Abstract
All activated sludge systems for removing phosphate microbiologically are configured so the biomass is cycled continuously through alternating anaerobic and aerobic zones. This paper describes a novel aerobic process capable of decreasing the amount of phosphate from 10 to 12 mg P liter−1 to less than 0.1 mg P liter−1 (when expressed as phosphorus) over an extended period from two wastewaters with low chemical oxygen demand. One wastewater was synthetic, and the other was a clarified effluent from a conventional activated sludge system. Unlike anaerobic/aerobic enhanced biological phosphate removal (EBPR) processes where the organic substrates and the phosphate are supplied simultaneously to the biomass under anaerobic conditions, in this aerobic process, the addition of acetate, which begins the feed stage, is temporally separated from the addition of phosphate, which begins the famine stage. Conditions for establishing this process in a sequencing batch reactor are detailed, together with a description of the changes in poly-β-hydroxyalkanoate (PHA) and poly(P) levels in the biomass occurring under the feed and famine regimes, which closely resemble those reported in anaerobic/aerobic EBPR processes. Profiles obtained with denaturing gradient gel electrophoresis were very similar for communities fed both wastewaters, and once established, these communities remained stable over prolonged periods of time. 16S rRNA-based clone libraries generated from the two communities were also very similar. Fluorescence in situ hybridization (FISH)/microautoradiography and histochemical staining revealed that “Candidatus Accumulibacter phosphatis” bacteria were the dominant poly(P)-accumulating organisms (PAO) in both communities, with the phenotype expected for PAO. FISH also identified large numbers of betaproteobacterial Dechloromonas and alphaproteobacterial tetrad-forming organisms related to Defluviicoccus in both communities, but while these organisms assimilated acetate and contained intracellular PHA during the feed stages, they never accumulated poly(P) during the cycles, consistent with the phenotype of glycogen-accumulating organisms.
High levels of phosphate in effluents from activated sludge systems not designed to remove it can lead to toxic cyanobacterial blooms in receiving bodies of water. Consequently, efforts have been directed towards removing phosphate during treatment by microbiological means with a process called enhanced biological phosphorus removal (EBPR), where phosphate is removed from the wasted biomass as intracellular poly(P) (5, 37, 45). Such treatment processes are based on the underlying principle that the biomass needs to be recycled repeatedly through alternating anaerobic and aerobic stages (37), a requirement regarded as crucial for successful EBPR operation. Only after repeated recycling are poly(P)-accumulating organisms (PAO) thought to have a selective advantage over other populations, eventually allowing them to become dominant (5, 37, 45). In the anaerobic (feed) stage, PAO are believed to assimilate preferentially volatile fatty acids, such as acetate entering the system in the influent and probably also being produced there by bacterial fermentation. These volatile fatty acids are not used for cell growth but for the synthesis of intracellular carbon/energy reserves of poly-β-hydroxyalkanoate (PHA), where intracellular poly(P) is degraded to provide energy, and phosphate is released into the bulk liquid. Then in the subsequent aerobic (famine) stage, where extracellular soluble substrates are scarce, bacteria containing carbon/energy reserves of PHA can grow and assimilate phosphate, which is converted to intracellular poly(P) stores (37, 45). As more phosphate is assimilated aerobically than is released anaerobically, a net removal of phosphate occurs. Evidence also suggests that glycogen provides energy and reducing power for anaerobic PHA synthesis in the PAO and is subsequently replenished in the aerobic phase (5, 37, 45).
Betaproteobacterial Rhodocyclus-related “Candidatus Accumulibacter phosphatis” organisms are numerically important PAO in laboratory-scale EBPR sequencing batch reactors (SBRs) fed synthetic wastewater with acetate as the sole carbon/energy source (12, 21) and also in some sewage-fed, full-scale systems (24) having ecophysiological features consistent with the chemical transformations described above (12, 24, 52). Evidence from other full-scale systems suggests that PAO may be Actinobacteria, possessing anaerobic ecophysiological features different from those of “Candidatus Accumulibacter phosphatis” organisms (25, 45).
However, because many existing systems around the world do not employ EBPR, the problem of phosphate pollution still remains. These systems produce effluents with a low chemical oxygen demand (COD), but they still contain an unacceptably high level of phosphate and are often treated with metal ions to precipitate the phosphate, a strategy that generates additional sludge for disposal and increases the salinity of the final effluent (16, 45).
One option would be to replace these systems with anaerobic/aerobic EBPR processes, although a more cost-effective strategy might be to develop a dedicated microbiological phosphate removal system which could be used as an “add on” downstream of the existing system. To this end, an SBR was used to develop a novel, aerobic, cyclic process to remove phosphate from low-COD wastewaters (34a).
Here we present in detail the chemical changes occurring during a typical complete cycle of the aerobic process carried out in a laboratory-scale SBR fed either synthetic phosphate-containing low-COD “wastewater” or clarified effluent from a conventional activated sludge system, with acetate being the separately added carbon/energy source with both feeds. Changes in the compositions of the microbial communities from inoculation to achievement of the climax population were followed with denaturing gradient gel electrophoresis (DGGE) profiling, 16S rRNA-based clone library analyses, and fluorescence in situ hybridization (FISH) to identify which were the PAO. The in situ ecophysiology features of the members of these communities are also described and compared to those identified in anaerobic/aerobic EBPR processes.
MATERIALS AND METHODS
Reactor feeds.
Concentrations of orthophosphate, acetate, and ammonium chloride are expressed in terms of phosphorus (P), carbon (C), and nitrogen (N), respectively.
Two types of phosphate-containing feeds were used. The first feed, a synthetic wastewater, was based on the chemically defined medium described by Bayly et al. (3), except that the phosphate level was reduced to 10 mg P liter−1, the ammonium chloride (the nitrogen source) was adjusted to a final feed level (see below) of 20 to 32 mg N liter−1, and the sodium acetate was omitted. The second feed was the clarified effluent from a non-EBPR activated sludge plant in Melton, Victoria, Australia. This effluent contained 11 to 12 mg P liter−1. With both feeds, the acetate was added separately at the appropriate time in the SBR cycle (see below) to give a concentration equivalent to 160 mg C liter−1 in the reactor-mixed liquor.
Sequencing batch reactors and control systems.
Each SBR consisted of a vessel and a stirrer drive unit (series 500 and model 502D, respectively; LH Fermentation, United Kingdom) and a control unit (see below). Each reactor was stirred (except during the settling of the biomass and when clarified phosphate-depleted liquor was pumped out) at 300 rpm and aerated at 710 ml min−1. The pH of the reactor-mixed liquor was maintained at 7.5 by using a pH controller (model 505; LH Fermentation, United Kingdom) and by adding 0.25 M HCl. The pO2 of the mixed liquor was monitored with a galvanic G2 oxygen electrode (Uniprobe, United Kingdom), and redox potential with an IJ64 redox probe (Ionde, Queensland). The sequence of events during a reactor cycle (pump-in and pump-out of liquids and shutdown of airflow and stirrer during both the settling of biomass and pump-out of clarified liquor) was controlled by a series of synchronized electronic timers (type PC787; Arlec, Australia).
The reactors were operated at room temperature (about 20°C) on a cycle time of 8 h, with each cycle consisting of five stages. Stage 1 (famine) allowed 99 min for phosphate uptake (phosphate-containing feed was pumped in during the first 5 min of this stage); stage 2 allowed 1 min for mixed liquor to be pumped out; stage 3 (feed) allowed 320 min for the utilization of acetate (acetate was pumped in during the first 4 min of this stage); stage 4 allowed 45 min for the settling of biomass; and stage 5 allowed 15 min for clarified phosphate-depleted liquor to be pumped out. During stages 4 and 5, the reactor stirrer and air supply were turned off. The maximum working volume of the reactor was 1,500 ml, the volume of phosphate-containing feed added to a reactor was 750 ml, and the volumes of mixed liquor and clarified, phosphate-depleted liquor removed from the reactor were 25 ml and 725 ml, respectively. These values gave a hydraulic retention time of 16 h and a mean cell retention time (sludge age) of 20 days.
