Abstract
In this paper we describe a rapid method for identifying bacteria which convert free linoleic acid to conjugated linoleic acid (CLA). This method is based on spectrophotometric detection of CLA and compares well with the standard gas-liquid chromatography method. This method should facilitate high-throughput screening of bacterial isolates for the ability to produce conjugated fatty acids.
Conjugated linoleic acid (CLA), a natural component of ruminant milk and tissue fat, is a mixture of positional and geometric conjugated isomers of the essential fatty acid linoleic acid. In recent years, there has been considerable interest in these biogenic isomers due to their potential health-promoting properties and the proposed positive effects that they have on many aspects of human health, most notably their anticarcinogenic, immune modulation, antiatherosclerotic, and antiobesity activities (3, 4, 8, 10, 14-16, 23-25). In addition to the increased interest in the possible physiological effects on humans following CLA consumption, there has been a concomitant increase in interest in the isolation of novel human-derived or dairy starter cultures with the ability to produce the bioactive fatty acid (1, 7, 12, 13, 17, 19, 21). Indeed, bacterial strains belonging to a few genera, such as Lactobacillus, Propionibacterium, and Bifidobacterium, have been reported to produce CLA in either synthetic media or milk (21); however, the ability to produce CLA can vary from strain to strain. The standard gas-liquid chromatography (GLC)-based screening process is laborious and time-consuming and can be a limiting factor when a large number of strains are to be tested. In this study we describe the use of a simple and straightforward spectrophotometric method for screening a large number of culture supernatants for CLA production; this method eliminates the need for GLC during the screening process.
Fifty-eight fecal samples were obtained from a diverse population. Twenty-eight of the samples were obtained from healthy full-term neonates who were 2 to 5 days old. The remaining samples were obtained from adults, 20 of whom were elderly patients infected with the bacterium Clostridium difficile. Following sampling, fecal samples were stored at 5°C and were processed in the laboratory within 5 h. Swabs were mixed by vortexing them in maximum-recovery diluents (Oxoid, Ltd., Hampshire, United Kingdom), serially diluted, spread plated on the appropriate medium, and incubated as described below. Fifteen colonies were randomly selected from each fecal sample and were screened spectrophotometrically at a wavelength of 233 nm for CLA production following incubation in medium containing linoleic acid (see below).
Serial dilutions of fecal samples were spread plated onto modified MRS (mMRS) (Difco Laboratories, Detroit, MI) medium supplemented with 0.05% (wt/vol) cysteine hydrochloride (Sigma, St. Louis, MO), 1% (wt/vol) agar (Oxoid), and 100 μg/ml of mupirocin (Oxoid) added as antimicrobial susceptibility disks as previously described (18) to preselect for bifidobacteria. The agar plates were incubated anaerobically (anaerobic jars with Anaerocult A gas packs [Merck, Darmstadt, Germany]) at 37°C for 5 days.
Linoleic acid and the internal standard tridecanoic acid were obtained from Sigma-Aldrich (St. Louis, MO). Isomers of CLA (cis-9,trans-11 and trans-10,cis-12) were obtained from Matreya Inc. (Pennsylvania). All other chemicals were obtained from Sigma-Aldrich or Labscan (Dublin, Ireland).
Fecal isolates were incubated anaerobically in mMRS broth containing free linoleic acid (0.5 mg/ml) and 2% (wt/vol) Tween 80 at 37°C for 48 h to determine the ability of strains to convert linoleic acid to CLA, using a modification of a previously described method (17). Following incubation, 1 ml of a culture was centrifuged at 20,800 × g for 1 min, the pellet was discarded, and the supernatant was mixed with 2 ml of isopropanol by vortexing and allowed to stand for 3 min. The fatty acids were extracted by vortexing the solution and allowing it to stand for 3 min following the addition of 1.5 ml of hexane. The presence of CLA in the culture supernatant was assayed spectrophotometrically by dispensing 230 μl of the fat-soluble hexane layer into a UV-transparent 96-well plate (Costar, Corning, NY) and determining the absorbance at 233 nm using a 96-well plate spectrophotometer (GENios Plus; Tecan, Medford, MA).
