquae NIES 81. The number of ndaF gene copies of N. spumigena AV1 was either 100 or 10,000 copies per qPCR reaction. Approximately 103, 104, 105, or 106 genome copies of either N. harveyana Hübel 1983/300 or A. flos-aquae NIES 81 DNA were used per qPCR reaction. Three replicates of each combination of concentration were analyzed in qPCR. Genome copy numbers of the background DNA samples were determined using the Nodularia sp. strain PCC 73104 genome size, (3.34 ± 0.17) × 109 Da, for N. harveyana Hübel 1983/300 and the mean, 3.365 × 109 Da, of the Anabaena sp. strain PCC 7122 genome size, (3.17 ± 0.18) × 109 Da, and the Anabaena sp. strain PCC 6309 genome size, (3.56 ± 0.22) × 109 Da, for A. flos-aquae NIES 81 (9). No Aphanizomenon genome sizes are available, so the genome size of its nearest relative, Anabaena (30), was used.
Sequencing.
To verify that only the ndaF gene was amplified, qPCR products of standards and several environmental samples were sequenced. qPCR products were purified using a Montage PCR kit (Millipore). Cycle sequencing was performed in 1× sequencing buffer (Big Dye Terminator v3.1 cycle sequencing kit; Applied Biosystems) containing 10 pmol of primer ndaF8452, 1 μl Big Dye Ready Reaction mix (Big Dye Terminator v3.1 cycle sequencing kit; Applied Biosystems), and 20 ng of template DNA (qPCR product) in a total volume of 10 μl. The amplification was performed using a PTC-200 PCR instrument (MJ Research) with the following program: 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. The amplification product was precipitated using 96% ethanol with 0.1 M sodium acetate, washed with 70% ethanol, resuspended in 12 μl of Template Suppression reagent (Applied Biosystems), and denatured for 3 min at 94°C. Sequencing was performed with an ABI PRISM 310 genetic analyzer (Applied Biosystems).
Nodularin analysis of Baltic seawater samples.
Nodularin concentrations in the Baltic seawater samples were determined by using a liquid chromatograph (LC)/mass spectrometer (MS). Nodularin was extracted from the polycarbonate filters by a methanol bath sonication method described previously by Spoof et al. (40), with certain modifications. The filters were first dried in a vacuum evaporator (the Heto vacuum centrifuge Maxi-dry plus; Jouan Nordic). Twelve hundred microliters of 75% methanol was added, and the samples were shaken with a FastPrep instrument (Savant Instruments) three times at a speed of 5.0 for 20 s and sonicated in a bath sonicator at efficacy 10 (Sonorex Super 10 P digital; Bandelin) for 30 min. The FastPrep and sonication treatments were repeated. The FastPrep treatment was repeated once more, and the sample tubes were centrifuged at 20,000 × g for 5 min. Six hundred microliters of the supernatant was transferred into a glass vial containing about 100 μl 0.5-mm glass beads (Scientific Industries). The samples were dried in a vacuum evaporator (the Heto vacuum centrifuge Maxi-dry plus; Jouan Nordic), dissolved in 100 μl 10% acetonitrile, shaken in a FastPrep instrument (Savant Instruments) at a speed of 5.0 for 10 s, and centrifuged at 10,000 × g for 3 min. The samples were transferred into high-performance liquid chromatography (HPLC) sample vials and analyzed by LC/MS.
Nodularin concentrations were analyzed by LC/MS using a method described previously by Spoof et al. (40), with slight modifications. LC/MS equipment from Agilent (1100 Series LC/MSD with Ion Trap XCT Plus and Electrospray Ionizator) was used. In a liquid chromatograph, a Zorbax SB-C18 column (75 mm by 2.1 mm; particle size, 3.5 μm) (Agilent) and a Phenomenex C18 precolumn (4.0 mm by 2.0 mm) were used. The columns were warmed to +40°C during the run. Water (purified with Milli-Q plus; Millipore) and acetonitrile (HPLC quality; Merck KGaA), both containing 0.10% formic acid (HPLC quality; Fluka Chemie GmbH), were used as eluents. During the first 7 min, the concentration of the acetonitrile was increased from 27% to 35.8%. From the 7th to the 11th minute, the column was washed with 85% acetonitrile, and from the 11th to the 15th minute, the column was washed with 100% acetonitrile. From the 15th to the 30th minute, the column was balanced with 27% acetonitrile. Between the sample injections, the injector needle was washed with 50% methanol (outside) and acetonitrile of various concentrations (inside and out). Positively charged ions with mass/charge ratios of 810 to 835 m/z were analyzed by MS. Ions with a mass/charge ratio of 826 ± 0.5 m/z and a retention time of about 4.9 min were defined as being nodularin. Nodularin was quantified using purified nodularin as a standard.