Startup of reactors.
The inoculum to start the reactors was obtained from a full-scale anaerobic/aerobic EBPR system (modified University of Cape Town type) located at Castlemaine, Victoria, Australia. The system produced an effluent with a phosphate concentration of less than 0.5 mg P liter−1. During the startup of the reactors, the SBR plus the synthetic wastewater containing 10 mg P liter−1 was operated in a batch mode until the phosphate concentration there was maintained at less than 0.1 mg P liter−1 for at least 24 h. Influent phosphate concentrations were then gradually increased stepwise to 10 mg P liter−1, and influent N concentrations (supplied as ammonium chloride, NH4Cl) increased in parallel from 8 to 32 mg N liter−1, with a corresponding increase in the amount of acetate provided from 40 to 160 mg C liter−1, as shown in Fig. 1a. An initial C-to-N ratio of 5:1 was adopted during startup and then gradually increased to 8:1 by decreasing the NH4Cl levels, where stable performance was obtained, and no N was detectable in the treated effluent. Stable SBR performance was achieved after 15 to 20 days (Fig. 1a), where 10 mg P liter−1 was almost always completely removed, as described below. The same startup protocol was used with the process to treat the clarified effluent from a non-EBPR activated sludge treatment plant, and the same stepwise increases in the acetate reactor concentration and phosphate levels in the appropriately diluted clarified effluent feed were used (Fig. 1b).
FIG. 1.
(a) Time profile showing the incremental increase of influent phosphate (—) and influent acetate (- - - - ) during the startup period for the SBR using synthetic wastewater. The levels of phosphate contained in the effluent (○) are also given. (b) Time profile showing the incremental increase of influent phosphate (—) and influent acetate (- - - - ) during the startup period for the SBR using clarified effluent from a non-EBPR activated sludge plant. The levels of phosphate contained in the effluent (○) are also given. d, days.
Chemical analyses of reactor samples.
Analyses of phosphate and ammonium nitrogen levels were all performed according to standard methods (9). Acetate was analyzed by ion chromatography (LC-10Ai; Shimadzu, Japan) and fitted with an anion column (Shodex KC-811; Showa Denko, Japan) and a diode array detector (SPD-M10AVP; Shimadzu, Japan).
To determine the intracellular levels of PHA and glycogen, biomass samples were collected at appropriate times during the SBR cycles (see Results) and immediately frozen in a mixture of dry ice and methanol, followed by lyophilization. For quantifying PHA, biomass was incubated at 100°C for 24 h with 1:1 chloroform-acidified methanol (10% sulfuric acid) (44). The identification and quantification of PHA methyl esters were conducted in a 1:10 split mode by a gas chromatography system (model 3900; Varian) equipped with a ChromPack capillary column (CP-Sil5CB; Varian) and a flame ionization detector. Poly(3-hydroxybutyric acid-co-3-hydroxyvaleric acid) (Aldrich) and sodium 3-hydroxybutyrate (Lancaster, United Kingdom) were used as standards. For quantifying glycogen, sludge samples were autoclaved with 0.6 N HCl at 121°C for 60 min. After cooling to room temperature and adjusting the pH to 7 with 0.6 M NaOH, We measured glycogen concentrations as glucose equivalents by using a hexokinase enzymatic glucose kit (Thermo, Australia).
Generation of 16S rRNA-based clone libraries of communities.
We followed the manufacturer's instructions (MoBio UltraClean soil DNA kit; GeneWorks, Adelaide, Australia) to extract DNA from samples taken at the end of the feed stage of the SBR cycle from both communities. Clone libraries containing almost complete 16S rRNA gene inserts were constructed with the 27f and 1525r primers of Lane (30). PCRs (total volume, 50 μl) were performed in 0.2-ml, thin-walled PCR tubes with an iCycler thermocycler (Bio-Rad) and a reaction mix containing 1× AmpliTaq Gold reaction buffer (Applied Biosystems), 200 μM deoxynucleoside triphosphates (Promega), 25 pmol of each primer, 1.5% (vol/vol) dimethyl sulfoxide (Sigma), 2.5 U AmpliTaq Gold Taq polymerase (Applied Biosystems), 2.5 mM MgCl2, and 5 μl of template DNA prepared from a 1:200 dilution of the extracted DNA. The following PCR protocol was applied to all reactions: 96°C for 10 min, followed by 30 cycles of denaturation at 96°C for 1 min, annealing at 52°C for 30 s, and extension at 72°C for 2 min. A 7-min extension was performed at the end of the final cycle. Negative controls containing no template DNA were included. A 16S rRNA clone library was also constructed with primers targeting populations of the betaproteobacterial “Candidatus Accumulibacter phosphatis” but only for the community in the reactor fed the synthetic wastewater. The primers PAO462f (GTTAATACCCTGWGTAGATGACGG) and PAO846r (GTTAGCTACGGCACTAAAAGG) were designed from the PAO FISH probes of Crocetti et al. (12), generating PCR products of about 380 nucleotides (nt). PCR conditions were the same as those for the universal primers described above.
All PCR products were purified with the Concert rapid PCR purification system (Life Technologies) and cloned into the p-GEM-T Easy vector system II (Promega) following the manufacturers' instructions. Plasmids were extracted with the QIAprep Spin Miniprep kit (QIAGEN), and the sizes of the DNA inserts were confirmed (approximately 1,500 nt for the universal primers and 380 nt for the “ Candidatus Accumulibacter”-targeted primers) after digestion with EcoRI. Sequencing of 16S rRNA gene fragments was as described previously (26), and completed reactions were analyzed by AgGenomics (Bundoora, Melbourne, Australia). For clone libraries generated with universal primers, partial, one-directional reads of about the first 500 bp of the 16S rRNA gene were obtained initially, using either pUC/m13f or pUC/m13r primers. Almost complete 16S rRNA gene sequences were then generated from each unique partial sequence by using the 16S rRNA gene internal primers (519r, 530f, and 927f) of Lane (30). With the “Candidatus Accumulibacter” primers, sequences of the expected length of about 380 nt were obtained and the m13f and m13r primers were used to sequence them. Sequences were constructed with DNAStar software (SeqMan, version 4.05 for Windows) and aligned with the NAST alignment tool (17), and chimera formation was checked with the Bellerophon program. Both NAST and Bellerophon were accessed through the Greengenes website (18). Bootstrapped neighbor-joining relationships were estimated with MEGA, version 3.1 (29). Clones sharing sequences with similarities greater than 97% were grouped into individual operational taxonomic units (OTUs) for representation in the phylogenetic tree.
DGGE profiling of communities.
DNA samples were acquired as described above. Amplification of the partial 16S rRNA gene sequences spanning the v3 region was achieved with the primers 341f and 534r, with the addition of a GC clamp to the forward primer, as detailed by Muyzer et al. (39). The PCR cycle parameters were also those of Muyzer et al. (39), except that an annealing temperature of 52°C and an extension period of 30 s were used. DGGE gels were cast and run essentially as described previously (26) using the DCode universal mutation detection system (Bio-Rad, Sydney, Australia). A denaturation potential of 30 to 70% (100% denaturation 10 M urea and 40% [vol/vol] formamide) was established in an 8% acrylamide solution (Sigma). Gels were stained for 30 min in 1× Tris-acetate-EDTA containing a 1× concentration of SYBR Gold (Invitrogen, Melbourne, Australia). Fragments in DGGE bands were tentatively identified (see Results) by comparing their migratory positions with those of bands generated from the completely sequenced and identified 16S rRNA clones as described earlier (4, 26).