The activity of fructose-6-phosphate phosphoketolase (F-6-PPK), an enzyme that indicates bifidobacterial carbohydrate metabolism, of each CLA-producing isolate was determined based on an assay described previously (5). 16S rRNA gene sequencing was performed by previously described methods (20, 22). An approximately 1.5-kb 16S rRNA gene fragment was generated using two 16S rRNA gene primers, CO1 (5′-AGTTTGATCCTGGCTCAG-3′) for the 5′ end and CO2 (5′-TACCTTGTTACGACT-3′) for the 3′ end or, alternatively, Im26-f (5′-GATTCTGGCTCAGGATGAACG-3′) for the 5′ end and Im3-r (5′-CGGGTGCTCCCACTTTCATG-3′) for the 3′ end. Alternatively, a segment of the gene coding for the 60-kDa heat shock protein HSP60 was used to characterize the strains (11). DNA sequencing was performed by Lark Technologies (United Kingdom). Strains were assigned to species following comparison of the 16S rRNA gene and HSP60 sequences using the BLAST program (2). The species identification results obtained for strains using the 16S rRNA gene and HSP60 sequences were confirmed using a modification of a previously described species-specific primer identification method for bifidobacteria (9). The only modification involved using an annealing temperature of 70°C when both primer mixtures were used.
High-molecular-weight chromosomal DNA was isolated from stationary-phase cultures by a previously described method (22). Restriction enzyme XbaI (New England Biolabs) was used to cleave chromosomal DNA, and the fragments were separated using a contour-clamped homogeneous electric field CHEF-DR III pulsed-field system (Bio-Rad Laboratories). Fragments were resolved with a linear ramp pulse time of 1 to 15 s for 18 h at 6 V/cm in a running buffer containing 0.5× Tris base-borate-EDTA maintained at 12°C. DNA fragment sizes were estimated by comparison with the lambda ladder PFG marker (New England Biolabs).
The ability of isolates to convert free linoleic acid to CLA was assayed by incubating cultures in mMRS broth containing free linoleic acid (0.5 mg/ml) at 37°C for 72 h and subsequently assessing the fatty acid profile of the culture supernatant by GLC as previously described (7).
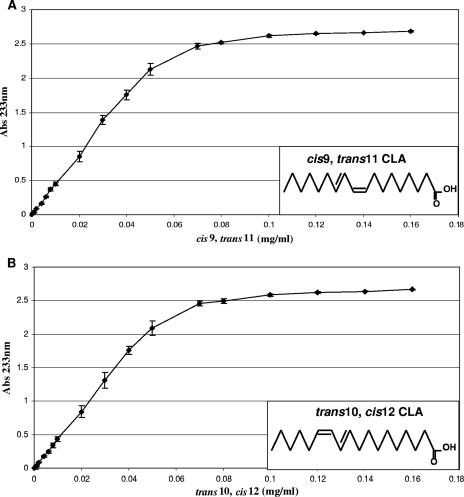
The rapid screening method employed in this study involved the use of a UV-transparent and colorless 96-well plate to detect CLA production by bacterial cultures at a wavelength of 233 nm. In order to verify the suitability of this method, a standard curve was constructed for the absorbance at 233 nm versus the CLA concentration (Fig. 1), using pure cis-9,trans-11 and trans-10,cis-12 CLA isomers. The graph demonstrated that an increase in the CLA concentration (from 0 to 0.05 mg/ml) coincided with a linear increase (R2 = 0.9985) in absorbance for the cis-9,trans-11 CLA isomer up to an absorbance of 2.1. Therefore, the CLA concentrations in culture supernatants with an absorbance at 233 nm less than or equal to 2.1 could be calculated from the linear trend line of the standard curve using the equation y = 43.431x + 0.0053. The standard curve constructed using the trans-10,cis-12 CLA isomer displayed similar results (Fig. 1). The assay was extremely sensitive and could detect cis-9,trans-11 CLA isomer concentrations as low as 0.002 mg/ml in the hexane layer with a corresponding absorbance at 233 nm of 0.0871 ± 0.004.
