Abstract
Numerous marine sponges harbor enormous amounts of as-yet-uncultivated bacteria in their tissues. There is increasing evidence that these symbionts play an important role in the synthesis of protective metabolites, many of which are of great pharmacological interest. In this study, genes for the biosynthesis of polyketides, one of the most important classes of bioactive natural products, were systematically investigated in 20 demosponge species from different oceans. Unexpectedly, the sponge metagenomes were dominated by a ubiquitously present, evolutionarily distinct, and highly sponge-specific group of polyketide synthases (PKSs). Open reading frames resembling animal fatty acid genes were found on three corresponding DNA regions isolated from the metagenomes of Theonella swinhoei and Aplysina aerophoba. Their architecture suggests that methyl-branched fatty acids are the metabolic product. According to a phylogenetic analysis of housekeeping genes, at least one of the PKSs belongs to a bacterium of the Deinococcus-Thermus phylum. The results provide new insights into the chemistry of sponge symbionts and allow inference of a detailed phylogeny of the diverse functional PKS types present in sponge metagenomes. Based on these qualitative and quantitative data, we propose a significantly simplified strategy for the targeted isolation of biomedically relevant PKS genes from complex sponge-symbiont associations.
With an age of more than 600 million years, sponges (Porifera) are the most ancient extant group of metazoans. They harbor a remarkable diversity of biologically active secondary metabolites and have gained considerable attention as one of the richest sources of new drug candidates (6). Often there is little correlation between sponge taxonomy and the presence or absence of specific natural products. Thus, in some cases species exist as distinct chemotypes, while in others identical compounds are found in distantly related sponges. Because many sponges contain enormous amounts of bacteria within their tissues, sometimes occupying 40 to 60% of the total biomass (equivalent to 108 to 1010 bacteria per gram) (9, 47, 48), it has frequently been proposed that bacterial symbionts might be the actual producers of many sponge-derived natural products (32). So far, however, attempts to cultivate bacterial producers have generally failed. As a consequence, only very limited data about their biology and chemistry exist in spite of their suspected great biomedical importance.
Much of our current knowledge about sponge symbionts stems from culture-independent studies. However, work on cultured bacteria has also contributed significantly to our understanding of the sponge-microbe association (16, 27). Several research groups have assessed prokaryotic diversities within various bacterium-rich hosts by 16S rRNA gene analysis (for a recent review, see reference 14). They uncovered large and highly diverse multispecies associations with almost no taxonomic overlap with cultivated microorganisms. The 16S rRNA gene-based studies provide a complex picture of microbial consortia composed of two distinct symbiont types: those found ubiquitously in unrelated bacteriosponges from different parts of the world's oceans and those living specialized in a few bacteriosponge hosts. It is so far unknown which microbial types mainly contribute to the rich natural product chemistry in sponges.
Culture-independent strategies can also provide valuable insights into the true origin of sponge-derived secondary metabolites, e.g., by isolating and analyzing biosynthetic genes from the collective genome of the animal and its associated microbiota (the “metagenome”). Importantly, if bacteria are the original producers, their secondary metabolite-encoding genes are usually clustered together. Therefore, this metagenomic approach might also allow heterologous expression of entire pathways in culturable bacteria and thus create sustainable sources of valuable drug candidates. Complex polyketides have been particularly often used as targets for metagenomic analyses (36, 40). Many of the biomedically most interesting compounds from sponges, such as peloruside A (50) or laulimalide (17), belong to this natural product group. Since the enzymes catalyzing complex polyketide biosynthesis, termed type I polyketide synthases (PKSs) (42), are so far known exclusively from microorganisms, it has long been suspected that bacteria are the true producers of these compounds. We recently obtained strong support for this hypothesis by cloning PKS genes for the biosynthesis of antitumor polyketides of the onnamide series from the sponge Theonella swinhoei and tracing them back to a prokaryote (35, 36). This result suggests that metagenomic strategies could ultimately lead to the creation of renewable production systems for many animal-derived drug candidates. Currently, however, such an approach suffers from technical problems associated with the genomic complexity of most pharmacologically relevant sponges. In T. swinhoei, identification of the onnamide genes was possible because genes for the structurally related polyketide pederin from a symbiont of Paederus sp. beetles were known before and could be used for phylogeny-based gene targeting (33, 35, 37). For most other sponges, however, such information is not available. Numerous homologous gene clusters from different pathways would then have to be isolated, at least partly sequenced, and possibly expressed before the correct candidate could be identified.
The aim of this study was to obtain more specific information about PKS diversity in sponge-associated microbial communities. Questions addressed in this study were as follows. Is bacterial polyketide biosynthesis, as shown for the onnamides, more common in marine sponges? How does the presence of a widely distributed and specific microflora translate into chemistry? Can a thorough metagenomic analysis of PKSs in different sponge species provide guidance in the targeted search for genes of pharmaceutically relevant natural products?
MATERIALS AND METHODS
Sponge collection.
All sponges were collected by scuba diving at a depth of 5 to 15 m: Aplysina aerophoba offshore from Banyuls sur Mer, Mediterranean Sea, France (global positioning system data: 42°29′N, 03°08′E); Aplysina cavernicola around Elba, Mediterranean Sea, Italy (42°43′N, 10°09′E); Verongula gigantea, Aiolochroia crassa, Xestospongia muta, Ectyoplasia ferox, Ircinia felix, Plakortis sp., Cribochalina vasculum, Siphonodictyon (Aka) coralliphagum, Amphimedon compressa, Chalinula molitba, Tedania ignis, Dysidea etheria, Ptilocaulis sp., Callyspongia vaginalis, and Agelas dilatata offshore from Little San Salvador Island, Caribbean Sea, Bahamas (24°34.39′N, 75°58.00′W); the two Theonella swinhoei chemotypes at Hachijo Island, Japan (33°06′N, 139°47′E); and Cacospongia mycofijiensis offshore from Vanuatu, southern Pacific Ocean (17°34′S, 168°19′E). Individuals were placed separately into plastic bags and brought to the surface. After collection, sponge tissues were cut into pieces and stored at −80°C or in 70% or 95% ethanol at 4°C until use. In addition, seawater samples from the sampling site of Banyuls sur Mer, France, were collected. Taxonomic identification of sponges was performed by Sven Zea, Universidad Nacional de Colombia.
DNA isolation, PCR amplification, and sequencing of KS gene fragments.