FISH analyses of EBPR communities.
Biomass samples taken at various stages of SBR cycles during their stable operational periods were fixed at 4°C for 3 h. For gram-negative bacteria, 4% formaldehyde in phosphate-buffered saline (PBS) was used and, for gram-positive bacteria, 50% ethanol in PBS was used (2). FISH analyses followed the protocol of Amann (2). Details on probe sequences and hybridization conditions are available upon request (33). The DECH454 (5′ ACCGTCATCCGCACAGGG 3′) and DECH472 (5′ CCGTCATCCACACCCTGTATTAAA 3′) probes, designed and validated in this study for use against the Dechloromonas clone sequences revealed in the clone libraries (Fig. 4a and b), both used 35% formamide (see Results). Hybridization conditions for the others were those given in their original descriptions. 16S rRNA-targeted probes targeting Dechloromonas-related organisms (see Results) were designed and validated as previously described by Kong et al. (26). Probes were labeled with Cy5, Cy3, or FLUOS [5(6)-carboxyfluorescein-N-hydroxy-succinimide ester] fluorochromes and purchased from Prooligo. Vectashield (Vector Laboratories) was used to mount samples, which were examined with either a Nikon Eclipse 800 epifluorescence microscope or a Leica TCS SP2 confocal scanning laser microscope (model DM IRE2). Populations were quantified with the confocal scanning laser microscope after FISH with Daime software (15), and each was expressed as a percentage of the total area fluorescing with the EUBmix FISH probes (13), based on examining at least 20 fields of view for each sample at a magnification of ×400.
FIG. 4.
(a) Neighbor-joining tree of the sequences (OTUs) obtained from the SBR community fed synthetic wastewater sampled after 28 days and all other similar sequences obtained from the Greengenes database. Bootstrap values are given as a percentage of 1,000 analyses. (b) Neighbor-joining tree of the sequences obtained from the SBR community sampled after 20 days and fed clarified effluent from a conventional activated sludge plant and all other similar sequences obtained from the Greengenes database. Bootstrap values are given as a percentage of 1,000 analyses.
Ecophysiology of aerobic EBPR communities using FISH/MAR.
Microautoradiography (MAR)-FISH was carried out as previously described by Lee et al. (31) and further detailed by Kong et al. (24), with some modifications. Biomass samples taken from the reactor at various times during the cycles were incubated with radiolabeled substrates under fully aerobic conditions. Experimental conditions for MAR were chosen to most closely resemble those existing in the reactor for each sample examined. The radiolabeled substrate was added to biomass samples to a final concentration of 0.5 μCi mg−1 mixed-liquor-suspended solids (MLSS), with [14C]acetate and 10 μCi mg−1 MLSS with 33P, where MLSS concentrations were adjusted to those levels present in the SBRs at that sample time. For experiments involving acetate assimilation, samples were taken from the reactor immediately before the acetate addition (feed) stage. They were then incubated with a mixture of cold sodium acetate and 1-14C-labeled acetic acid (Amersham Pharmacia Biotech) at a final concentration of 6.6 mM, which is the same as the reactor concentration after acetate addition. Samples were incubated for 45 min and 4 h, to mimic the time taken for all influent acetate to be consumed and for the duration of the feed phase before the addition of P to the reactor, respectively.
To identify populations assimilating P, biomass samples were taken immediately before the P addition (famine) stage and incubated with sodium dihydrogen orthophosphate and 33P-labeled orthophosphate (Amersham Biosciences) to a final concentration of 0.322 mM, which is the reactor concentration after the addition of P. Samples were incubated for 45 and 90 min, reflecting the time taken for all P in the system to be consumed and the duration of the famine phase before the settling phase and subsequent acetate addition, respectively. All MAR incubations were carried out on an orbital mixer at room temperature. Controls allowing for possible chemography (24) were incorporated into all experiments, with biomass samples pasteurized at 70°C for 10 min before the addition of substrates. Samples were fixed as described above, washed three times with 1× PBS for acetate incubations and 0.1 M sodium citrate-HCl buffer (pH 2) for 33P incubations (24), and subsequently stored at −20°C. MAR samples were counted with a liquid scintillation counter (Wallac 1450 Microbeta Plus) to confirm the absence of substrate uptake in control samples (19).
FISH probing of biomass after MAR incubations was as described by Kong et al. (24). Samples were homogenized to break up clustered cells by briefly rubbing a gelatin-coated coverslip and a glass slide together with 30 μl sample in between and then dehydrated with ethanol (50%, 80%, and absolute for 3 min each) (31) before hybridization with FISH probes (2). Slides were coated with LM-1 emulsion (Amersham Biosciences), incubated for 3, 6, and 9 days at 4°C for [1-14C]acetate and 33P incubations (24), and developed by immersion in Kodak D-19 developer for 2 to 3 min, followed by 1 min in water and 4 min in sodium thiosulfate, with a final 10-min wash in water (31). Slides were dried and viewed as described above. Cells giving a positive MAR response with 33P after 3 or 6 days were considered putative PAO, following suggestions of Kong et al. (24).
Cytochemical staining.
Staining for intracellular poly(P) was carried out with 4′,6-diamidino-2-phenylindole (DAPI) simultaneously with FISH, as previously described by Kawaharasaki et al. (23) and Liu et al. (32). After FISH hybridization, cells were immersed in 10 ppm DAPI for 10 min before being rinsed in water and air dried. Viability staining with the Live/Dead system (Molecular Probes) followed the method of Carr et al. (7). Staining for PHA was performed by the method of Ostle and Holt (41), with some modifications. Fixed biomass samples were submerged in prewarmed Nile blue A (100 mg liter −1) in ethanol at 55°C for 10 min, followed by rinsing with water and washing for 1 min in 8% acetic acid to remove excess stain. Slides were viewed as described before and then destained by immersion in absolute ethanol for 30 min. FISH was then performed as described above, with cells that were identified earlier as being PHA positive or negative being relocated on the same slide.
RESULTS
Startup and stabilization of the aerobic phosphorus removal process.
Data from the startup phase of this process (feeding with synthetic wastewater eventually containing 10 mg P liter−1) showed that once established, the phosphate removal efficiency was always close to 100% (Fig. 1a). The data given are for the first 60 days of operation, during which the initial phosphate and acetate levels were gradually increased to allow the EBPR community to become functionally established, but similar stable operation was maintained for more than 3 months. The highest measured effluent phosphate level was 0.06 mg P liter−1, but in most daily samples, no phosphate could be detected. Similar results were obtained for the SBR treating the clarified plant effluent (Fig. 1b). GC analyses of biomass samples from both systems revealed that poly-β-hydroxybutyrate (PHB) was the only PHA synthesized by the microbial communities under these conditions (data not shown).
Chemical transformations occurring during the aerobic SBR cycle. (i) Using synthetic wastewater.
A typical profile showing the chemical transformations occurring during both feed and famine stages of the aerobic SBR cycle after 28 days of operation is given in Fig. 2a. The system was operating steadily at that time, and DGGE profiles (see Fig. 5) suggested that the community composition was stable. Repeated sample analyses at regular intervals always gave the same patterns, and standard errors (SE) for replicates for each individual parameter were always within ±2%. During the feed stage, the acetate was rapidly assimilated within 44 min after addition and used for the synthesis of intracellular PHB, whose levels rose (Fig. 2a). A carbon mass balance based on existing stoichiometric equations (46) showed that about 98% of the acetate assimilated by this biomass was converted to PHB. During acetate assimilation, phosphate was released from the biomass into the bulk liquid (0.12 mmol P released/mmole acetate utilized), which also happens in the feed stage of anaerobic/aerobic EBPR processes, but at a slightly lower P-to-acetate ratio (48). Complete release of intracellular phosphate did not occur (Fig. 2a).