Statistical analyses.
Pearson correlation coefficients between the ndaF gene copy number and the nodularin concentration were calculated with SPSS 12.0.1 statistical software for Windows (Chicago, IL).
Nucleotide sequence accession numbers.
All sequences produced in this study have been deposited in GenBank under accession numbers EF215830 to EF215837.
RESULTS AND DISCUSSION
Specificities of primers ndaF8452 and ndaF8640.
To test the specificities of the targeting of primers ndaF8452 and ndaF8640 to the nodularin synthetase gene ndaF, DNA from 63 nodularin- or microcystin-producing or nonproducing cyanobacterial strains were amplified in conventional PCR. A PCR product was amplified only from the nodularin-producing Nodularia strains (Table 1), and therefore, the primers were specific for the ndaF gene and nodularin-producing Nodularia. It is possible that the nda gene cluster involving non-nodularin-producing Nodularia strains exists in the environment, since inactive mcy gene clusters (closely related to the nda gene cluster) of Planktothrix have previously been detected in alpine lakes (3, 18). However, among Nodularia strains, such inactive nda genotypes have thus far not been observed, and thus, determining ndaF gene copy numbers can be considered to be a reliable way to quantify toxic Nodularia genotypes.
Based on amplification in conventional PCR, 20 strains were further selected to be tested by qPCR. In qPCR, only the nodularin-producing Nodularia strains gave CT values over the detection limit (Table 1), and therefore, the qPCR was also specific for nodularin-producing Nodularia strains. All the cyanobacterial DNA samples tested gave an amplification product in conventional PCR with cyanobacterium-specific 16S rRNA gene primers, indicating that there were no PCR inhibitors present in the DNA samples. A gene bank was explored to find out whether the primer sequences also pair with sequences other than ndaF, but in a BLAST search, no close hits from genes other than ndaF were found. Thus, primers ndaF8452 and ndaF8640 were specific to the ndaF gene and the nodularin-producing Nodularia strains based on both conventional PCR and qPCR and BLAST searches.
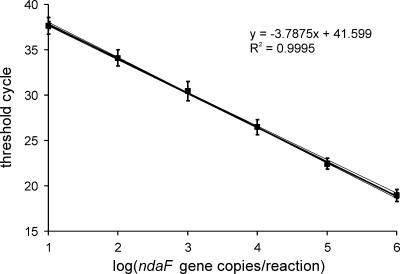
Standard curve.
A standard curve was determined for primers ndaF8452 and ndaF8640 in qPCR. To estimate the variation between the qPCR amplification efficiencies of different Nodularia strains, a dilution series of DNA from three separate nodularin-producing Nodularia strains, N. spumigena AV1 (isolated from the Baltic Sea), NSOR-12 (Australia), and PCC 9350 (Baltic Sea), was analyzed by qPCR, each in triplicate. Since the ndaF gene copy numbers were similar among these three strains, it was possible to define a common average standard curve for the three N. spumigena strains (Fig. 2). It was also deduced that separate N. spumigena strains probably do not differ from each other substantially in their genome sizes or in qPCR amplification efficiencies. Therefore, the qPCR method developed in this study could be applicable to all N. spumigena strains and to nodularin-producing Nodularia-containing environmental samples worldwide. The detection range of the qPCR method was at least 101 to 106 ndaF copies per 10-μl reaction mixture. When unknown samples were analyzed by qPCR, a standard curve was determined separately for each run to avoid the effect of the variability of the amplification between separate qPCR runs.
FIG. 2.
Standard curve (bold line) for qPCR determined with N. spumigena AV1, NSOR-12, and PCC 9350. The three thin curves are separate curves for N. spumigena strains AV1, NSOR-12, and PCC 9350. The error bars indicate standard deviations among the three strains in three parallel runs.
By analyzing the ndaF gene copy numbers, the number of Nodularia genomes could be determined, since there is only one copy of the nda gene cluster per one Nodularia genome (the data are based on an unpublished N. spumigena genome sequence). By contrast, the number of Nodularia cells cannot be deduced from the number of ndaF genes or Nodularia genomes with complete accuracy, since the number of genomes per cell varies, depending on the growth phase (45) and possibly the differentiation (42) of the cell.