Genomic DNA was isolated from freshly collected sponges as described in the work of Fieseler et al. (7), applying the Fast DNA spin kit for soil (Q-Biogene, Heidelberg, Germany). Genomic DNA from the collected seawater samples was isolated as follows: 1 to 3 liters of seawater were prefiltered through folded filter papers and then filtered through 0.2-μm-pore-size bottle-top SFCA membrane filters (Nalgene, Rochester, NY). After thermal lysis in a boiling water bath for 10 min, DNA was subsequently precipitated in ethanol. Amplification of KS gene fragments was performed as described previously (33, 40) using combinations of the following degenerate KS primer pairs: KSDPQQF (5′-MGN GAR GCN NWN SMN ATG GAY CCN CAR CAN MG-3′)/KSHGTGR (5′-GGR TCN CCN ARN SWN GTN CCN GTN CCR TG-3′), KSF2.i (5′-GCI ATG GAY CCI CAR CAR MGI VT-3′)/KS5R.i (5′-GTI CCI GTI CCR TGI SCY TCI AC-3′), and KSF2.gc (5′-GCS ATG GAY CCS CAR CAR CGS VT-3′)/KS5R.gc (5′-GTS CCS GTS CCR TGS SCY TCS AC-3′). For sequencing, PCR products of ca. 750 bp in size were cloned into pGEM T-easy (Promega) or pCR2.1-TOPO (Invitrogen) and transformed into electrocompetent Escherichia coli XL1-Blue cells. Plasmid minipreparations were done by standard alkaline lysis procedures (39). Sequencing was performed on an ABI 377XL and an ABI 3730 automated sequencer (Applied Bioscience).
Metagenomic large-insert library construction, identification of PKS-encoding clones, and sequencing.
Construction of the cosmid library of total DNA preparations of T. swinhoei (onnamide type) using the vector pWEB (Epicenter) was performed according to the manufacturer's protocols and has been described previously (36). The library consisted of 60,000 clones and covered ca. 2.1 Gb of mixed sponge-bacterial DNA. A second library of T. swinhoei (theonellamide type) was freshly prepared in the same way and encompassed ca. 29,000 clones covering an estimated 1.2 Gb. For A. aerophoba, preparation of a 29,108-member fosmid library from enriched bacteria has been described previously (8). Briefly, microbial cells were separated from the sponge matrix after mechanical disruption followed by filtration through folded filter papers in order to remove tissue fibers. Remaining eukaryotic cells were removed by centrifugation at 4°C at 100 × g for 10 min. Microbial cells were harvested by centrifugation at 4°C at 12,000 × g before they were embedded in agarose plugs for metagenomic DNA isolation (109 cells ml−1 in 0.5% low-melting-point agarose [SeaPlaque; FMC Bioproducts]). In addition, a second library was constructed from A. aerophoba after the method of Piel et al. (36) with the following modifications: microbial cells were collected by centrifugation at 13,000 × g for 15 min and then resuspended in 0.5 ml of lysis buffer containing 100 mM Tris-HCl (pH 8), 1.4 M NaCl, 20 mM EDTA, 200 μl of cetyltrimethylammonium bromide solution at 55°C, 10 μl of 10% sodium dodecyl sulfate, 35 μl of 100 mM diethyldithiocarbamate, 10 μl of mercaptoethanol, 60 μl of 10% polyvinylpyrrolidone, 10 μl of 100-mg/ml lysozyme, and 25 μl of 20-mg/ml proteinase K. The mixture was incubated at 30°C for 1.5 h and extracted once with 10 ml of chloroform, three times with equal volumes of phenol-chloroform, and twice with equal volumes of chloroform. DNA was precipitated from the aqueous phase by the addition of 1.2 volumes of isopropanol, washed with 10 ml of ice-cold ethyl alcohol, air dried, and dissolved in water. The library was constructed using an E. coli-Streptomyces shuttle cosmid vector, pAY1 (22). Both A. aerophoba-derived libraries contained a total of ca. 2.4 Gb of sponge-associated microbial DNA.
To isolate the fosmids pAE27P20 and pAPKS18, degenerate primer pairs targeting KS genes (KSDPQQF and KSHGTGR; see sequences above) were used in PCRs on library pools according to the procedure described previously (36). The cosmid pSW1H8 was isolated from the T. swinhoei library in a similar way with the specific primers sponge11f (5′-GCA TGA TGC TGG AGA CGA GCT G-3′) and sponge11r (5′-CGT CGA ACG CCT TGC ACT GC-3′) derived from the sequence of a selected KS amplicon. Positive clones were subcloned into pBluescriptII SK(−) and end sequenced. Sequencing of pAE27P20 and pAPKS18 was performed by Agowa GmbH, Berlin, Germany. The insert of pAPKS18 was sequenced entirely, while the insert of pAE27P20 was only partially sequenced by primer walking until the PKS gene was covered. To sequence pSW1H8, the cosmid was sheared using a standard nebulizer (Octurno) and the fragment ends were repaired with T4 DNA polymerase and Klenow fragment. Fragments of 1 to 1.5 kb were isolated by agarose electrophoresis, cloned into the pUC18 vector, and end sequenced using the BigDye Terminator Ready Mix (Applied Biosystems) and an ABI 3700 sequencer (Applied Biosystems). Sequence data were assembled using GAP4 software (34) and analyzed using the BLASTX, PSI-BLAST, FramePlot, and InterProScan algorithms.
Phylogenetic analyses.
Phylogenetic tree constructions on partial KS amino acid sequences (at least 166 positions) and on full-length NuoG amino acid sequences (at least 490 positions) were conducted via automated sequence alignments applying ClustalX followed by the ClustalX (46) tree calculation function for neighbor-joining trees (1,000 replicates) and by Phylip analyses applying neighbor-joining, maximum parsimony (100 replicates each), and maximum likelihood algorithms on the Jones-Taylor-Thornton amino acid replacement model. KS sequences were also analyzed based on Bayesian statistics with the MrBayes program version 3.1.1 (15), applying the Jones-Taylor-Thornton model and a gamma distribution with four categories. The Markov chain Monte Carlo analysis was run over seven million generations and four independent chains. The Markov chains were sampled every 100 generations. Convergence was judged when a standard deviation of less than 0.01 was achieved. KS amino acid sequences were manually truncated as described previously (35).
Nucleotide sequence accession numbers.
The KS gene sequences were deposited into GenBank under the accession numbers DQ996313 to DQ996391. The PKS sequences were deposited into GenBank under the accession numbers DQ438986 to DQ438988.
RESULTS
Assessment of PKS gene diversity in marine sponges.