FIG. 2.
(a) Chemical profile of a typical SBR cycle from the stable operational period of the system using synthetic wastewater showing the changes in acetate levels (▴), P content of biomass (□), phosphate content of mixed liquor (○) PHB content of biomass (▵), and glycogen content of biomass (▿) over a feed/famine cycle. The downward facing arrows indicate where acetate and the synthetic wastewater were added to the reactor. (b) Dissolved oxygen (—) and redox potential (- - - - ) profiles over a feed/famine period for the SBR fed synthetic wastewater.
FIG. 5.
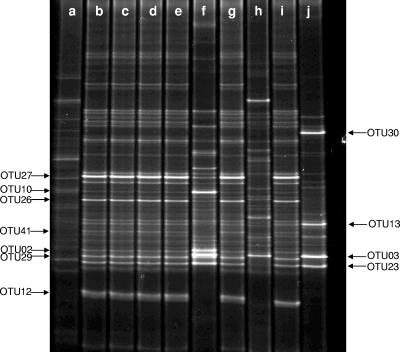
DGGE profiles of partial 16S rRNA gene, PCR-amplified fragments using universal primers with DNA extracted from biomass samples collected from the reactor fed synthetic wastewater at (a) day 0, (b) day 11 with a feed containing 5 mg P liter−1 (c) day 14 with a feed containing 8 mg P liter−1, (d) day 28 with a feed containing 10 mg P liter−1, (e) day 36 with a feed containing 10 mg P liter−1, (f) day 55 where the effluent contained over 1 mg P liter−1, (g) day 65 where the P-removing capacity of the system recovered; and samples collected from the reactor supplemented with clarified effluent from a conventional activated sludge plant with a feed containing 10 mg P liter−1 at (h) day 0 and (i) under stable operation at day 20, and (j) day 28 where the effluent contained over 1 mg P liter−1.
With the exhaustion of acetate and with phosphate now available in the medium, populations containing PHB grew by respiring it aerobically as their carbon and energy sources, as happens in the famine stage of anaerobic/aerobic EBPR processes (6). Intracellular PHB levels fell until all of the released phosphate had been reutilized (Fig. 2a). Then its rate of utilization slowed and eventually ceased, presumably because of eventual phosphate exhaustion from the medium. Crucially, not all of the intracellular PHB was exhausted during this feed phase (Fig. 2a).
FIG. 6.
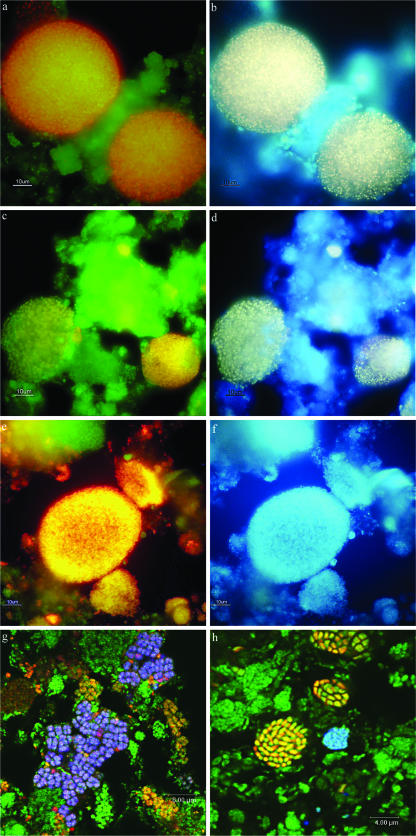
Micrographs of in situ analyses of aerobic phosphorus removal reactor sludge communities. (a and c) Images showing cells hybridizing with both EUBmix (FLUOS) and PAOmix (Cy3) probes, from the reactors fed the synthetic wastewater and the clarified effluent, respectively. Cells that are yellow have hybridized with both probes. (b and d) Same fields of view where PAOmix FISH-positive clustered cells also fluoresce white/yellow with DAPI showing that they contain poly(P). (e) Images showing cells from communities fed synthetic wastewater hybridizing with both EUBmix (FLUOS) and DECH454 (Cy3) probes, where Dechloromonas cells fluoresce yellow/orange. (f) Same field of view where the Dechloromonas cells fluoresce blue with DAPI, showing they contain no poly(P). (g) Images of cells with typical TFO morphologies from communities fed clarified treated effluent after FISH with EUBmix (FLUOS), ALF1b (Cy3), and DF988 (Cy5) probes. Cells appearing green have hybridized with the EUBmix probe only, yellow/orange cells have hybridized with both the EUBmix and ALF1b probes, and purple cells have hybridized with all three probes. Note the presence of some TFO appearing yellow/orange, suggesting that they are phylogenetically distinct. (h) Image of cells from communities fed synthetic wastewater after FISH with EUBmix (FLUOS), Nso1225 (Cy3), and Ntspa 662 (Cy5) probes, showing yellow clusters of Nitrosomonas and blue clusters of Nitrospira cells.
Thus, when phosphate was added to the reactor in the next stage of the cycle (the carbon famine stage, corresponding to the aerobic phase of a conventional EBPR process), it was completely and rapidly assimilated by the phosphate-starved cells (Fig. 2a). Because biomass PHB levels fell during the phosphate uptake phase, it is likely that it was used as the energy source for phosphate assimilation and poly(P) storage (Fig. 2a). The transformation of glycogen appeared to play little role in phosphate removal (Fig. 2a) since only minimal changes in biomass levels were detected across the SBR cycle. The P content of the harvested biomass after the famine stage was about 4.5% ± 0.1% during stable operation (Fig. 2a).
During the feed stage, the addition of acetate and phosphate release corresponded to a fall in the level of dissolved oxygen, which fell eventually to about 5 to 12% saturation (Fig. 2b) and rose rapidly to about 80% saturation when all of the acetate was utilized (44 min after addition). It remained at this level during the rest of the cycle, except for the settling stage, where as expected, it fell to about 6 to 10% saturation (Fig. 2b). The redox potential showed a similar trend, falling to about 100 mV during the acetate feed period, confirming that the reactor environment was always an oxidizing one (Fig. 2b). So this process never becomes anaerobic.
(ii) Using clarified effluent.
When these analyses were repeated with samples taken after 20 days of stable operation from the SBR fed the clarified effluent, very similar profiles were obtained. A typical profile is shown in Fig. 3a, but again, replicate weekly analyses gave almost identical profiles. Thus, the relationships and trends described for the SBR fed synthetic wastewater were seen with this community fed clarified effluent. However, less PHB appeared to be metabolized during the famine stage, and the ratio of P released to acetate utilized in the feed stage was lower (0.048 mmol P released/mmole acetate assimilated). Biomass P content at harvest time was very similar to that with the synthetic wastewater (again about 4.5%, wt/wt), and glycogen again appeared to be incidental to the process, since its biomass content changed little across the cycle. Likewise, the dissolved oxygen and redox profiles (Fig. 3b) were in agreement with those with the synthetic wastewater and consistent with these chemical transformations occurring in an oxidizing environment.
FIG. 3.
(a) Chemical profile of a typical SBR cycle from the stable operational period of the system using clarified effluent from a non-EBPR activated sludge plant showing the changes in acetate levels (▴), P content of biomass (□), phosphate content of mixed liquor (○), PHB content of biomass (▵), and glycogen content of biomass (▿) over a feed/famine cycle. The downward facing arrows indicate where acetate and the clarified effluent were added to the reactor. (b) Dissolved oxygen (—) and redox potential (- - - - ) profiles over a feed/famine period for the SBR fed clarified effluent from a non-EBPR activated sludge plant.
16S rRNA-based clone libraries of aerobic SBR communities.
