Abstract
International drilling projects for the study of microbial communities in the deep-subsurface hot biosphere have been expanded. Core samples obtained by deep drilling are commonly contaminated with mesophilic microorganisms in the drilling fluid, making it difficult to examine the microbial community by 16S rRNA gene clone library analysis. To eliminate mesophilic organism contamination, we previously developed a new method (selective phylogenetic analysis [SePA]) based on the strong correlation between the guanine-plus-cytosine (G+C) contents of the 16S rRNA genes and the optimal growth temperatures of prokaryotes, and we verified the method's effectiveness (H. Kimura, M. Sugihara, K. Kato, and S. Hanada, Appl. Environ. Microbiol. 72:21-27, 2006). In the present study we ascertained SePA's ability to eliminate contamination by archaeal rRNA genes, using deep-sea hydrothermal fluid (117°C) and surface seawater (29.9°C) as substitutes for deep-subsurface geothermal samples and drilling fluid, respectively. Archaeal 16S rRNA gene fragments, PCR amplified from the surface seawater, were denatured at 82°C and completely digested with exonuclease I (Exo I), while gene fragments from the deep-sea hydrothermal fluid remained intact after denaturation at 84°C because of their high G+C contents. An examination using mixtures of DNAs from the two environmental samples showed that denaturation at 84°C and digestion with Exo I completely eliminated archaeal 16S rRNA genes from the surface seawater. Our method was quite useful for culture-independent community analysis of hyperthermophilic archaea in core samples recovered from deep-subsurface geothermal environments.
The temperature of a subsurface environment increases steadily with depth at a fairly uniform rate. The thermal gradient is approximately 15 to 30°C per km of depth in nonvolcanic regions (14). Deep-subsurface crusts are therefore considered extremely hot environments. To date, a number of thermophilic and hyperthermophilic prokaryotes have been isolated from geothermal and hydrothermal environments, such as terrestrial hot springs and deep-sea hydrothermal vent fields (16, 32, 39, 41). Pyrolobus fumarii, on record as the prokaryote having the highest growth temperature, is known to have a maximum growth temperature of 113°C (1). In addition, an archaeal strain that is closely related to Pyrodictium occultum and Pyrobaculum aerophilum has the highest recorded maximum growth temperature, 121°C (21). Since prokaryotes can survive at temperatures up to 121°C, the prokaryotic habitat could reach a depth of 8 km in regions of crust that exhibit low thermal gradients (15°C per km or less) and a depth of 4 km where thermal gradients are high (30°C per km). Many microbiologists therefore take great interest in the deep-subsurface biosphere, because it is a vast space that can support an immense number and variety of thermophilic prokaryotes (13, 27, 30).
In recent years, international deep drilling projects for scientific study, including microbiological research, have been expanded. Geothermal core samples have already been recovered from extremely hot subsurface environments, such as that examined in the German Continental Deep Drilling Project “KTB” (8, 10) and that examined during the Ocean Drilling Program Leg 193 cruise (23, 25, 33). In these studies, the molecular biology of subsurface microbial communities was investigated using core samples obtained by drilling. Drilling fluid is indispensable for obtaining rock cores, and seawater and river water are commonly used as drilling fluids during ocean and continental deep drilling, respectively. Thus, it is almost inevitable that the rock cores are exposed to drilling fluid during excavation. The cell density of mesophiles in drilling fluid is clearly higher than the cell densities of thermophiles and hyperthermophiles in core samples recovered from deep-subsurface geothermal environments. This contamination by mesophilic microbes invalidates culture-independent community analysis based on rRNA gene sequences, because PCRs targeting rRNA genes amplify genes that originate in contaminating mesophiles, as well as in indigenous thermophiles and hyperthermophiles. Tracer monitoring with perfluorocarbon chemicals or fluorescent microspheres has recently been used to test for microbial contamination (35, 36), and the indigenous microbial community has been analyzed using internal parts of rock cores that are thought to be uncontaminated. This type of monitoring is, however, applicable only to complete and dense core samples. Since deep-subsurface materials are frequently altered and cracked by heat from magma, the tracer monitoring technique is probably less useful for determining microbial contamination in deep drilling.