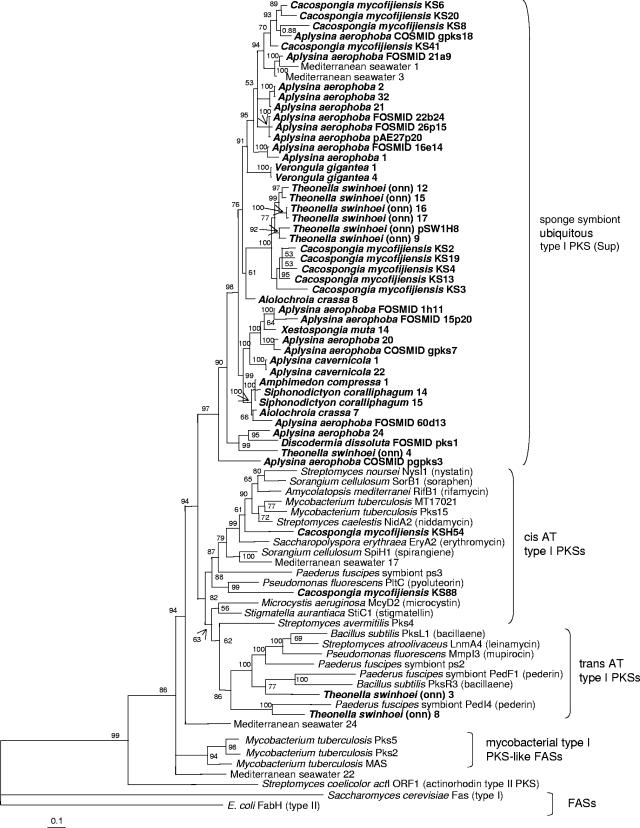
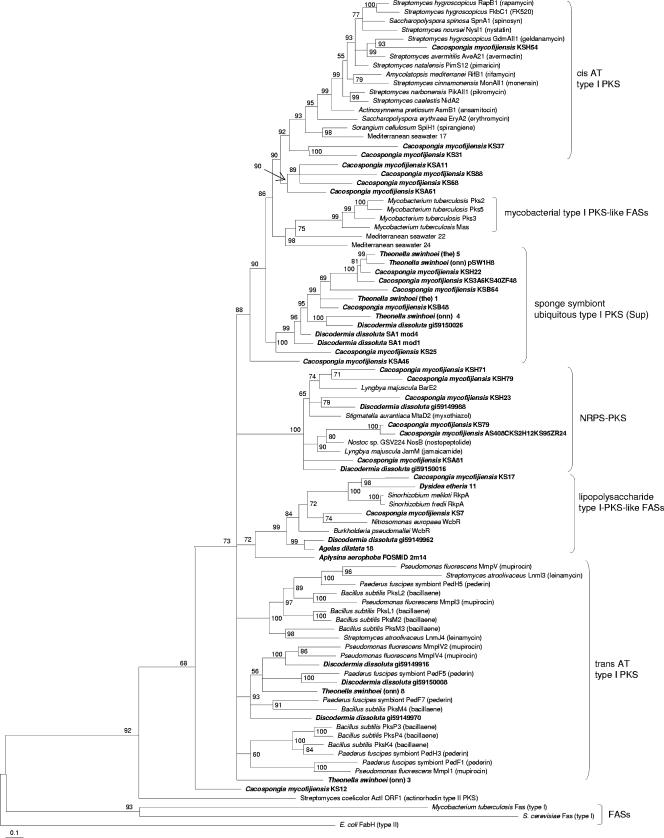
Bacterial type I PKSs are giant proteins consisting of repeated modules, each of which usually adds a single building block to the growing polyketide chain (42). Each module carries a set of catalytic domains for chain extension and modification. The architecture of individual PKS modules resembles that of type I fatty acid synthases (FASs) of animals and some bacteria. However, while FASs usually act iteratively and use a constant set of domains to produce a fully reduced carbon chain after each extension cycle, the domain structure of PKS modules is variable and gives rise to a wide range of optional intermediates. In PKSs, the KS domain, which is present in each module, exhibits the highest degree of conservation among all domains and is therefore especially well suited for phylogenetic analyses, as studies by us (35) and others (12, 18, 20, 23, 28) have shown. In order to investigate PKS diversity in sponges, an extensive phylogenetic analysis of KS amplicons from the metagenomic DNA of Aplysina aerophoba, Cacospongia mycofijiensis, and the theonellamide chemotype (25) of Theonella swinhoei was performed. Furthermore, 16 additional sponge species were screened for the presence of KS genes by PCR, and positive samples were added to the analyses (Table 1). The animals were collected from five different geographic locations and belonged to eight taxonomic orders. For comparison, sequences previously amplified from another Japanese T. swinhoei specimen of the onnamide chemotype were also included (35). Altogether, 498 amplicons were analyzed. Sequences that were less than 97% similar to each other were regarded as unique, resulting in 150 different amplicons. To allow for a phylogenetic classification into enzymatic groups, known sequences of various modular PKS types were retrieved from the GenBank database and used for the analysis. These included cis-acyltransferase (cis-AT) and trans-acyltransferase (trans-AT) PKSs (35), which are the two major groups of bacterial multimodular PKSs involved in the production of bioactive polyketides, and PKS-like monomodular FASs from pathogenic mycobacteria (27). As outgroups, the KS domain from the type I FAS of Saccharomyces cerevisiae, the free-standing type II KS from the E. coli FAS, and the ActI ORF1 product from the type II actinorhodin PKS of Streptomyces coelicolor were selected. Clade formation and branching patterns were best supported by applying Bayesian statistics (Fig. 1). Neighbor-joining, parsimony, and maximum likelihood analyses resulted in basically the same clade topology, although with less support by the bootstrap values. The analyses consistently revealed that 133, that is, 88%, of the 150 sponge-derived PKS amplicons did not group together with previously described PKS or FAS types. Remarkably, 127 of these 133 sequences formed an independent clade, members of which were dominant in amplicon collections of 11 of the 20 examined phylogenetically diverse sponges from different geographic regions. A report on PKS-like sequences in the sponge Discodermia dissoluta has previously revealed an unprecedented multimodular FAS gene cluster among numerous mostly unassigned KS amplicons (40). To relate these findings to the present study, a more detailed phylogenetic analysis of the “ubiquitous” sponge-derived KS sequences was conducted (Fig. 2). Most of the D. dissoluta KSs, including those of the multimodular FAS, clustered together with the predominant sponge amplicon type. Two related KS sequences were also identified in water samples collected from the Mediterranean sampling site of Banyuls sur Mer, France.
TABLE 1.
Sponges used in this study and results of KS amplification
| Sponge species | Order | Location of collection | No. of KS amplicons (total/ubiquitous)a |
|---|---|---|---|
| Aplysinaaerophoba | Verongida | France, Mediterranean Sea | 32/31 |
| Theonella swinhoei (onnamide chemotype) | Lithistida | Japan | 18/15 |
| Theonella swinhoei (theonellamide chemotype) | Lithistida | Japan | 19/19 |
| Cacospongiamycofijiensis | Dictyoceratida | Vanuatu, southern Pacific Ocean | 68/52 |
| Aplysinacavernicola | Verongida | Italy | 2/2 |
| Verongulagigantea | Verongida | Bahamas | 2/2 |
| Aiolochroiacrassa | Verongida | Bahamas | 2/2 |
| Xestospongiamuta | Haplosclerida | Bahamas | 1/1 |
| Siphonodictyoncoralliphagum | Haplosclerida | Bahamas | 2/2 |
| Amphimedoncompressa | Haplosclerida | Bahamas | 1/1 |
| Dysideaetheria | Dendroceratida | Bahamas | 2/0 |
| Agelasdilatata | Agelasida | Bahamas | 1/0 |
No KS amplicons were obtained with Ircinia felix (Dictyoceratida); Plakortis sp. (Homosclerophorida); Cribochalina vasculum, Callyspongia vaginalis, Ectyoplasia ferox, and Tedania ignis (all Poecilosclerida); or Ptilocaulis sp. and Chalinula molitba (both Haplosclerida) from the Bahamas.