16S rRNA trees generated with universal bacterial primers from biomass samples again taken after 28 and 20 days, respectively, from reactors fed synthetic wastewater and clarified effluent (Fig. 4a and b) revealed that members of the Betaproteobacteria were strongly represented in both communities. Smaller numbers of Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes, and Thermus-related clones were also detected. Surprisingly, bearing in mind the FISH data described below, none of the 120 clones analyzed from the community fed synthetic wastewater were “Candidatus Accumulibacter phosphatis,” and only a single clone from the 144 sequenced from the community fed the clarified effluent emerged as a related member of this population.
A large proportion of clones in both libraries (>50%) clustered closely with Dechloromonas spp., with OTU27 dominant in each (Fig. 4a and b). Even when “Candidatus Accumulibacter”-targeted primers were used for PCR from the community fed synthetic wastewater (data not shown), of 24 clones partially sequenced (about 380 nt), all were identified as being from the genus Dechloromonas and 21 derived from the same Dechloromonas OTU27 detected with the universal primers. The membership profiles of both libraries were very similar (Fig. 4a and b), except that the community treating the clarified effluent was more diverse and contained several novel clones of Alphaproteobacteria (e.g., OTU36) and Gammaproteobacteria (e.g., OTU38). Many clones from both communities had high sequence similarities to those recovered from other activated sludge systems, including anaerobic/aerobic EBPR processes.
DGGE profiles of the communities treating synthetic sewage and treated effuent.
The DGGE profiles from samples taken at the end of the feed stages during startup and stable operating phases of both systems are given in Fig. 5 (lanes a to j). Few changes in the profiles of the community fed synthetic wastewater were seen once P removal capacity had stabilized (Fig. 5, lanes b to e). Prominent DGGE fragments tentatively identified were again from populations closely related to Dechloromonas spp. (bands from OTU23, -26, -27, and -29) and from “Candidatus Accumulibacter phosphatis” (OTU41 with 99% sequence similarity). Once, on day 55, P removal failed (for reasons still unclear) and the DGGE profile taken then (Fig. 5, lane f) showed that while bands from OTU26 and -27 from Dechloromonas spp. became less prominent, others gave intensified fluorescent signals. Of these, two were identified as Pseudoxanthomonas spp. (OTU2 and -3) and one (OTU10) was an alphaproteobacterium distantly related (93% sequence similarity) to Defluviicoccus vanus. Also showing increased fluorescence was the fragment from OTU23 from Dechloromonas-related organisms. The fragment from OTU41 from “Candidatus Accumulibacter phosphatis” was no longer visible. After day 65, when the system had recovered its EBPR capacity, the DGGE profile (Fig. 5, lane g) had reverted to those seen earlier during stable high P removal (lanes b to e).
After changing during startup (Fig. 5, lanes h and i), almost identical DGGE profiles to these were generated from communities in the reactor fed clarified effluent during its stable operational periods (e.g., in biomass taken at 20 days [Fig. 5, lane i]). However, marked changes were again noted when this reactor also suddenly failed after 27 days of operation (Fig. 5, lane j). As before, fragments from OTU3 and -23 fluoresced more strongly. One prominent band seen only in the DGGE profile from the failed system was from OTU30, with a sequence identical to that of a Thermus sp., which was detected in the clone library from that plant (Fig. 4b).
Community composition of the aerobic EBPR community by FISH analyses.
Data from FISH-probed samples taken at the end of the feed stages at days 28 and 20 for the systems fed synthetic wastewater and clarified effluent, respectively, were very similar. In both, the Betaproteobacteria dominated, appearing as either rods or coccobacilli, with the latter mainly arranged in clusters of different sizes, which varied more in the biomass treating clarified effluent. Viability staining indicated that each individual cluster contained mainly viable cells (data not shown).
Most Betaproteobacteria in the large, encapsulated clusters responded to the PAOmix and RHC439 probes designed against the putative PAO “Candidatus Accumulibacter phosphatis” (Fig. 6a and c), contributing 22.6% (SE, ±4.6%) and 15.6% (SE, ±1.8%) of the total EUBmix-positive area to the communities fed the synthetic wastewater and clarified effluent, respectively. The RHC175 probe lit up the same clusters, but smaller and more scattered clustered cells also responded to it (data not shown). Combining FISH with DAPI staining showed that most cells in both communities taken at the end of the famine phase, which stained positive using the PAOmix and RHC439 FISH probes, also stained positive (Fig. 6a to d) using DAPI for poly(P) (>85% of the PAOmix/RHC439-positive cell area for both). Essentially the same data were obtained after repeated analyses of samples taken later from both reactors. A few tiny coccoid entities, occasionally appearing in small clusters, were also positive by DAPI staining, but could not be identified with any of the FISH probes used.
Most of the other clustered Betaproteobacteria in both communities fluoresced with the DECH454 and DECH472 probes. Those responding to the DECH454 probe targeting Dechloromonas OTU27 clones (Fig. 6e) (16.2% ± 2.5% SE and 10% ± 1% SE of the total EUBmix-positive area in communities treating synthetic wastewater and clarified effluent, respectively, in samples taken after 28 and 20 days, respectively) formed mainly large clusters. Clusters fluorescing with the DECH472 probe targeting OTU24 clones were always much smaller and less spherical in their appearance (data not shown) and far less frequently seen. None of the clustered cells of Dechloromonas contained poly(P) after DAPI staining at any time during either SBR cycle (Fig. 6f). However, cells fluorescing with the PAOmix FISH probes and DECH454-probed Dechloromonas cells in the large clusters contained PHA after Nile blue A staining (data not shown).
Most Alphaproteobacteria detected with the ALF968 (28) probe were distinctive tetrad-forming organisms (TFO) that were abundant in samples from both communities taken throughout their SBR cycles. These TFO did not respond to either the SBR9-1a or the AMAR839 probe (4, 34), each designed to detect alphaproteobacterial TFO populations reported in other EBPR communities. Nor did they respond to the TFO-DF862 or DEF438 probes targeting Defluviicoccus-related TFO in EBPR communities (26, 50), although a few clusters of coccobacilli and TFO fluoresced with the TFO-DF218/618 probe mix (50) (data not shown). Instead about 7.9% (SE, ±1.0%) and 9.8% (SE, ±0.7%) of the cell volume fluorescing with the EUBmix probes in samples taken after 28 and 20 days from communities fed the synthetic wastewater and clarified effluent, respectively, fluoresced with the DF988 probe (e.g., Fig. 6g) targeting other Defluviicoccus-related organisms (36) reported in anaerobic/aerobic EBPR systems. This probe appeared to have a slightly broader specificity than the DF1020 probe did, agreeing with the data of Meyer et al. (36), where all cells hybridizing with the DF1020 probe also hybridized with the DF988 probe, but not vice versa. A few TFO in both communities also fluoresced with the SPH120 probe (40) targeting the sphingomonads (data not shown), and other TFO not responding to either of the DF FISH probes were also seen in both communities (Fig. 6g). No TFO from either community (data not shown) ever stained positive for poly(P) with DAPI in samples taken throughout the SBR cycles. While some appeared to contain PHB, a few stained Nile blue A negative in samples taken at the start of the acetate feed stage (data not shown). No cells were seen responding to the GAO mix probes targeting the gammaproteobacterial “Candidatus Competibacter phosphatis” (11, 27) in the community treating the synthetic waste, although a few positive cell clusters (<1%) were present in that treating the clarified effluent.
Small numbers (<1% of the area of cells fluorescing with the EUBmix probes) of ammonia-oxidizing Nitrosomonas and nitrite-oxidizing Nitrospira bacteria were also seen in both communities after the use of the Nso1225 and Ntspa 662 FISH probes, respectively (Fig. 6h). Both were present in clusters, as reported for other activated sludge systems (14, 49), and were sometimes seen immediately adjacent to each other (22, 38). Their very low numbers would suggest that little nitrification was occurring in either community.