FIG. 1.
Standard curves for the absorbance of the fat-soluble hexane layer versus CLA concentration. (A) Absorbance at 233 nm versus concentration of the cis-9,trans-11 CLA isomer. The chemical structure of the cis-9,trans-11 CLA isomer is also shown. (B) Absorbance at 233 nm versus concentration of the trans-10,cis-12 CLA isomer. The chemical structure of the trans-10,cis-12 CLA isomer is also shown.
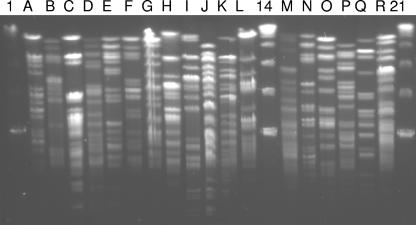
In order to determine whether this method could be used to test a large number of strains, 15 isolates were randomly selected from each of the 58 fecal samples. All of these isolates were then screened spectrophotometrically for CLA production following growth in the presence of linoleic acid. Using this approach, a total of 88 CLA-producing isolates (∼10% of the 870 isolates screened) were identified for 15 of the 58 fecal samples, while no CLA-producing isolates were detected in the remaining 43 samples using the 96-well plate assay. Pulsed-field gel electrophoresis (PFGE) was performed for all of the CLA-producing isolates following genomic restriction with the enzyme XbaI, and a total of 18 distinct CLA-producing strains were identified among the 88 initial isolates (Fig. 2). Subsequently, each strain was identified to the genus and species levels by partial sequencing of either the 60-kDa heat shock protein HSP60 or the 16S rRNA gene (Table 1). All of the CLA-producing bacteria exhibited levels of homology of ≥97% with different species belonging to the genus Bifidobacterium, and all of them were found to be F-6-PPK positive, a characteristic biomarker for the genus (Table 1). Nine of the CLA-producing strains belonged to B. longum, six strains belonged to B. breve, and a single strain each belonged to B. infantis, B. dentium, and B. catenulatum. These results were confirmed by Bifidobacterium species-specific PCRs using 16S rRNA gene-based primers (data not shown).
FIG. 2.
PFGE macrorestriction patterns following genomic DNA digestion with restriction enzyme XbaI. Lanes A through L, PFGE types A to L, respectively; lanes M through R, PFGE types M to R, respectively. Lanes 1, 14, and 21 contained molecular size markers (lambda ladder PFG marker).
TABLE 1.