FIG. 1.
Phylogenetic tree of partial KS sequences amplified from sponge metagenomes (in bold) and seawater. The tree was analyzed via Bayesian statistics. “Theonella swinhoei (onn)” is the onnamide chemotype of T. swinhoei. Probability values given in percents are shown at the nodes.
FIG. 2.
Fine structure of the sup clade, as analyzed by Bayesian statistics. Sequences labeled “Discodermia dissoluta SA1…” belong to the previously described multimodular FAS from the D. dissoluta metagenome; the architecture of the remaining D. dissoluta genes is unknown (40). “Theonella swinhoei (the)” is the theonellamide chemotype of T. swinhoei. Genomic regions belonging to the sequences marked with an asterisk were isolated in this study.
In a third Bayesian calculation (Fig. 3), we attempted to predict the function of the 23 PKS amplicons that did not group into the dominant sponge PKS clade. Seventeen sequences could be attributed to either trans-AT PKSs (three sequences from one sponge), cis-AT PKSs (three sequences from one sponge), hybrid PKS/nonribosomal peptide synthetases (NRPS) (six sequences from one sponge), or FASs of the RkpA type involved in lipopolysaccharide biosynthesis (30) (five sequences from four sponges). No amplicon exhibited a close relationship with type I mycobacterial FASs or other PKS/FAS types, such as bacterial type I polyunsaturated FASs (26), enediyne PKSs (1), cyanobacterial glycolipid FASs (4), or iterative bacterial type I PKSs (10). Taken together, with only 12 of 150 sequences falling into the cis- and trans-AT PKS groups, a remarkably low number of amplicons could be attributed to enzymes that are commonly involved in the biosynthesis of pharmacologically active polyketides.
FIG. 3.
Bayesian tree showing the phylogenetic affiliation of amplicons with PKSs and FASs of known function.
Characterization of three genomic regions harboring members of the ubiquitous sponge PKS group.
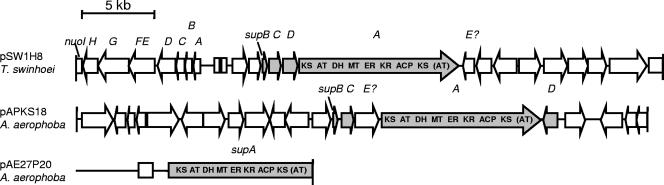
To gain further insights into the organismal origin and function of the ubiquitous PKS type, we isolated and sequenced individual PKS-positive clones of the constructed large insert metagenome libraries. One PKS-encoding genomic region was derived from a T. swinhoei specimen (onnamide chemotype, cosmid pSW1H8, 39.1 kb entirely sequenced), and two were from A. aerophoba (fosmid pAE27P20, 15.6 kb partially sequenced, whole insert of ca. 38 kb, and cosmid pAPKS18, 37.9 kb entirely sequenced) (Fig. 4 and Tables 2 to 4). These PKSs were selected from different subbranches of the clade to ensure that the widest possible range of PKS architectures and taxonomies would be sampled. In clear contrast to other DNA regions previously isolated from the same libraries (36), positive clones were detected with high frequencies. Thus, a T. swinhoei library of 60,000 members contained at least 50 clones harboring the common PKS genes, while onnamide genes were found in the same library in a single clone only. Among 70,000 clones of A. aerophoba, 30 positive cosmids/fosmids were detected. This suggests that the ubiquitous PKS either belongs to a dominant cell type in the sponge-microbial assemblage or is cloned preferentially during DNA isolation and library construction. Sequence analysis revealed similar G+C contents of 64.01% (pSW1H8, T. swinhoei), 66.95% (pAE27P20, A. aerophoba), and 64.99% (pAPKS18, A. aerophoba).
FIG. 4.
Genetic organization of the DNA regions isolated from the T. swinhoei and A. aerophoba metagenomes. Genes putatively belonging to the sup cluster are shaded in gray.
TABLE 2.
Putative genes identified on the genomic fragment pAPKS18 (A. aerophoba)
| CDS | Putative function | Most similar homolog (protein, accession no., and origin) | Similarity/identity (%) | No. of aab |
|---|---|---|---|---|
| ORF1 | Peptidase S15 | NOC_2221 (YP_344211), Nitrosococcusoceani ATCC 19707 | 65/51 | 675 |
| ORF2 | Agmatinase | RNGR00257 (AAQ87384), Rhizobium sp. strain NGR234 | 57/41 | 321 |
| ORF3 | Putative methyltransferase | RoseDRAFT_3522 (AAFG02000001), Silicibacter sp. strain TM1040 | 53/39 | 162 |
| ORF4 | TetR-type regulator | ArthDRAFT_2587 (AAHG01000004), Arthrobacter sp. strain FB24 | 45/28 | 195 |
| ORF5 | Cytochrome c family | RB8450 (CAD78645), Rhodopirellula baltica SH 1 | 54/37 | 679 |
| ORF6 | FADa-dependent oxidoreductase | BB2693 (BX640445), Bordetellabronchiseptica | 60/45 | 429 |
| ORF7 | Sulfatase | BambDRAFT_1967 (ZP_00689010), Burkholderiaambifaria AMMD | 62/50 | 503 |
| ORF8 | Conserved hypothetical protein | RxylDRAFT_3044 (ZP_00599827), Rubrobacter xylanophilus DSM 9941 | 62/51 | 310 |
| ORF9 | γ-Glutamyl transferase | BradDRAFT_6380 (ZP_00857491), Bradyrhizobium sp. strain BTAi1 | 68/55 | 598 |
| ORF10 | d-3-Phosphoglycerate dehydrogenase | RNGR00097 (AY316746), Rhizobium sp. strain NGR234 | 73/65 | 346 |
| ORF11 | Erythritol kinase | BradDRAFT_0972 (ZP_00863500), Bradyrhizobium sp. strain BTAi1 | 67/54 | 527 |
| ORF12 | Epimerase | BcenDRAFT_5477 (AAHI01000003), Burkholderiacenocepacia AU 1054 | 48/34 | 446 |
| SupB | ACP | AcpP (BA000028), Oceanobacillusiheyensis | 47/29 | 76 |
| SupC | Phosphopantetheinyl transferase | PT_SA1 (AAY00023), Discodermiadissoluta symbiont | 68/54 | 250 |
| SupE | Permease | CP000124 (YP_332786), Burkholderiapseudomallei 1710b | 46/27 | 597 |
| SupA | PKS (KS AT DH MT ER KR ACP KS [AT]) | SA1_PKSB (AAY00026), Discodermia dissoluta symbiont | 64/50 | 3,403 |
| SupD | Hydrolase | PoxA (AAT09607), Pseudomonas aeruginosa PAO1 | 41/28 | 326 |
| ORF18 | Aminotransferase | PA0299 (NP_248990), Pseudomonas aeruginosa PAO1 | 70/55 | 444 |
| ORF19 | Membrane protein | Magn03009209 (ZP_00054563), Magnetospirillum magnetotacticum MS-1 | 68/52 | 517 |
| ORF20 | Permease | Rsph17025DRAFT_3795 (ZP_00914910), Rhodobactersphaeroides ATCC 17025 | 74/54 | 275 |
| ORF21 | Permease | Rrub02002558 (ZP_00269018), Rhodospirillumrubrum | 72/56 | 298 |
FAD, flavin adenine dinucleotide.
aa, amino acids.