A melting curve analysis was made for the qPCR products of different concentrations of N. spumigena AV1, NSOR-12, and PCC 9350 DNA to ensure that the products were specific and no primer dimers were formed in the qPCR. All the qPCR products had similar melting curves with only one specific peak at 80.9 ± 0.4°C. No primer dimer peaks (at about 76°C) were observed. The melting curve analysis and sequencing showed that primers ndaF8452 and ndaF8640 amplified only the ndaF gene.
Effect of background DNA.
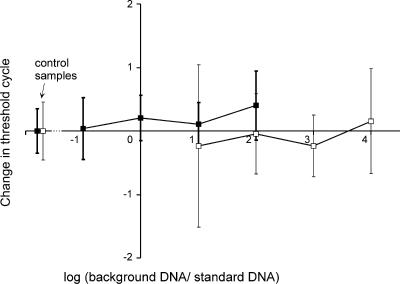
The effect of background DNA (DNA containing no ndaF gene sequences) on the reliability of the qPCR quantification was studied to determine whether the qPCR method is also applicable to samples containing DNA from strains other than nodularin-producing Nodularia strains. Approximately 102 or 104 copies of nodularin-producing N. spumigena AV1 DNA were analyzed and mixed with different concentrations of background DNA of non-nodularin-producing N. harveyana Hübel 1983/300 or A. flos-aquae NIES 81. N. harveyana and A. flos-aquae strains were used as background DNA, since they are closely related to N. spumigena (7, 11, 24, 25), and they are found in the Baltic Sea (16, 20, 41). The addition of background DNA changed the mean CT values by −0.24 to 0.4 and slightly increased the standard deviations of the CT values (Fig. 3). Thus, the background DNA had little effect on the CT values, even though the amount of target DNA, or the ndaF-containing genomes, was only 1/10,000 of the amount of total DNA in the sample.
FIG. 3.
Effect of background DNA on CT values in qPCR analysis with primers ndaF8452 and ndaF8640. Different concentrations of DNA of non-nodularin-producing N. harveyana Hübel 1983/300 or A. flos-aquae NIES 81 were added to a reaction mixture with 102 (□) or 104 (▪) copies of nodularin-producing N. spumigena AV1 DNA. The effect of the background DNA on the qPCR analysis is reported as changes in the CT values. The results are averages of the results with two different background strains (N. harveyana Hübel 1983/300 or A. flos-aquae NIES 81). The amount of background DNA is expressed as a logarithm of the ratio of the background DNA (DNA of N. harveyana Hübel 1983/300 or A. flos-aquae NIES 81) and the target DNA (DNA of N. spumigena AV1) concentrations. The error bars indicate standard deviations of 12 to 15 replicates, and in control samples, which contained only target DNA, error bars indicate standard deviations of 18 replicates.
In previous studies, even lesser amounts of background DNA have disturbed qPCR quantification. Wawrik et al. (44) developed a qPCR method for the rbcL gene of eukaryotic algae, in which a 1,000-fold surplus of background DNA hindered the quantification of the target DNA. In a study of Microcystis cyanobacteria with the mcyB and phycocyanin gene qPCR reported previously by Kurmayer and Kutzenberger (19), environmental background DNA caused at most a 100% overestimation or a 68% underestimation of the copy numbers compared to the respective values of 13% and 18% in this study. The qPCR method developed here was demonstrably less sensitive to the effect of background DNA than the methods described previously by Wawrik et al. (44) and Kurmayer and Kutzenberger (19).
The results of this study were similar to the results described previously by Becker et al. (2), although those authors used a TaqMan method. A 104-fold surplus of background DNA (phylogenetically closely related to the target DNA) did not disturb the qPCR analysis of the 16S rRNA gene of Synechococcus sp. strain BO 8807 (2). Based on this study and previous studies (2, 19, 44), it seems obvious that it is important to study the effect of background DNA on the efficiency of qPCR in order to determine the reliable detection range of the qPCR method. Since the qPCR method for the ndaF gene was insensitive to background DNA, a study of the early stages of blooms, where only a small proportion of the plankton is formed by Nodularia and the proportion of background DNA may be high, is possible.
Baltic Sea samples.