Description, PFGE patterns, 16S rRNA gene and HSP60 partial sequencing, F-6-PPK detection, subject age, and CLA conversion by the strains
| % Conversion | A233 | PFGE type | 16S rRNA gene-HSP60 partial sequencing and Bifidobacterium species-specific PCR identification | F-6-PPK assay | % Homology | Age of donor | Sample(s) | Subject description |
|---|---|---|---|---|---|---|---|---|
| 76.65 ± 1.75 | 2.39 | A | B. breve | Positive | 98 | 81 yr | 1, 3, 43 | C. difficile positive |
| 61.12 ± 3.85 | 2.38 | C | B. breve | Positive | 99 | 50 yr | 16 | C. difficile positive |
| 60.12 ± 5.14 | 2.40 | E | B. longum | Positive | 99 | 25 yr | 22 | Healthy adult |
| 53.08 ± 2.51 | 2.38 | B | B. longum | Positive | 99 | 64 yr | 7 | C. difficile positive |
| 44.65 ± 2.57 | 2.41 | H | B. breve | Positive | 99 | 63 yr | 26 | C. difficile positive |
| 38.50 ± 0.96 | 2.41 | I | B. longum | Positive | 99 | 3 days | 30 | Healthy baby |
| 27.20 ± 8.81 | 2.37 | G | B. breve | Positive | 97 | 73 yr | 23 | C. difficile positive |
| 21.53 ± 2.37 | 2.30 | O | B. breve | Positive | 99 | 81 yr | 43 | C. difficile positive |
| 20.16 ± 3.77 | 2.27 | J | B. longum | Positive | 100 | 3 days | 30, 35 | Healthy baby |
| 18.66 ± 0.26 | 2.27 | K | B. longum | Positive | 99 | 5 days | 34 | Healthy baby |
| 18.11 ± 1.40 | 2.23 | D | B. infantis | Positive | 98 | 31 yr | 17 | Healthy adult |
| 12.55 ± 0.42 | 2.05 | L | B. dentium | Positive | 99 | 4 days | 35 | Healthy baby |
| 6.31 ± 0.92 | 1.19 | R | B. longum | Positive | 100 | 4 days | 52 | Healthy baby |
| 5.24 ± 4.79 | 1.42 | N | B. longum | Positive | 99 | 4 days | 36 | Healthy baby |
| 4.12 ± 0.68 | 0.86 | P | B. breve | Positive | 99 | 81 yr | 43 | C. difficile positive |
| 3.98 ± 2.64 | 0.54 | F | B. longum | Positive | 99 | 25 yr | 22 | Healthy adult |
| 3.68 ± 0.41 | 0.57 | Q | B. longum | Positive | 100 | 37 yr | 47 | Healthy adult |
| 2.60 ± 1.00 | 0.55 | M | B. pseudocatenulatum | Positive | 99 | 4 days | 36 | Healthy baby |
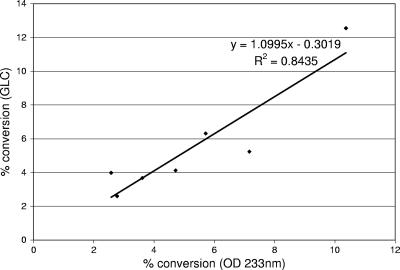
In addition to the standard curve, the suitability of the 96-well plate assay for determining the CLA content of a culture broth was compared with the suitability of GLC. The percentage of linoleic acid converted to CLA by each individual stain, as determined by GLC, was compared with the absorbance at 233 nm obtained by analysis of the hexane layer during the initial step of fatty acid extraction prior to drying under nitrogen, and the data are shown in Table 1. Strains are listed in Table 1 in descending order based on the percentage of linoleic acid converted to CLA. The CLA conversion values for the seven strains with an absorbance at 233 nm of less than 2.2 (Table 1) were calculated from the linear trend line equation (y = 43.431x + 0.0053) of the standard curve for the cis-9,trans-11 CLA isomer. The conversion values calculated from the standard curve were plotted against the conversion values determined by GLC (Fig. 3).
FIG. 3.
Plot of CLA conversion as determined by GLC versus CLA conversion calculated from the standard curve for bacterial samples with an absorbance at 233 nm (OD 233nm) of less than 2.1.
In all cases, the cis-9,trans-11 CLA isomer was the predominant isomer in the culture supernatant; the trans-9,trans-11 CLA isomer was also generated, however, at much lower levels, as determined by GLC (data not shown). Interestingly, we have recently shown that the latter isomer also has very potent antiproliferative activity against a human colonic cancer cell line (6). A number of non-CLA-producing bacteria were also assayed to determine their CLA production; using both GLC and the spectrophotometric assay, no CLA production was detected in these strains (data not shown). Importantly, it should be emphasized that the spectrophotometric method does not distinguish between isomers of CLA since it is based on measurement of the conjugated double bond in the fatty acid.