TABLE 4.
Putative genes identified on the genomic fragment pAE27P20 (A. aerophoba)
| CDS | Putative function | Most similar homolog (protein, accession no., and origin) | Similarity/identity (%) | No. of aaa |
|---|---|---|---|---|
| ORF1 | Transcriptional regulator, LuxR family | Bcepa03003049 (ZP_00215451), Burkholderiacepacia | 54/31 | 297 |
| SupA (partial) | PKS (KS AT DH MT ER KR ACP KS [AT]) |
aa, amino acids.
Each of the three isolated regions harbored a PKS gene cluster embedded in distinct genomic environments (Fig. 4). While the PKS clusters on pSW1H8 from T. swinhoei and pAPKS18 from A. aerophoba were cloned in their entirety, parts of the A. aerophoba pAE27P20 cluster were located outside of the fosmid insert. All genes present on the fosmid/cosmid inserts exhibited the highest similarity to bacterial genes; were preceded by putative Shine-Dalgarno sequences, as judged by strong similarities to the consensus sequence AGGAGG; and lacked discernible introns. With the exception of a noncoding 2-kb region in pSW1H8 (T. swinhoei) and a 5.8-kb region on pAPKS18, the genes on these cosmids/fosmids were densely packed with an average distance below 100 bp, indicating the presence of polycistronic operons, as is typical for prokaryotes. On the sequenced portion of pAE27P20 (A. aerophoba), the PKS gene was the only coding sequence (CDS) that appeared intact. In addition, one putative pseudogene was identified. In summary, these features clearly implicate a bacterial origin of at least pSW1H8 and pAPKS18.
Phylogenetic analysis to identify the PKS host.
The attribution of metagenomic DNA regions to a specific organism can be challenging. In this study, no ribosomal operons could be identified that could serve as taxonomic markers. We therefore attempted to extract host information from essential genes. pSW1H8 carried a large gene cluster putatively encoding nine components (NuoA to NuoI) of the NADH:ubiquinone oxidoreductase machinery from the respiration chain. In a BLAST search, all deduced proteins exhibited high similarities (60% to 94%) to homologs from bacteria of the Deinococcus-Thermus phylum. However, as the closest BLAST hit is not always identical with the nearest actual neighbor, a phylogenetic analysis of NuoG sequences from various bacteria was performed (Fig. 5). In the inferred tree, the symbiont protein was part of a well-supported clade together with homologs from Deinococcus radiodurans, Deinococcus geothermalis, and Thermus thermophilus, thus corroborating the BLAST results. On the other two fosmids, only putative nonessential genes were present with the nearest homologs being in taxonomically diverse bacteria. Therefore, a tetranucleotide analysis (43) was conducted to determine whether the three DNA segments belonged to organisms that are closely related to each other or to members of the Deinococcus-Thermus phylum. This resulted in correlation coefficients in the range of 0.55 to 0.63, which were too low to be taxonomically significant.
FIG. 5.
Neighbor-joining phylogenetic analyses of NADH:ubiquinone oxidoreductase (Nuo) G-chain proteins. The sequence from the T. swinhoei metagenome is shown in bold. Bootstrap values are given at the nodes.
Analysis of the PKS architecture.
Gene analysis of the three cosmid/fosmid inserts revealed the presence of exceptionally small PKS clusters that resembled neither the typical multimodular genes responsible for complex polyketide biosynthesis nor the previously characterized giant D. dissoluta FAS (Fig. 4). On both pSW1H8 (T. swinhoei) and pAPKS18 (A. aerophoba), a large open reading frame, designated supA for sponge symbiont ubiquitous PKS, with identical architecture was found. The deduced PKS contains a single complete module with a KS, an AT, a dehydratase (DH), a methyltransferase (MT), an enoylreductase (ER), a ketoreductase (KR), and an acyl carrier protein (ACP) domain. In addition, the C terminus consists of a short region with a KS and an AT domain, the latter of which lacks the conserved serine residue crucial for activity and contains a glycine instead. In contrast to almost all other PKSs and FASs, no thioesterase domain was present at the C terminus that normally releases the assembled carbon chain from the enzyme. Adjacent to supA, four further CDSs common to the two genome regions were identified. SupB strongly resembles free-standing ACPs, SupC is homologous to 5′-phosphopantetheinyl transferases involved in the conversion of apo-ACPs to the holo proteins, and SupD exhibits similarity to enzymes of the αβ-hydrolase superfamily. Inserted within the sup cluster of pAPKS18 lies an additional gene, the protein product of which exhibits similarity to permease proteins of ABC transporters. Although the location suggests an involvement in the sup pathway, pSW1H8 (T. swinhoei) lacked an obvious counterpart. On this cosmid, a putative permease gene was positioned directly downstream of supA, but since it exhibited only low similarity to the pAPKS18 (A. aerophoba) gene, its participation in the sup pathway remains a matter of speculation.
The PKS encoded on pAE27P20 (A. aerophoba) seemed to be highly similar to these two PKSs. A deduced protein was again found with the domain set KS AT DH MT ER KR ACP KS (AT) and an identity of 60% and 57% to SupA from pSW1H8 (T. swinhoei) and pAPKS18 (A. aerophoba), respectively. Since the downstream end of this gene was located outside of the cloned region, it is currently unknown whether the protein also ends with the AT domain or contains additional modules. Similarly, homologs of the other sup genes were missing that would indicate an orthologous function of the PKS.
DISCUSSION
In this study, PKS diversities in 20 different sponges were assessed to obtain insights into the elusive chemistry of uncultivated sponge bacteria. Unexpectedly, metagenomic libraries in bacteriosponges were found to contain a large number of clones harboring an architecturally unusual type of PKS that has no previously published close homologs in other biological sources. The sup genes were detected in 11 species of the class Demospongiae, regardless of subclass-level taxonomy or locality of collection. Close relatives of these PKSs were previously also found in D. dissoluta (40), and Kim and Fuerst recently reported additional members of this group from the Australian bacteriosponge Pseudoceratina clavata, thus further extending the geographic distribution of the sup genes (19). Notable was the unusually shallow branching topology of the subclade in spite of the taxonomic heterogeneity of the examined sponges. This finding is inconsistent with the possibility that the sequences originate from the animals themselves but would be expected if a group of closely related, ubiquitous sponge symbionts are the source organisms. Indeed, it is noteworthy that the sponges from which the ubiquitous PKSs were recovered all belong to the “high-microbial-abundance” group (U. Hentschel et al., unpublished data). A bacterial source of the sup pathway was confirmed by sequencing three extended genomic regions, which exhibited distinctly prokaryotic features. The fact that sup genes were also detected in one seawater sample could be explained if the producing bacteria also exist as a free-living form. Alternatively, members of the sponge microbiota may be released accidentally, for example, through grazing and predation by snails, fish, or turtles. Sequencing efforts have provided extensive DNA data on marine bacteria (49); however, sup homologs from sources other than sponges have not yet been found in the GenBank database. Provided that there is no experimental bias, it can therefore be assumed that if unassociated bacteria carrying the sup genes exist, these organisms form only a minor portion of the free-living microbial community.