Changes in population ecophysiology.
Staining properties of samples taken at different times during both SBR cycles agreed generally with the chemical profile data described earlier. Thus, DAPI staining showed that PAOmix FISH-positive cells contained poly(P) during both the famine and feed stages, confirming that complete P release did not occur during acetate assimilation. Not all individual cells in each cluster were DAPI positive, as noted in similar populations from anaerobic/aerobic EBPR systems (52). PHB was detected by Nile blue A staining in cells responding to the PAOmix probes throughout the cycles with both feeds. However, by the end of the feed stage, fewer PAOmix FISH-positive cells appeared to stain strongly for PHB, suggesting that these are the populations utilizing their PHB stores during uptake of the released phosphate, and even fewer contained PHB at the end of the famine phase. Thus, it would appear that the “Candidatus Accumulibacter phosphatis” PAO in these aerobic EBPR processes share some ecophysiological features with the same PAO populations in anaerobic/aerobic EBPR systems in terms of their storage polymer cycling. However, they appear to retain some poly(P) during the aerobic feed/famine cycle (37, 45).
FISH/MAR studies.
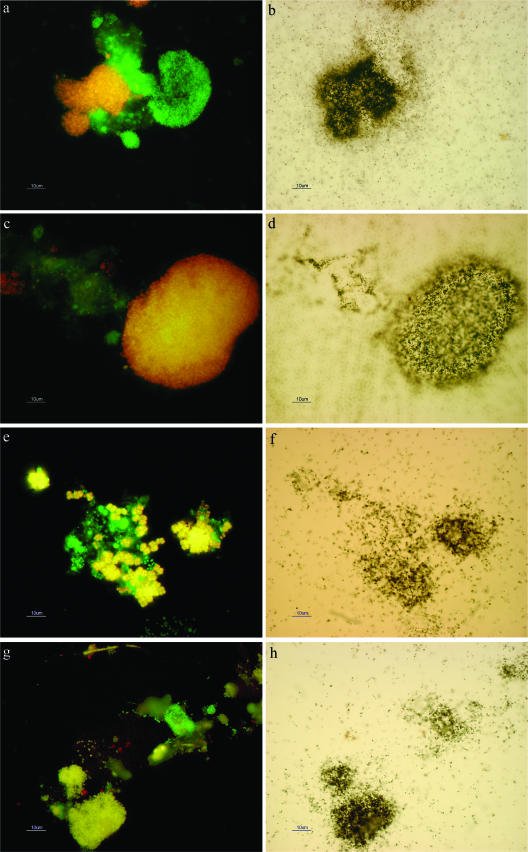
Under conditions set up to mimic those existing in the SBRs at the time samples were taken, FISH/MAR data suggest that under aerobic conditions, the PAOmix FISH-probed positive cells behaved in a manner expected of PAO. Thus, only these populations could both assimilate 14C-acetate and 33P in the feed and famine stages, respectively (Fig. 7a to d). All clusters responding to the PAOmix probes assimilated 33P in the famine stage, suggesting that all were capable of EBPR (Fig. 7a and b), but from silver grain deposition intensity, some appeared to assimilate more 33P than others. Furthermore, the majority of PAO clusters assimilated 14C-acetate in the feed stage (Fig. 7c and d). MAR evidence supported uptake of 14C-acetate by both the Defluviicoccus-related TFO (Fig. 7e and f) and clustered Dechloromonas cells (Fig. 7g and h) and other unidentified populations in samples taken immediately prior to acetate addition. Subjective assessment based on the intensity of silver grain deposition suggested that the “Candidatus Accumulibacter phosphatis” PAO and Dechloromonas were the most active acetate assimilators. No evidence for 33P assimilation was seen with either Defluviicoccus-related TFO or Dechloromonas in biomass samples taken immediately before phosphate addition, agreeing with the earlier data obtained with DAPI staining (Fig. 6e and f).
FIG. 7.
FISH/MAR images of populations from the aerobic SBR reactor fed synthetic wastewater to illustrate their substrate uptake abilities. (a) FISH images showing yellow cells hybridizing with both the EUBmix (FLUOS) and PAOmix (Cy3) probes. (b) The same field of view after MAR, showing the ability of the PAO to assimilate 33Pi. (c) FISH images showing yellow cells hybridizing with both the EUBmix (FLUOS) and PAOmix (Cy3-labeled) probes. (d) The same field of view after MAR, showing the ability of the PAO to assimilate acetate. (e) FISH images showing yellow cells hybridizing with both the EUBmix (FLUOS labeled) and DF988 (Cy3) probes. (f) The same field of view after MAR, showing the ability of the Defluviicoccus-related TFO to assimilate acetate. (g) FISH images showing yellow cells hybridizing with both the EUBmix (FLUOS) and DECH454 (Cy3) probes. In panel h, the same field of view after MAR, showing the ability of the Dechloromonas-related organisms to assimilate acetate aerobically.
DISCUSSION
This paper describes a process for the microbiological removal of phosphate (up to 10 mg P liter−1) in an aerobic SBR system fed either synthetic wastewater or clarified effluent from a non-EBPR activated sludge process, with acetate being added separately as the sole carbon/energy source with both feeds. That is, the process operates as an aerobic EBPR system. Selective pressures favoring the ultimately dominant PAO populations were imposed in a way fundamentally different from those thought to be essential for anaerobic/aerobic EBPR processes. Crucial to the success was the temporal separation of the additions to the reactor of the carbon/energy source (acetate in the feed stage) and the P-containing, low-COD feed (the famine stage).
This ensures that at the time that acetate is added, no exogenous phosphate is present, and acetate is absent when the phosphate-containing low-COD feed is added.
This cyclic aerobic process resulted in selective enrichment of bacteria which could alternately accumulate poly(P) and PHA aerobically, presumably because (i) in the feed stage, in the absence of an external source of P, the populations utilized their poly(P) reserves to assimilate acetate primarily for PHA synthesis and not cell growth and (ii) in the famine stage when phosphate-containing low-COD feed was added to the reactor in the absence of an external carbon/energy supply, the bacteria used their internal reserves of PHA as a source of energy to take up phosphate and convert it to poly(P). Thus, whereas the anaerobic/aerobic cycle of conventional EBPR systems produces low-phosphate effluents when treating raw sewage with a high COD, this aerobic cyclic process operates to produce a low-phosphate effluent from a phosphate-containing wastewater with a low COD.
The critical importance of such a feed strategy becomes apparent when the fate of EBPR in communities fed phosphate and acetate simultaneously under aerobic conditions is considered (20, 42). Under these conditions, EBPR capacity rapidly diminishes, but the reasons are still unclear. There appears to be no thermodynamic reason for why the chemical transformations so characteristic of anaerobic/aerobic EBPR systems should not be achievable aerobically. The role of the anaerobic phase is generally viewed as providing conditions favorable for the PAO capable of anaerobic substrate assimilation to synthesize PHA, thus allowing their subsequent aerobic growth (5, 37, 45). PHA synthesis is known to be induced in activated sludge by oxygen limitation (47), which might explain this need for anaerobic conditions. However, the limitations of N and phosphate (the latter occurring in the aerobic process) may also induce aerobic PHA synthesis to high levels in bacteria (8, 43, 48) and thus provide selective pressures similar to those encountered in anaerobic/aerobic EBPR systems.
Equally, it is generally considered that prevailing aerobic or anoxic (O2 absent but NO3 present) conditions in EBPR systems, when metabolizable substrates are available, would allow aerobic or anaerobic respiring organisms, respectively, to out-compete the PAO for these substrates, thus ensuring that the PAO would no longer dominate the community and EBPR would fail (37, 45). This opinion is based on the (untested) hypothesis that other aerobic and anaerobic (denitrifying) respiring populations are better able to accumulate available substrates like acetate than the PAO are. The FISH/MAR data presented here suggest that this is not obviously the case with this aerobic process.