Altogether, nine strains converted more than 20% of the linoleic acid to CLA in the culture supernatant, and one of these strains converted more than 75% of the linoleic acid to CLA. Five of the CLA-producing strains which converted more than 20% of the linoleic acid to CLA belong to the species B. breve, and the remaining four strains were identified as B. longum strains.
The source of the CLA-producing bifidobacteria in this study was a further point of interest. In previous studies workers have isolated CLA-producing bifidobacteria from the fecal material of neonates (17, 19); however, the best source of CLA-producing bifidobacteria in this study, in terms of both conversion of linoleic acid to CLA and prevalence of CLA-producing strains within a group, was the fecal material of elderly patients infected with the bacterium C. difficile. In this study only 5 of the 28 healthy infants screened for CLA-producing bifidobacteria harbored CLA-producing strains, while 7 of the 20 C. difficile patients screened harbored CLA-producing strains and 3 of the 10 healthy adults harbored CLA-producing bifidobacteria. Indeed, in the diverse population screened in this study, one in four subjects harbored a CLA-producing Bifidobacterium strain.
Currently, there is a need in intestinal microbiology to develop high-throughput methods to identify specific bioactivities in certain subpopulations. Therefore, a high-throughput spectrophotometric method for analyzing the CLA production capabilities of gut microfloras was developed. This method allows rapid screening of large numbers of bacterial isolates for CLA production. Using the method which we developed, in this study the prevalence and diversity of CLA-producing Bifidobacterium species in the gastrointestinal tracts of human subjects were highlighted.
Acknowledgments
This work was funded by Science Foundation Ireland and the Irish Government under National Development Plan 2000-2006 and by the European Research and Development Fund (EU project QLK1-2002-02362).
The technical assistance of Alan Hennessy, Mairead Coakley, and Seamus Aherne is gratefully acknowledged.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Alonso, L., E. P. Cuesta, and S. E. Gilliland. 2003. Production of free conjugated linoleic acid by Lactobacillus acidophilus and Lactobacillus casei of human intestinal origin. J. Dairy Sci. 86:1941-1946. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bassaganya-Riera, J., R. Hontecillas, and D. C. Beitz. 2002. Colonic anti-inflammatory mechanisms of conjugated linoleic acid. Clin. Nutr. 21:451-459. [DOI] [PubMed] [Google Scholar]
- 4.Belury, M. A. 2002. Inhibition of carcinogenesis by conjugated linoleic acid: potential mechanisms of action. J. Nutr. 132:2995-2998. [DOI] [PubMed] [Google Scholar]
- 5.Bibiloni, R., P. F. Perez, and G. L. De Antoni. 2000. An enzymatic-colorimetric assay for the quantification of Bifidobacterium. J. Food Prot. 63:322-326. [DOI] [PubMed] [Google Scholar]
- 6.Coakley, M., M. C. Johnson, E. McGrath, S. Rahman, R. P. Ross, G. F. Fitzgerald, R. Devery, and C. Stanton. 2006. Intestinal bifidobacteria that produce trans-9,trans-11 CLA: a fatty acid with anti-proliferative activity against SW480 and HT-29 colon cancer cells. Nutr. Cancer 56:95-102. [DOI] [PubMed] [Google Scholar]
- 7.Coakley, M., R. P. Ross, M. Nordgren, G. Fitzgerald, R. Devery, and C. Stanton. 2003. Conjugated linoleic acid biosynthesis by human-derived Bifidobacterium species. J. Appl. Microbiol. 94:138-145. [DOI] [PubMed] [Google Scholar]