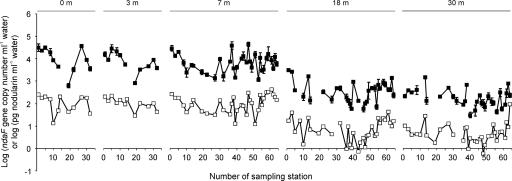
The qPCR method developed in this study was tested for its applicability to environmental water samples. We monitored the development of Nodularia blooms in the Baltic Proper and in the Gulf of Finland in July 2004 through ndaF gene copy numbers and nodularin concentrations. In all but six of the samples, the ndaF gene copy numbers exceeded the detection limit, 15 to 75 copies ml−1 seawater (the variation in detection limits was caused by the differences in parallel qPCR runs and the original sample volumes). The copy numbers varied 1,500-fold, between 30 and 45,000 ndaF gene copies ml−1 water (Fig. 4). Melting curve analysis (for all the samples) and sequencing (for five representative samples) showed that only the ndaF gene was amplified. The qPCR method for primers ndaF8452 and ndaF8640 was fully applicable to the environmental samples, since the ndaF gene copy numbers could be identified from the water samples.
FIG. 4.
ndaF gene copy numbers (▪) and nodularin concentrations (□) in the Baltic Proper and the Gulf of Finland in July 2004. The samples were taken during a Nodularia bloom from depths of 0, 3, 7, 18, and 30 m. The error bars indicate standard deviations defined by at least three parallel measurements. The ndaF gene copy numbers were under the detection limit (the logarithm of the copy number was under 1.18 to 1.88) in 30-m samples from sampling stations 10, 19, 39, and 55 and in 18-m samples from sampling stations 19 and 44.
In the nodularin analysis, the detection limit was 0.1 to 0.4 pg nodularin ml−1 water (depending on the volume of the filtered water sample), and in all the samples, nodularin concentrations exceeded the detection limit. Nodularin concentrations in the samples varied between 0.6 and 420 pg nodularin ml−1 seawater (Fig. 4). Both the ndaF gene copy numbers and the nodularin concentrations were highest in the upper water layers (at 0-, 3-, and 7-m depths). However, toxic Nodularia and nodularin were also detected in the deeper water layers (18 and 30 m). Satellite images reported in previous studies (12) have shown the massive horizontal distribution of cyanobacterial blooms in the Baltic Sea. The present study shows that toxic Nodularia blooms are also vertically widely distributed. This may imply that during the blooms, water fauna in the Baltic Sea can hardly escape exposure to hepatotoxin nodularin. This is in agreement with the previous studies, which have shown that nodularin is frequently found in the Baltic fauna (e.g., zooplankton [13], mussels [35], fish [35, 37], and waterfowl [36]).
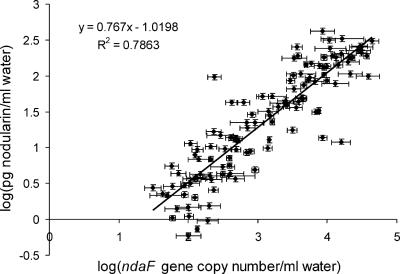
The ndaF copy numbers were found to correlate with the nodularin concentrations (correlation coefficient, 0.89; P < 0.01) (Fig. 5), which refer to a relatively constant production of nodularin by Nodularia in the Baltic Sea plankton during the period studied. The amount of nodularin per one ndaF gene copy was on average 28 fg (the standard deviation was 42 fg per copy). In total, the nodularin content per ndaF copy varied between 0.8 and 406 fg nodularin per gene copy (500-fold), so it is not possible to predict ndaF gene copy numbers accurately based on nodularin concentrations.
FIG. 5.
Correlation between ndaF gene copy numbers and nodularin concentrations in water samples collected from the Baltic Proper and the Gulf of Finland in July 2004 from sampling stations 1 to 64 from depths of 0, 3, 7, 18, and 30 m (a total of 115 samples).