FISH analyses showed that the same betaproteobacterial Rhodocyclus-related “Candidatus Accumulibacter phosphatis” PAO populations detected in most anaerobic/aerobic EBPR communities (12, 21, 24) were the major PAO in both aerobic EBPR communities, contributing almost all of the DAPI-positive cells seen. Awareness of their increasing importance as PAO under denitrifying conditions (1, 51) confirms that these are metabolically very adaptable organisms, able to adjust to and cope with fluctuations in nutrient availability under a wide range of conditions. From FISH/MAR data, their ecophysiological features were also those expected of PAO (24), being able to utilize acetate when available and store it as intracellular PHB and consequently assimilate phosphate into poly(P), probably using stored PHB as an energy source. Surprisingly, in light of the FISH data and that “Candidatus Accumulibacter phosphatis” has been detected as a dominant clone in most libraries generated from conventional acetate-fed SBR EBPR systems operating with anaerobic/aerobic cycling (12, 21, 52), clones of “Candidatus Accumulibacter phosphatis” were rarely detected in the libraries generated from the community fed the synthetic wastewater with either universal bacterial or Rhodocyclus-targeted primers. Only one was seen (OTU41) 99% similar to clone AF502228 described by McMahon et al. (35) when clarified effluent was used as feed. This was the result despite using the same DNA extraction methods, primers, and PCR conditions successful in detecting these PAO populations elsewhere. Yet DGGE profiles from both communities (Fig. 5) contained 16S rRNA fragments identifiable as “Candidatus Accumulibacter phosphatis.” This suggests that the method used for DNA extraction was not responsible, although it was clear from microscopic examination of the biomass that the heavily capsulated clusters, presumably of the PAO, were largely intact after the extraction protocol. Other studies have often shown poor agreement between activated sludge community compositions revealed by clone library data and FISH analyses (21, 36), and a combination of different DNA extraction methods is probably required (22) for studies with EBPR communities.
Dechloromonas clones were much more abundant in these libraries from both communities than in those analyzed previously from anaerobic/aerobic EBPR systems (12, 21). They shared the highest sequence homology (97 to 99%) with the Dechloromonas clones reported by Coates et al. (10) and those of Zilles et al. (52), which were recovered from anaerobic/aerobic EBPR communities. FISH probes targeting these Dechloromonas clones confirmed their presence in situ in large numbers. The clones were usually organized in clusters, similar in appearance to those formed by the PAO, but none stained positively for poly(P) with DAPI in samples taken at the end of the famine stage or assimilated 33P in samples removed at the end of the acetate feed stage. Hence, they appear to play no positive role in this aerobic EBPR process. However, FISH/MAR data showed that the clones also assimilated acetate, presumably in competition with “Candidatus Accumulibacter phosphatis” in the feed stage, and Nile blue A staining showed that they contained PHB in samples taken throughout the SBR cycle. Their ecophysiological features in anaerobic/aerobic EBPR communities where they have been detected are unknown. The alphaproteobacterial TFO also seen in large numbers in both communities were identified by FISH as Defluviicoccus-related TFO and were detected in large numbers in an EBPR plant with poor EBPR capacity (36). They too failed to assimilate 33P under simulated reactor famine conditions and always stained negatively with DAPI. Like Dechloromonas, these TFO contained PHB and assimilated acetate under conditions mimicking the feed stage of the process. Consequently, both the TFO and Dechloromonas appear to have the phenotype of a glycogen-accumulating organism (GAO) (4, 11, 28, 50) in assimilating acetate and synthesizing PHB during the feed stage, but with no subsequent poly(P) production during the famine stage. The possible impact of both on the performance of this aerobic process is unclear since biomass glycogen levels changed very little over the SBR cycles in both communities. Knowing which operational factors might encourage their presence is important, since from FISH/MAR data based on silver grain deposition, the putative GAO Dechloromonas appeared to compete more strongly than the Defluviicoccus-related TFO for acetate with “Candidatus Accumulibacter phosphatis.”
The results presented here show that aerobic EBPR is achievable, and from our experiences, the process behaves extremely reliably in producing an effluent with little or no detectable phosphate when fed both synthetic wastewater and clarified effluent containing about 10 mg P liter−1. Its future is not to replace anaerobic/aerobic EBPR systems where COD and phosphate are supplied simultaneously, but to be an add-on unit to remove phosphate from low-COD effluents. The process still needs optimization, especially to see whether acetate feed levels can be reduced, making it cheaper to operate, and to determine what final clarified plant effluent phosphate levels it can cope with, how the SBR configuration may influence its performance, and how robust and stable it is under changes in operational parameters like temperature, pH, and effluent chemical composition. This work is under way.
Acknowledgments
This research was funded by an Australian Research Council Discovery research grant (DP0557646DS) and the Smart Water Fund from the Victorian State Government. J. Ahn was supported by a La Trobe University Postdoctoral Fellowship, and S. McIlroy was supported by an Australian Postgraduate Award. We thank Land and Water Resources Research Development Corporation (now Land and Water) and Sydney Water Corporation for financial support.
R. W. Cole is thanked for the design and construction of the control unit of the sequencing batch reactor.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Ahn, J., T. Daidou, S. Tsuneda, and A. Hirata. 2002. Characterization of denitrifying phosphate-accumulating organisms cultivated under different electron acceptor conditions using polymerase chain reaction-denaturing gradient gel electrophoresis assay. Water Res. 36:403-412. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruin (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Bayly, R. C., A. Duncan, J. W. May, M. Schembri, A. Semerjis, G. Vasiliadis, and W. G. C. Raper. 1991. Microbiological and genetic aspects of synthesis of polyphosphate by species of Acinetobacter. Water Sci. Technol. 23:747-754. [Google Scholar]
- 4.Beer, M., Y. H. Kong, and R. J. Seviour. 2004. Are some putative glycogen accumulating organisms (GAO) in anaerobic:aerobic activated sludge systems members of the α-Proteobacteria? Microbiology 150:2267-2275. [DOI] [PubMed] [Google Scholar]
- 5.Blackall, L. L., G. R. Crocetti, A. M. Saunders, and P. L. Bond. 2002. A review and update of the microbiology of enhanced biological phosphorus removal in wastewater treatment plants. Antonie Leeuwenhoek 81:681-691. [DOI] [PubMed] [Google Scholar]
- 6.Bond, P. L., P. Hugenholtz, J. Keller, and L. L. Blackall. 1995. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl. Environ. Microbiol. 61:1910-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, E. L., K. Eales, J. Soddell, and R. J. Seviour. 2005. Improved permeabilization protocols for fluorescence in situ hybridization (FISH) of mycolic-acid-containing bacteria found in foams. J. Microbiol. Methods 61:47-54. [DOI] [PubMed] [Google Scholar]
- 8.Chinwetkitvanich, S., C. W. Randall, and T. Panswad. 2004. Effects of phosphorus limitation and temperature on PHA production in activated sludge. Water Sci. Technol. 50:135-143. [PubMed] [Google Scholar]
- 9.Clescerl, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1996. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, DC.