- 8.Gaullier, J. M., J. Halse, K. Hoye, K. Kristiansen, H. Fagertun, H. Vik, and O. Gudmundsen. 2004. Conjugated linoleic acid supplementation for 1 y reduces body fat mass in healthy overweight humans. Am. J. Clin. Nutr. 79:1118-1125. [DOI] [PubMed] [Google Scholar]
- 9.Germond, J. E., O. Mamin, and B. Mollet. 2002. Species specific identification of nine human Bifidobacterium spp. in feces. Syst. Appl. Microbiol. 25:536-543. [DOI] [PubMed] [Google Scholar]
- 10.Ip, M. M., P. A. Masso-Welch, and C. Ip. 2003. Prevention of mammary cancer with conjugated linoleic acid: role of the stroma and the epithelium. J. Mammary Gland Biol. Neoplasia 8:103-118. [DOI] [PubMed] [Google Scholar]
- 11.Jian, W., L. Zhu, and X. Dong. 2001. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 51:1633-1638. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, J., L. Bjorck, and R. Fonden. 1998. Production of conjugated linoleic acid by dairy starter cultures. J. Appl. Microbiol. 85:95-102. [DOI] [PubMed] [Google Scholar]
- 13.Kishino, S., J. Ogawa, Y. Omura, K. Matsumura, and S. Shimizu. 2002. Conjugated linoleic acid production from linoleic acid by lactic acid bacteria. J. Am. Oil Chem. Soc. 79:159-163. [Google Scholar]
- 14.Kritchevsky, D. 2000. Antimutagenic and some other effects of conjugated linoleic acid. Br. J. Nutr. 83:459-465. [PubMed] [Google Scholar]
- 15.Lee, K. N., D. Kritchevsky, and M. W. Pariza. 1994. Conjugated linoleic acid and atherosclerosis in rabbits. Atherosclerosis 108:19-25. [DOI] [PubMed] [Google Scholar]
- 16.Miller, A., C. Stanton, J. Murphy, and R. Devery. 2003. Conjugated linoleic acid (CLA)-enriched milk fat inhibits growth and modulates CLA-responsive biomarkers in MCF-7 and SW480 human cancer cell lines. Br. J. Nutr. 90:877-885. [DOI] [PubMed] [Google Scholar]
- 17.Oh, D. K., G. H. Hong, Y. Lee, S. Min, H. S. Sin, and S. K. Cho. 2003. Production of conjugated linoleic acid by isolated Bifidobacterium strains. World J. Microbiol. Biotechnol. 19:907-912. [Google Scholar]
- 18.Rada, V. 1997. Detection of Bifidobacterium species by enzymatic methods and antimicrobial susceptibility testing. Biotechnol. Tech. 11:909-912. [Google Scholar]
- 19.Rosberg-Cody, E., R. P. Ross, S. Hussey, C. A. Ryan, B. P. Murphy, G. F. Fitzgerald, R. Devery, and C. Stanton. 2004. Mining the microbiota of the neonatal gastrointestinal tract for conjugated linoleic acid-producing bifidobacteria. Appl. Environ. Microbiol. 70:4635-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieber, R., M. Collomb, A. Aeschlimann, P. Jelen, and H. Eyer. 2004. Impact of microbial cultures on conjugated linoleic acid in dairy products—a review. Int. Dairy J. 14:1-15. [Google Scholar]
- 22.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2003. Genomic diversity and relatedness of bifidobacteria isolated from a porcine cecum. J. Bacteriol. 185:2571-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terpstra, A. H. 2004. Effect of conjugated linoleic acid on body composition and plasma lipids in humans: an overview of the literature. Am. J. Clin. Nutr. 79:352-361. [DOI] [PubMed] [Google Scholar]
- 24.Thom, E., J. Wadstein, and O. Gudmundsen. 2001. Conjugated linoleic acid reduces body fat in healthy exercising humans. J. Int. Med. Res. 29:392-396. [DOI] [PubMed] [Google Scholar]
- 25.Wahle, K. W., S. D. Heys, and D. Rotondo. 2004. Conjugated linoleic acids: are they beneficial or detrimental to health? Prog. Lipid Res. 43:553-587. [DOI] [PubMed] [Google Scholar]