These data on metabolic genes in sponge metagenomes remarkably reflect previous 16S rRNA-based studies that detected globally distributed but highly sponge-specific microorganisms in a wide range of host species (13, 14). Analyses of nuo housekeeping genes strongly indicate that at least the T. swinhoei symbiont belongs to the Deinococcus-Thermus phylum. A tetranucleotide analysis did not provide further evidence for a relationship to such bacteria. However, since tetranucleotide usage can vary even within the same bacterial genome (38), the predictive power of such an analysis is limited. Bacteria of the Deinococcus-Thermus phylum have indeed been detected in several sponges, e.g., in D. dissoluta (40) and Halichondria panicea (16), although not as ubiquitously as suggested by our results. This apparent underrepresentation of Deinococcus-Thermus 16S rRNA gene sequences might be caused by biases introduced during DNA preparation, PCR amplification, or cloning conducted with the extremely complex cell mixtures. On the other hand the sup genes might have been transferred horizontally into a Deinococcus/Thermus member from a more common sponge bacterium and might thus not generally be present in this phylum. Finally, it cannot be excluded that a lateral transfer (5) of the nuo genes between unrelated bacteria might have occurred, resulting in tree topologies that are incongruent with bacterial phylogeny. Although a transfer of housekeeping genes is generally rare, instances of horizontally acquired genes of bacterial respiration are indeed known (3, 31). To allow for an unequivocal taxonomic classification, sampling of additional symbiont genome regions will therefore be necessary.
Sequencing of three extended DNA regions from two sponges revealed unusually small PKS genes encoding a single intact module. Monomodular bacterial PKSs have rarely been documented. Such an architecture is typical for animal FASs (41), to which SupA also bears architectural resemblance. In bacteria, fatty acid biosynthesis is usually catalyzed by free-standing individual enzymes, the type II FASs (24). Among the few known cases of type I, monomodular FASs or PKSs from bacteria are enzymes from pathogenic mycobacteria, many of which are involved in the biosynthesis of methyl-branched fatty acid components of the cell wall, such as mycocerosic acid (27). The architecture of SupA strongly suggests a biosynthetic product with a similar structure: the presence of KR, DH, and ER domains on the module indicates that the resulting compound is a fully reduced, fatty-acid-type molecule, which should be methyl branched to account for the MT domain. Kim and Fuerst have isolated a further monomodular member of the sup group (19). Although this member exhibits high sequence similarity to the PKSs of our study, an MT domain was not reported. However, closer examination of the sequence data also revealed such a domain at the corresponding enzymatic region. In spite of the close similarity of these enzymes, the unusual fatty acids in sponges can apparently be biosynthesized by more than one type of FAS. The D. dissoluta metagenome harbors a giant PKS cluster that comprises 15 modules and has been proposed to encode the production of an octamethyl-branched long-chain fatty acid (40). Like SupA, the last module terminates with an AT domain with the active-site serine replaced by a glycine residue, indicating that the domain is more than a mere decayed evolutionary remnant. Current data do not allow a prediction whether it plays a catalytic role or is rather present for structural reasons.
Methyl-branched fatty acids are indeed present in numerous sponges (44, 45), and it is generally assumed that the true producers are bacteria (11). Aplysina (syn. Verongia) aerophoba and other verongid sponges display an unusually high level of fatty acid diversity, many of the fatty acids being methyl branched, indicating a microbial origin (29). Taking the common presence of mid-chain-branched alkanes in fossil sediments and fuels into account, the intriguing hypothesis has been put forward that microbial producers of methylated fatty acids were widespread in the past but are largely extinct today (45). Sponges, which have changed little during evolution, might therefore provide one of the few extant habitats for such microbes.
The biological functions of methyl-branched fatty acids in sponge symbionts are unknown. Interestingly, Mycobacterium tuberculosis deletion mutants lacking mycocerosate and other cell wall lipids exhibited an attenuated growth in various animal hosts, indicating that the compounds are crucial for the infection process (2). It is an interesting question whether the sup genes play a similar role in that they might be important for the establishment and maintenance of symbiosis in sponges. Studies to address this issue and to elucidate the chemical structures of the metabolites are under way.
Unexpectedly, only 8% of the amplified sequences could be attributed to PKS families usually involved in the production of bioactive polyketides, i.e., cis-AT PKSs, trans-AT PKSs, and hybrid NRPS-PKSs. This result bears important implications for biotechnology. Previous to this study, the biggest challenge in the identification of pharmacologically relevant PKS genes in sponges has been to identify the correct target genes among the enormous number of PKS homologs present in the metagenome. This problem had so far been solved only in the case of the onnamides by exploiting sequence information on the related pederin gene cluster (34). However, the present qualitative and quantitative investigation of sponge PKSs should permit cloning of natural product genes even without detailed a priori knowledge of the target genes. After amplification of KS fragments with degenerate PCR primers, the few “drug-like” amplicons belonging to the cis-AT and trans-AT type will be easily distinguishable from the FAS-type majority by phylogenetic analysis. These sequences can then serve as the basis for library screening and ultimately heterologous expression to create environmentally sound sources of drug candidates. In addition to the known PKS types, similar phylogenetic strategies might in future also reveal other, previously unrecognized PKS groups with novel architectures and pharmacological potential.
TABLE 3.