We recently proposed a new method for eliminating 16S rRNA genes of contaminating mesophiles in drilling core samples (24). This method is based on the following findings: the optimal growth temperatures (Topt) of prokaryotes are strongly correlated with the guanine-plus-cytosine (G+C) contents of 16S rRNA sequences (11, 12, 22); the rRNA sequences of thermophiles and hyperthermophiles tend to have high G+C contents, and the high-G+C-content rRNA gene fragments are indicative of melting temperatures (Tm) that are higher than those of mesophiles; and the clear difference in Tm values allows selective denaturation of PCR-amplified 16S rRNA gene fragments of contaminating mesophiles by a moderate heat treatment and then elimination of the fragments by digestion with exonuclease I (Exo I), an enzyme that is strictly specific for single-stranded DNAs (3).
In a previous paper (24), using a mixture of terrestrial hot spring water (76°C) and river water (14°C), we demonstrated that this technique, called selective phylogenetic analysis (SePA), could feasibly eliminate almost all 16S rRNA gene fragments originating from mesophilic bacteria. In the present study, we examined whether SePA can differentiate archaeal 16S rRNA gene fragments. To ascertain whether SePA eliminates all the fragments amplified from mesophilic archaea, deep-sea hydrothermal fluid (117°C) and surface seawater (29.9°C) were used as simulated deep-subsurface geothermal samples and drilling fluid, respectively.
MATERIALS AND METHODS
Sample collection.
Samples of deep-sea hydrothermal fluid were collected during the YK05-09 cruise, which was conducted in July and August 2005 and during which the deep-sea submersible vehicle Shinkai 6500 and R/V Yokosuka were used to investigate active submarine hydrothermal systems in the southern Mariana Trough. A hydrothermal fluid sample was collected during dive 903 from an active mound in the Archaean field (12o56.336′N, 143o37.894′E; depth, 3,002 m), where fluid was venting vigorously both from the surface of the mound and from the top of chimney structures. Prior to sampling, the temperature of the hydrothermal fluid was determined with a platinum resistance thermometer by putting it into venting orifices. While higher-temperature fluid (343°C) was found at the top of the chimney, the temperature of the shimmering fluid at the mound surface was 117°C. The fluid sample used in this study was collected from the mound surface. About 10 liters of the hydrothermal fluid was drawn into a clean plastic bag using an impeller pump installed in a sample basket of the deep-sea submersible vehicle. At the same time, surface seawater (29.9°C) was collected in the southern Mariana Trough area from the deck of R/V Yokosuka. The surface seawater was obtained using a clean plastic bucket prewashed with sterilized water and rerinsed with surface seawater. Exactly 4 liters of each water sample was each aseptically filtered with a Sterivex-GV filter unit (pore size, 0.22 μm; Millipore, Bedford, MA) using sterilized silicon tubes and tubing pumps. The microbial cells trapped in the filter units were immediately frozen at −80°C in the shipboard laboratory until DNA was extracted.
DNA extraction, real-time PCR, and melting curve analysis.
The bulk DNAs of microbes trapped by the filter units were extracted using a method described by Somerville et al. (37). The filters were washed with 10 ml of SET buffer (20% [wt/vol] sucrose, 50 mM EDTA, 50 mM Tris-HCl; pH 8.0), and 1.8 ml of SET buffer was added to each filter unit. The microbial cells were lysed with a solution of lysozyme and proteinase K in the filter units. The bulk DNAs were extracted with a phenol-chloroform-isoamyl alcohol mixture (25:24:1, vol/vol/vol; pH 8.0) and were concentrated by ethanol precipitation.
We first tried to amplify archaeal 16S rRNA genes with a standard primer set (primers 8aF [4, 9] and 1512uR [26] or primers 8aF and 1406uR [26]), but a sufficient amount of PCR products could not be obtained. Therefore, the 16S rRNA genes were PCR amplified with another archaeon-specific primer set, primers Arch109F (15) and Arch915R (38), and real-time PCR reagents (SYBR green PCR master mixture; Applied Biosystems, Foster City, CA). To determine the Tm values of the PCR amplicons, melting curve analyses were performed immediately after the real-time PCR. The fluorescence signals during the real-time PCR and melting curve analysis were monitored by using a 7300 real-time PCR system (Applied Biosystems).
PCR, cloning, and sequencing.
The archaeal 16S rRNA genes obtained from bulk DNAs extracted from the deep-sea hydrothermal fluid and the surface seawater were PCR amplified using KOD DNA polymerase (Toyobo, Osaka, Japan) and the archaeon-specific primer set that was used in the real-time PCRs (primers Arch109F and Arch915R). The resultant PCR amplicons were cloned using a Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA). Clone libraries of archaeal 16S rRNA genes were constructed separately.