- 10.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crocetti, G. R., J. F. Banfield, J. Keller, P. L. Bond, and L. L. Blackall. 2002. Glycogen-accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology 148:3353-3364. [DOI] [PubMed] [Google Scholar]
- 12.Crocetti, G. R., P. Hugenholtz, P. L. Bond, A. Schuler, J. Keller, D. Jenkins, and L. L. Blackall. 2000. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66:1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 14.Daims, H., J. L. Nielsen, P. H. Nielsen, K. H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daims, H., S. Lucker, and M. Wagner. 2006. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200-213. [DOI] [PubMed] [Google Scholar]
- 16.de-Bashan, L. E., and Y. Bashan. 2004. Recent advances in removing phosphorus from wastewater and its future use as fertilizer (1997-2003). Water Res. 38:4222-4246. [DOI] [PubMed] [Google Scholar]
- 17.DeSantis, T. Z., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eales, K., J. L. Nielsen, C. Kragelund, R. J. Seviour, and P. H. Nielsen. 2005. The in situ physiology of pine tree like organisms (PTLO) in activated sludge foams. Acta Hydrochim. Hydrobiol. 33:203-209. [Google Scholar]
- 20.Guisasola, G., M. Pijuan, J. A. Baeza, J. Carrera, C. Casas, and J. Lafuente. 2004. Aerobic phosphorus release linked to acetate uptake in bio-P sludge: process modelling using oxygen uptake rate. Biotechnol. Bioeng. 85:721-733. [DOI] [PubMed] [Google Scholar]
- 21.Hesselmann, R. P., C. Werlen, D. Hahn, J. R. van der Meer, and A. J. Zehnder. 1999. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 22:454-465. [DOI] [PubMed] [Google Scholar]
- 22.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analysed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 23.Kawaharasaki, M., A. Manome, T. Kanagaw, and K. Nakamura. 2002. Flow cytometric sorting and RFLP analysis of phosphate accumulating bacteria in an enhanced biological phosphorus removal system. Water Sci. Technol. 46:139-144. [PubMed] [Google Scholar]
- 24.Kong, Y., J. L. Nielsen, and P. H. Nielsen. 2004. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 70:5383-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong, Y., J. L. Nielsen, and P. H. Nielsen. 2005. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 71:4076-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong, Y. H., M. Beer, R. J. Seviour, K. C. Lindrea, and G. N. Rees. 2001. Structure and functional analysis of the microbial community in an aerobic:anaerobic sequencing batch reactor (SBR) with no phosphorus removal. Syst. Appl. Microbiol. 24:597-609. [DOI] [PubMed] [Google Scholar]
- 27.Kong, Y. H., S. L. Ong, W. J. Ng, and W.-T. Liu. 2002. Diversity and distribution of a deeply branched novel proteobacterial group found in anaerobic:aerobic activated sludge processes. Environ. Microbiol. 4:753-757. [DOI] [PubMed] [Google Scholar]
- 28.Kong, Y. H., Y. Xia, J. L. Nielsen, and P. H. Nielsen. 2006. Ecophysiology of a group of uncultured Gammaproteobacterial glycogen-accumulating organisms in full-scale enhance biological phosphorus removal wastewater treatment plants. Environ. Microbiol. 8:479-489. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991.16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Modern microbial methods: nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 31.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K. H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, W. T., A. T. Nielsen, J. H. Wu, C. S. Tsai, Y. Matsuo, and S. Molin. 2001. In situ identification of polyphosphate- and polyhydroxyalkanoate-accumulating traits for microbial populations in a biological phosphorus removal process. Environ. Microbiol. 3:110-122. [DOI] [PubMed] [Google Scholar]
- 33.Loy, A., M. Horn, and M. Wagner. 2003. probeBase—an online resource for rRNA targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maszenan, A., R. J. Seviour, B. K. Patel, and J. Wanner. 2000. A fluorescently-labelled r-RNA targeted oligonucleotide probe for the in situ detection of G-bacteria of the genus Amaricoccus in activated sludge. J. Appl. Microbiol. 88:826-835. [DOI] [PubMed] [Google Scholar]
- 34a.May, J. W., R. C. Bayly, G. Vasiliadis, R. W. Cole, P. R. Meyers, P. Romas, and J. K. Davies. 1997. Enhanced biological removal of phosphate by Acinetobacter. Land and Water Resources Research and Development Corporation, Braddon, Australia. http://www.med.monash.edu.au/microbiology/research/lwrrdc/project-umo20/report1997.pdf.
- 35.McMahon, K. D., M. A. Dojka, N. R. Pace, D. Jenkins, and J. D. Keatling. 2002. Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal. Appl. Environ. Microbiol. 68:4971-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer, R. L., A. M. Saunders, and L. L. Blackall. 2006. Putative glycogen-accumulating organisms belonging to the Alphaproteobacteria identified through rRNA-based stable isotope probing. Microbiology 152:419-429. [DOI] [PubMed] [Google Scholar]
- 37.Mino, T., M. C. M. van Loosdrecht, and J. J. Heinen. 1998. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32:3192-3207. [Google Scholar]
- 38.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muyzer, G., E. C. D. Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neef, A., R. Witzenberger, and P. Kampfer. 1999. Detection of sphingomonads and in situ identification in activated sludge using 16S rRNA-targeted oligonucleotide probes. J. Ind. Microbiol. Biotechnol. 23:261-267. [DOI] [PubMed] [Google Scholar]
- 41.Ostle, A. G., and J. G. Holt. 1982. Nile blue A as a fluorescent stain for poly-β-hydroxybutyrate. Appl. Environ. Microbiol. 44:238-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pijuan, M., A. Guisasola, J. A. Baeza, J. Carrera, C. Casas, and J. Lafuente. 2006. Net P-removal deterioration in enriched PAO sludge subjected to permanent aerobic conditions. J. Biotechnol. 123:117-126. [DOI] [PubMed] [Google Scholar]
- 43.Ryu, H. W., S. K. Hahn, Y. K. Chang, and H. N. Chang. 1997. Production of poly(3hydroxybutyrate) by high cell density fed-batch culture of Alcaligenes eutrophus with phosphate limitation. Biotechnol. Bioeng. 55:28-32. [DOI] [PubMed] [Google Scholar]
- 44.Satoh, H., W. D. Ramey, F. A. Koch, W. K. Oldham, T. Mino, and T. Matsuo. 1996. Anaerobic substrate uptake by the enhanced biological phosphorus removal activated sludge treating real sewage. Water Sci. Technol. 34:9-16. [Google Scholar]
- 45.Seviour, R. J., T. Mino, and M. Onuki. 2003. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol. Rev. 27:99-127. [DOI] [PubMed] [Google Scholar]
- 46.Smolders, G. J. F., J. van der Meij, M. C. M. van Loosdrecht, and J. J. Heijnen. 1994. Model of the anaerobic metabolism of the biological phosphorus removal process. Biotechnol. Bioeng. 43:461-470. [DOI] [PubMed] [Google Scholar]
- 47.Third, K. A., M. Newland, and R. Cord-Ruwisch. 2003. The effect of dissolved oxygen on PHB accumulation in activated sludge cultures. Biotechnol. Bioeng. 82:238-252. [DOI] [PubMed] [Google Scholar]
- 48.van Loosdrecht, M. C. M., C. M. Hooijmans, D. Brdjanovic, and J. J. Heihnen. 1997. Biological phosphorus removal processes. Appl. Environ. Biotechnol. 48:289-296. [Google Scholar]
- 49.Wagner, M., G. Rath, R. Amann, H.-P. Kops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 50.Wong, M. T., F. M. Tan, W. J. Ng, and W. T. Liu. 2004. Identification and occurrence of tetrad-forming Alphaproteobacteria in anaerobic-aerobic activated sludge processes. Microbiology 150:3741-3748. [DOI] [PubMed] [Google Scholar]
- 51.Zeng, R. J., A. M. Saunders, Z. Yuan, L. L. Blackall, and J. Keller. 2003. Identification and comparison of aerobic and denitrifying polyphosphate-accumulating organisms. Biotechnol. Bioeng. 83:140-148. [DOI] [PubMed] [Google Scholar]
- 52.Zilles, J. L., J. Peccia, M. W. Kim, C. H. Hung, and D. R. Noguera. 2002. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl. Environ. Microbiol. 68:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]