Putative genes identified on the genomic fragment pSW1H8 (T. swinhoei)
| CDS | Putative function | Most similar homolog (protein, accession no., and origin) | Similarity/identity (%) | No. of aab |
|---|---|---|---|---|
| NuoI (partial) | NADH:ubiquinone oxidoreductase, I subunit | NuoF (AE000513), Deinococcusradiodurans | 90/79 | |
| NuoH | NADH:ubiquinone oxidoreductase, H subunit | DR1498 (AAF11065), Deinococcusradiodurans | 77/65 | 337 |
| NuoG | NADH:ubiquinone oxidoreductase, G subunit | DR1499 (AAF11066), Deinococcusradiodurans | 60/48 | 811 |
| NuoEF | NADH:ubiquinone oxidoreductase, E+F subunit | NuoF (AAHE01000001), Deinococcusgeothermalis | 62/76 | 639 |
| NuoD | NADH:ubiquinone oxidoreductase, D subunit | DR1503 (AAF11069), Deinococcusradiodurans | 77/64 | 404 |
| NuoC | NADH:ubiquinone oxidoreductase, C subunit | NQO5 (AAA97940), Thermusthermophilus | 68/53 | 201 |
| NuoB | NADH:ubiquinone oxidoreductase, B subunit | DR1505 (AAF11070), Deinococcusradiodurans | 94/85 | 178 |
| NuoA | NADH:ubiquinone oxidoreductase, A subunit | DR1506 (AAF11072), Deinococcusradiodurans | 72/59 | 108 |
| ORF9a | Proteic killer suppression protein | PSPTO0423 (AAO53967), Pseudomonas syringae | 67/43 | |
| ORF10a | Putative transcriptional regulator | R00504 (CAC41941), Rhizobiummeliloti | 67/50 | |
| ORF11 | ABC transporter, substrate binding protein | ALR0543 (EAL75729), Anabaena sp. strain PCC7120 | 66/46 | 304 |
| ORF12 | Putative ATP binding protein | NE1564 (CAD85475), Nitrosomonaseuropaea | 66/49 | 251 |
| SupB | ACP | AcpP (AAC07567), Aquifexaeolicus | 51/30 | 76 |
| SupC | Phosphopantetheinyl transferase | SLR0495 (BAA10326), Synechocystis sp. strain PCC6803 | 52/39 | 252 |
| SupD | Hydrolase or acyltransferase? | PA5513 (AAG08898), Pseudomonas aeruginosa | 37/24 | 307 |
| SupA | PKS (KS AT DH MT ER KR ACP KS [AT]) | 3,461 | ||
| ORF17 | Permease protein | BtuC (AAQ59234), Chromobacteriumviolaceum | 30/47 | 222 |
| ORF18 | ABC transporter, iron binding protein | ALR1382 (BAB73339), Anabaena sp. strain PCC7120 | 68/47 | 332 |
| ORF19 | Peptide amidase precursor | PAM (CAC93616), Xanthomonasmaltophilia | 67/53 | 498 |
| ORF20 | Sulfatase | AtsG (CAD74202), Rhodopirellulabaltica | 65/52 | 515 |
| ORF21 | Sugar permease | BH1865 (BAB05584), Bacillus halodurans | 65/43 | 442 |
| ORF22 | Sugar permease | BH1866 (BAB05585), B. halodurans | 66/45 | 288 |
| ORF23 | Sugar permease | ABC1215 (BAD63753), Bacillus clausii | 71/51 | 288 |
| ORF24 | Sarcosine dehydrogenase | SAV6951 (BAC74662), Streptomycesavermitilis | 58/44 | 823 |
| ORF25a | SnoK-like protein | BC5091 (AAP11960), Bacillus cereus | 49/26 | |
| ORF26 | X-Pro dipeptidase | ST2323 (BAB67434), Sulfolobustokodaii | 50/30 |
Belongs to a pseudogene.
aa, amino acids.
Acknowledgments
We thank S. Matsunaga and N. Fusetani (University of Tokyo) for obtaining T. swinhoei; P. Crews and P. Wenzel (University of Santa Cruz) for the C. mycofijiensis samples; J. R. Pawlik (University of North Carolina, Wilmington) for excellent cruise organization; S. Zea (Universidad Nacional de Colombia, INVEMAR) for sponge identification; J. P. Schülke, S. Proksch (University of Würzburg), and D. Hui (MPI for Chemical Ecology, Jena, Germany) for assistance in the laboratory; and E. Dittmann (Humboldt University, Berlin, Germany) for discussion of PKS phylogeny.
This study was supported by grants of the DFG (EvoMet: PI430 5/1), the bmb+f (BIOTECmarin: 03F0414F), and the JSPS (S02275) to J.P. and by grants of the DFG (SFB630 TP A5) and the bmb+f (BIOTECmarin: 03F0414E) to U.H.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Ahlert, J., E. Shepard, N. Lomovskaya, E. Zazopoulos, A. Staffa, B. O. Bachmann, K. X. Huang, L. Fonstein, A. Czisny, R. E. Whitwam, C. M. Farnet, and J. S. Thorson. 2002. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297:1173-1176. [DOI] [PubMed] [Google Scholar]
- 2.Azad, A. K., T. D. Sirakova, N. D. Fernandes, and P. E. Kolattukudy. 1997. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272:16741-16745. [DOI] [PubMed] [Google Scholar]
- 3.Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. R. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, E. L., M. F. Cohen, and J. C. Meeks. 1997. A polyketide-synthase-like gene is involved in the synthesis of heterocyst glycolipids in Nostoc punctiforme strain ATCC 29133. Arch. Microbiol. 167:251-258. [DOI] [PubMed] [Google Scholar]
- 5.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 6.Faulkner, D. J. 2000. Highlights of marine natural products chemistry (1972-1999). Nat. Prod. Rep. 17:1-6. [DOI] [PubMed] [Google Scholar]
- 7.Fieseler, L., M. Horn, M. Wagner, and U. Hentschel. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fieseler, L., A. Quaiser, C. Schleper, and U. Hentschel. 2006. Analysis of the first genome fragment from the marine sponge-associated, novel candidate phylum Poribacteria by environmental genomics. Environ. Microbiol. 8:612-624. [DOI] [PubMed] [Google Scholar]
- 9.Friedrich, A. B., I. Fischer, P. Proksch, J. Hacker, and U. Hentschel. 2001. Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 10.Gaitatzis, N., B. Silakowski, B. Kunze, G. Nordsiek, H. Blocker, G. Höfle, and R. Müller. 2002. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J. Biol. Chem. 277:13082-13090. [DOI] [PubMed] [Google Scholar]
- 11.Gillan, F. T., I. L. Stoilov, J. E. Thompson, R. W. Hogg, C. R. Wilkinson, and C. Djerassi. 1988. Phospholipid studies of marine organisms. 21. Fatty acids as biological markers for bacterial symbionts in sponges. Lipids 23:1139-1145. [DOI] [PubMed] [Google Scholar]
- 12.Ginolhac, A., C. Jarrin, B. Gillet, P. Robe, P. Pujic, K. Tuphile, H. Bertrand, T. M. Vogel, G. Perriere, P. Simonet, and R. Nalin. 2004. Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Appl. Environ. Microbiol. 70:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentschel, U., K. M. Usher, and M. W. Taylor. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55:167-177. [DOI] [PubMed] [Google Scholar]
- 15.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 16.Imhoff, J. F., and R. Stöhr. 2003. Sponge-associated bacteria: general overview and special aspects of bacteria associated with Halichondria panicea, p. 35-57. In W. E. G. Müller (ed.), Marine molecular biotechnology. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 17.Jefford, C. W., G. Bernardinelli, J. Tanaka, and T. Higa. 1996. Structures and absolute configurations of the marine toxins, latrunculin A and laulimalide. Tetrahedron Lett. 37:159-162. [Google Scholar]