The ratios of “nodularin per ndaF gene copy” of environmental samples measured in this study (0.8 to 406 fg nodularin per ndaF gene copy) were comparable to cellular nodularin concentrations reported previously in a study by Repka et al. (32), with laboratory cultures of N. spumigena (100 to 440 fg nodularin cell−1). In addition to sample types, the slight differences in cellular nodularin concentrations observed between these studies may arise from differences in the units of measure. In this study, nodularin concentrations were defined per ndaF gene copy (or the genome of nodularin-producing Nodularia), while in the study described previously by Repka et al. (32), the unit of measure was the amount of nodularin per cell. The values might therefore differ, for the nodularin content per cell is higher than that per ndaF gene copy if there are several genomes, and, thus, several ndaF gene copies, per cell. There might also be some inaccuracy in the genome size of N. spumigena, since the unpublished N. spumigena sequence has not yet been finalized, and thus, no exact data on the genome size of N. spumigena are available.
A qPCR method was developed in this study to enable the quantification of hepatotoxin nodularin-producing Nodularia in the Baltic Sea. By this highly sensitive and specific method, toxic Nodularia blooms can be monitored, and bloom development can be studied in the Baltic Sea and other water environments worldwide.
Acknowledgments
This work was supported financially by the Academy of Finland (grants 202441 [the Bireme project], 214457, and 211494).
We thank the personnel of the Finnish Institute of Marine Research on the R/V Aranda for help in sampling onboard. Matti Wahlsten is gratefully acknowledged for his assistance with nodularin analysis by LC/MS.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Bartram, J., W. W. Carmichael, I. Chorus, G. Jones, and O. M. Skulberg. 1999. Introduction, p. 1-14. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water. A guide to their public health consequences, monitoring and management. E & FN Spon, London, United Kingdom.
- 2.Becker, S., M. Fahrbach, P. Böger, and A. Ernst. 2002. Quantitative tracing, by Taq nuclease assays, of a Synechococcus ecotype in a highly diversified natural population. Appl. Environ. Microbiol. 68:4486-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christiansen, G., R. Kurmayer, Q. Liu, and T. Börner. 2006. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl. Environ. Microbiol. 72:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edler, L., S. Fernö, M. G. Lind, R. Lundberg, and P. O. Nilsson. 1985. Mortality of dogs associated with a bloom of the cyanobacterium Nodularia spumigena in the Baltic Sea. Ophelia 24:103-109. [Google Scholar]
- 5.Ergashev, A. E. 1979. The origin and typology of the Central Asian lakes and their algal flora. Int. Rev. Ges. Hydrobiol. 64:629-642. [Google Scholar]
- 6.Galat, D. L., J. P. Verdin, and L. L. Sims. 1990. Large-scale patterns of Nodularia spumigena blooms in Pyramid Lake, Nevada, determined from Landsat imagery: 1972-1986. Hydrobiologia 197:147-164. [Google Scholar]
- 7.Gugger, M., C. Lyra, P. Henriksen, A. Couté, J.-F. Humbert, and K. Sivonen. 2002. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int. J. Syst. Evol. Microbiol. 52:1867-1880. [DOI] [PubMed] [Google Scholar]
- 8.Gußmann, H. J., J. Molzahn, and B. Bicks. 1985. Vergiftungen bei Jungrinden durch die Blaualge Nodularia spumigena. Monatsh. Veterinarmed. 40:76-79. [Google Scholar]
- 9.Herdman, M., M. Janvier, R. Rippka, and R. Y. Stanier. 1979. Genome size of cyanobacteria. J. Gen. Microbiol. 111:73-85. [Google Scholar]
- 10.Heresztyn, T., and B. C. Nicholson. 1997. Nodularin concentrations in lakes Alexandrina and Albert, South Australia, during a bloom of the cyanobacterium (blue-green alga) Nodularia spumigena and degradation of the toxin. Environ. Toxicol. Water 12:273-282. [Google Scholar]
- 11.Iteman, I., R. Rippka, N. T. de Marsac, and M. Herdman. 2002. rDNA analyses of planktonic cyanobacteria, including members of the genera Anabaenopsis and Cyanospira. Microbiology 148:481-496. [DOI] [PubMed] [Google Scholar]
- 12.Kahru, M., U. Horstmann, and O. Rud. 1994. Satellite detection of increased cyanobacteria blooms in the Baltic Sea: natural fluctuation or ecosystem change? Ambio 23:469-472. [Google Scholar]
- 13.Karjalainen, M., M. Reinikainen, F. Lindvall, L. Spoof, and J. A. O. Meriluoto. 2003. Uptake and accumulation of dissolved, radiolabeled nodularin in the Baltic Sea zooplankton. Environ. Toxicol. 18:52-60. [DOI] [PubMed] [Google Scholar]
- 14.Karjalainen, M., M. Reinikainen, L. Spoof, J. A. O. Meriluoto, K. Sivonen, and M. Viitasalo. 2005. Trophic transfer of cyanobacterial toxins from zooplankton to planktivores: consequences for pike larvae and mysid shrimps. Environ. Toxicol. 20:354-362. [DOI] [PubMed] [Google Scholar]
- 15.Kolmonen, E., K. Sivonen, J. Rapala, and K. Haukka. 2004. Diversity of cyanobacteria and heterotrophic bacteria in cyanobacterial blooms in Lake Joutikas, Finland. Aquat. Microb. Ecol. 36:201-211. [Google Scholar]
- 16.Kononen, K., J. Kuparinen, K. Mäkelä, J. Laanemets, J. Pavelson, and S. Nõmmann. 1996. Initiation of cyanobacterial blooms in a frontal region at the entrance to the Gulf of Finland, Baltic Sea. Limnol. Oceanogr. 41:98-112. [Google Scholar]
- 17.Kuiper-Goodman, T., I. Falconer, and J. Fitzgerald. 1999. Human health aspects, p. 113-153. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water. A guide to their public health consequences, monitoring and management. E & FN Spon, London, United Kingdom.