- 18.Jenke-Kodama, H., A. Sandmann, R. Müller, and E. Dittmann. 2005. Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22:2027-2039. [DOI] [PubMed] [Google Scholar]
- 19.Kim, T. K., and J. A. Fuerst. 2006. Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: culture-dependent and culture-independent approaches. Environ. Microbiol. 8:1460-1470. [DOI] [PubMed] [Google Scholar]
- 20.Kim, T. K., A. K. Hewavitharana, P. N. Shaw, and J. A. Fuerst. 2006. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl. Environ. Microbiol. 72:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafi, F. F., M. J. Garson, and J. A. Fuerst. 2005. Culturable bacterial symbionts isolated from two distinct sponge species (Pseudoceratina clavata and Rhabdastrella globostellata) from the Great Barrier Reef display similar phylogenetic diversity. Microb. Ecol. 50:213-220. [DOI] [PubMed] [Google Scholar]
- 22.Li, A. Y., and J. Piel. 2002. A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem. Biol. 9:1017-1026. [DOI] [PubMed] [Google Scholar]
- 23.Lopez, J. V. 2003. Naturally mosaic operons for secondary metabolite biosynthesis: variability and putative horizontal transfer of discrete catalytic domains of the epothilone polyketide synthase locus. Mol. Genet. Genomics 270:420-431. [DOI] [PubMed] [Google Scholar]
- 24.Lu, Y. J., Y. M. Zhang, and C. O. Rock. 2004. Product diversity and regulation of type II fatty acid synthases. Biochem. Cell Biol.-Biochim. Biol. Cell. 82:145-155. [DOI] [PubMed] [Google Scholar]
- 25.Matsunaga, S., N. Fusetani, K. Hashimoto, and M. Walchli. 1989. Bioactive marine metabolites. 26. Theonellamide F—a novel antifungal bicyclic peptide from a marine sponge Theonella sp. J. Am. Chem. Soc. 111:2582-2588. [Google Scholar]
- 26.Metz, J. G., P. Roessler, D. Facciotti, C. Levering, F. Dittrich, M. Lassner, R. Valentine, K. Lardizabal, F. Domergue, A. Yamada, K. Yazawa, V. Knauf, and J. Browse. 2001. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290-293. [DOI] [PubMed] [Google Scholar]
- 27.Minnikin, D. E., L. Kremer, L. G. Dover, and G. S. Besra. 2002. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 9:545-553. [DOI] [PubMed] [Google Scholar]
- 28.Moffitt, M. C., and B. A. Neilan. 2003. Evolutionary affiliations within the superfamily of ketosynthases reflect complex pathway associations. J. Mol. Evol. 56:446-457. [DOI] [PubMed] [Google Scholar]
- 29.Nechev, J., W. W. Christie, R. Robaina, F. de Diego, S. Popov, and K. Stefanov. 2002. Lipid composition of the sponge Verongia aerophoba from the Canary Islands. Eur. J. Lipid Sci. Technol. 104:800-807. [Google Scholar]
- 30.Parada, M., J. M. Vinardell, F. J. Ollero, A. Hidalgo, R. Gutierrez, A. M. Buendia-Claveria, W. Lei, I. Margaret, F. J. Lopez-Baena, A. M. Gil-Serrano, M. A. Rodriguez-Carvajal, J. Moreno, and J. E. Ruiz-Sainz. 2006. Sinorhizobium fredii HH103 mutants affected in capsular polysaccharide (KPS) are impaired for nodulation with soybean and Cajanus cajan. Mol. Plant-Microbe Interact. 19:43-52. [DOI] [PubMed] [Google Scholar]
- 31.Pereira, M. M., T. M. Bandeiras, A. S. Fernandes, R. S. Lemos, A. M. P. Melo, and M. Teixeira. 2004. Respiratory chains from aerobic thermophilic prokaryotes. J. Bioenerg. Biomembr. 36:93-105. [DOI] [PubMed] [Google Scholar]
- 32.Piel, J. 2004. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 21:519-538. [DOI] [PubMed] [Google Scholar]
- 33.Piel, J. 2002. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 99:14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piel, J., D. Butzke, N. Fusetani, D. Q. Hui, M. Platzer, G. P. Wen, and S. Matsunaga. 2005. Exploring the chemistry of uncultivated bacterial symbionts: antitumor polyketides of the pederin family. J. Nat. Prod. 68:472-479. [DOI] [PubMed] [Google Scholar]
- 35.Piel, J., D. Hui, N. Fusetani, and S. Matsunaga. 2004. Targeting polyketide synthases with iteratively acting acyltransferases from metagenomes of uncultured bacterial consortia. Environ. Microbiol. 6:921-927. [DOI] [PubMed] [Google Scholar]
- 36.Piel, J., D. Hui, G. Wen, D. Butzke, M. Platzer, N. Fusetani, and S. Matsunaga. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 101:16222-16227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piel, J., G. Wen, M. Platzer, and D. Hui. 2004. Unprecedented diversity of catalytic domains in the first four modules of the putative pederin polyketide synthase. Chembiochem 5:93-98. [DOI] [PubMed] [Google Scholar]
- 38.Reva, O., and B. Tummler. 2005. Differentiation of regions with atypical oligonucleotide composition in bacterial genomes. BMC Bioinformatics 6:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning—a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Schirmer, A., R. Gadkari, C. D. Reeves, F. Ibrahim, E. F. Delong, and C. R. Hutchinson. 2005. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 71:4840-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer, E., and J. Hofmann. 2004. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol. Mol. Biol. Rev. 68:501-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staunton, J., and K. J. Weissman. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380-416. [DOI] [PubMed] [Google Scholar]
- 43.Teeling, H., J. Waldmann, T. Lombardot, M. Bauer, and F. O. Glockner. 2004. TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics 5:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiel, V., M. Blumenberg, J. Hefter, T. Pape, S. Pomponi, J. Reed, J. Reitner, G. Wörheide, and W. Michaelis. 2002. A chemical view of the most ancient metazoa—biomarker chemotaxonomy of hexactinellid sponges. Naturwissenschaften 89:60-66. [DOI] [PubMed] [Google Scholar]
- 45.Thiel, V., A. Jenisch, G. Wörheide, A. Löwenberg, J. Reitner, and W. Michaelis. 1999. Mid-chain branched alkanoic acids from “living fossil” demosponges: a link to ancient sedimentary lipids? Org. Geochem. 30:1-14. [Google Scholar]
- 46.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vacelet, J. 1975. Étude en microscopie électronique de l'association entre bactéries et spongiaires du genre Verongia (Dictyoceratida). J. Microsc. Biol. Cell. 23:271-288. [Google Scholar]
- 48.Vacelet, J., and C. Donadey. 1977. Electron-microscope study of association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30:301-314. [Google Scholar]
- 49.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 50.West, L. M., P. T. Northcote, and C. N. Battershill. 2000. Peloruside A: a potent cytotoxic macrolide isolated from the New Zealand marine sponge Mycale sp. J. Org. Chem. 65:445-449. [DOI] [PubMed] [Google Scholar]