- 18.Kurmayer, R., G. Christiansen, J. Fastner, and T. Börner. 2004. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 6:831-841. [DOI] [PubMed] [Google Scholar]
- 19.Kurmayer, R., and T. Kutzenberger. 2003. Application of real-time PCR for quantification of microcystin genotypes in a population of the toxic cyanobacterium Microcystis sp. Appl. Environ. Microbiol. 69:6723-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehtimäki, J., C. Lyra, S. Suomalainen, P. Sundman, L. Rouhiainen, L. Paulin, M. Salkinoja-Salonen, and K. Sivonen. 2000. Characterisation of Nodularia strains, cyanobacteria from brackish waters, by genotypic and phenotypic methods. Int. J. Syst. Evol. Microbiol. 50:1043-1053. [DOI] [PubMed] [Google Scholar]
- 21.Lehtimäki, J., P. Moisander, K. Sivonen, and K. Kononen. 1997. Growth, nitrogen fixation, and nodularin production by two Baltic Sea cyanobacteria. Appl. Environ. Microbiol. 63:1647-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehtimäki, J., K. Sivonen, R. Luukkainen, and S. I. Niemelä. 1994. The effects of incubation time, temperature, light, salinity, and phosphorus on growth and hepatotoxin production by Nodularia strains. Arch. Hydrobiol. 130:269-282. [Google Scholar]
- 23.Lepére, C., A. Wilmotte, and B. Meyer. 2000. Molecular diversity of Microcystis strains (Cyanophyceae, Chrococcales) based on 16S rDNA sequences. Syst. Geogr. Plants 70:275-283. [Google Scholar]
- 24.Lyra, C., M. Laamanen, J. M. Lehtimäki, A. Surakka, and K. Sivonen. 2005. Benthic cyanobacteria of the genus Nodularia are non-toxic, without gas vacuoles, able to glide and genetically more diverse than planktonic Nodularia. Int. J. Syst. Evol. Microbiol. 55:555-568. [DOI] [PubMed] [Google Scholar]
- 25.Lyra, C., S. Suomalainen, M. Gugger, C. Vezie, P. Sundman, L. Paulin, and K. Sivonen. 2001. Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis and Planktothrix genera. Int. J. Syst. Evol. Microbiol. 51:513-526. [DOI] [PubMed] [Google Scholar]
- 26.Main, D. C., P. H. Berry, R. L. Peet, and J. P. Robertson. 1977. Sheep mortalities associated with the blue green alga. Aust. Vet. J. 53:578-581. [DOI] [PubMed] [Google Scholar]
- 27.Moffitt, M. C., and B. A. Neilan. 2004. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl. Environ. Microbiol. 70:6353-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norwegian Institute for Water Research. 1990. Catalogue of strains, edition 1990, culture collection of algae. Norwegian Institute for Water Research, Oslo, Norway.
- 29.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajaniemi, P., P. Hrouzek, K. Kaštovská, R. Willame, A. Rantala, L. Hoffmann, J. Komárek, and K. Sivonen. 2005. Phylogenetic and morphological evaluation of the genera Anabaena, Aphanizomenon, Trichormus and Nostoc (Nostocales, Cyanobacteria). Int. J. Syst. Evol. Microbiol. 55:11-26. [DOI] [PubMed] [Google Scholar]
- 31.Rantala, A., D. Fewer, M. Hisbergues, L. Rouhiainen, J. Vaitomaa, T. Börner, and K. Sivonen. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 101:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Repka, S., J. Mehtonen, J. Vaitomaa, L. Saari, and K. Sivonen. 2001. Effects of nutrients on growth and nodularin production of Nodularia strain GR8b. Microb. Ecol. 42:606-613. [DOI] [PubMed] [Google Scholar]
- 33.Rinehart, K. L., K. Harada, M. Namikoshi, C. Chen, and C. A. Harvis. 1988. Nodularin, microcystin, and the configuration of Adda. J. Am. Chem. Soc. 110:8557-8558. [Google Scholar]
- 34.Seaman, M. T., P. J. Ashton, and W. D. Williams. 1991. Inland salt waters of southern Africa. Hydrobiologia 210:75-91. [Google Scholar]
- 35.Sipiä, V. O., H. T. Kankaanpää, S. Pflugmacher, J. Flinkman, A. Furey, and K. J. James. 2002. Bioaccumulation and detoxication of nodularin in tissues of flounder (Platichthys flesus), mussels (Mytilus edulis, Dreissenia polymorpha), and clams (Macoma balthica) from the Northern Baltic Sea. Ecotoxicol. Environ. Saf. 53:305-311. [DOI] [PubMed] [Google Scholar]
- 36.Sipiä, V. O., K. M. Karlsson, J. A. O. Meriluoto, and H. Kankaanpää. 2004. Eiders (Somateria mollissima) obtain nodularin, a cyanobacterial hepatotoxin, in Baltic Sea food web. Environ. Toxicol. Chem. 23:1256-1260. [DOI] [PubMed] [Google Scholar]
- 37.Sipiä, V. O., K. Lahti, H. T. Kankaanpää, P. J. Vuorinen, and J. A. O. Meriluoto. 2002. Screening for cyanobacterial hepatotoxins in herring and salmon from the Baltic Sea. Aquat. Ecosyst. Health Manage. 5:451-456. [Google Scholar]
- 38.Sivonen, K., and G. Jones. 1999. Cyanobacterial toxins, p. 41-111. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water. A guide to their public health consequences, monitoring and management. E & FN Spon, London, United Kingdom.
- 39.Sivonen, K., K. Kononen, W. W. Carmichael, A. M. Dahlem, K. L. Rinehart, J. Kiviranta, and S. I. Niemelä. 1989. Occurrence of the hepatotoxic cyanobacterium Nodularia spumigena in the Baltic Sea and structure of the toxin. Appl. Environ. Microbiol. 55:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spoof, L., P. Vesterkvist, T. Lindholm, and J. Meriluoto. 2003. Screening for cyanobacterial hepatotoxins, microcystins and nodularin in environmental water samples by reversed-phase liquid chromatography-electrospray ionisation mass spectrometry. J. Chromatogr. A 1020:105-119. [DOI] [PubMed] [Google Scholar]
- 41.Stal, L. J., P. Albertano, B. Bergman, K. von Bröckel, J. R. Gallon, P. K. Hayes, K. Sivonen, and A. E. Walsby. 2003. BASIC: Baltic Sea cyanobacteria. An investigation of the structure and dynamics of water blooms of cyanobacteria in the Baltic Sea—responses to a changing environment. Cont. Shelf Res. 23:1695-1714. [Google Scholar]
- 42.Tandeau de Marsac, N. 1994. Differentiation of hormogonia and relationships with other biological processes, p. 825-842. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 43.Van Halderen, A., W. R. Harding, J. C. Wessels, D. J. Schneider, E. W. P. Heine, J. van der Merwe, and J. M. Fourie. 1995. Cyanobacterial (blue-green algae) poisoning of livestock in the Western Cape province of South Africa. J. S. Afr. Vet. Assoc. 66:260-264. [PubMed] [Google Scholar]
- 44.Wawrik, B., J. H. Paul, and F. R. Tabita. 2002. Real-time PCR quantification of rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes. Appl. Environ. Microbiol. 68:3771-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zyskind, J. W., and D. W. Smith. 1992. DNA replication, the bacterial cell cycle, and cell growth. Cell 69:5-8. [DOI] [PubMed] [Google Scholar]