The sequences of insert PCR amplicons selected from recombinant colonies were determined with a capillary DNA sequencer (RISA-384 system; Shimadzu, Kyoto, Japan). The vector-specific primers T7 and T3 were used for the sequencing reactions. All of the sequences of archaeal 16S rRNA genes obtained were checked for chimera formation with Bellerophon (17, 18) and RDP_Chimera_Check (5). Nonchimeric sequences were aligned, and pairwise similarity values were calculated by using Genetyx-Mac (Genetyx, Tokyo, Japan). A level of similarity of 99% was used as the cutoff value for grouping the sequences into different operational taxonomic units (OTUs). A representative sequence in each OTU was homology searched using the FASTA program (28, 31) of the DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp/).
Heat treatment and digestion with Exo I.
Archaeal 16S rRNA genes were PCR amplified from bulk DNAs in the deep-sea hydrothermal fluid and the surface seawater using the archaeon-specific primers Arch109F and Arch915R. In addition, a DNA mixture was prepared by blending equal amounts of bulk DNAs extracted from the two environmental samples. The archaeal 16S rRNA genes in the DNA mixture were then amplified using the same primer set (primers Arch109F and Arch915R). The three PCR products were purified with a QIAquick PCR purification kit (QIAGEN, Valencia, CA) and were resuspended in sterilized water. Then the purified PCR products were heat denatured and digested with Exo I using the method described previously (24).
The PCR products that were amplified from the DNA mixture and survived the heat treatment and digestion with Exo I were cloned by using a Zero Blunt TOPO PCR cloning kit (Invitrogen), and they were sequenced with a capillary DNA sequencer (RISA-384 system; Shimadzu). Sequencing was performed using the vector-specific primer T7, which determined approximately 600 bp of sequence from the T7 priming site. The sequences obtained were aligned using Genetyx-Mac, and pairwise similarity values were calculated in order to group the sequences into OTUs. The level of similarity mentioned above was used as a cutoff value for grouping the sequences into different OTUs. At least one representative sequence in each OTU was homology searched using FASTA, and the sequences were compared with the sequences obtained from the original deep-sea hydrothermal fluid and surface seawater.
The bulk DNAs extracted from the deep-sea hydrothermal fluid and the surface seawater were blended at ratios of 1:1, 1:10, 1:100, and 1:1,000 separately. The archaeal 16S rRNA genes in the four DNA mixtures were amplified using KOD DNA polymerase and the universal primer set for archaea (primers Arch109F and Arch915R). The PCR products were purified, heat denatured at 84°C, and digested with Exo I. The archaeal 16S rRNA genes that survived the heat denaturation and digestion were PCR amplified again with the same primer set (Arch109F and Arch915R) to determine their Tm values by melting curve analysis. The fluorescence signals during the second PCR and the melting curve analysis were monitored by using a 7300 real-time PCR system (Applied Biosystems).
Nucleotide sequence accession numbers.
The archaeal 16S rRNA gene sequences obtained from deep-sea hydrothermal fluid and surface seawater have been deposited in the DDBJ/EMBL/GenBank database under the following accession numbers: AB257406 to AB257411 for ARCS-01 to ARCS-06 from deep-sea hydrothermal fluid and AB257412 to AB257418 for SURF-01 to SURF-07 from surface seawater.
RESULTS AND DISCUSSION
Correlation between the Topt values of species of archaea and the G+C contents of the 16S rRNA gene sequences.
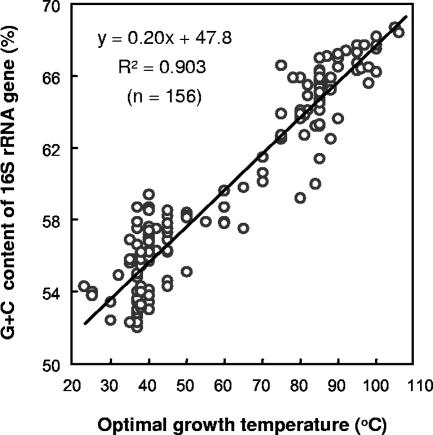
The Topt values of 156 archaeal species are plotted against the G+C contents of the 16S rRNA genes in Fig. 1. The Topt values of the archaeal strains were strongly correlated with the G+C contents of the 16S rRNA gene sequences (R2 = 0.903). Although there were a few exceptions in some groups of halophilic archaea (e.g., the genera Halococcus, Halorubrum, and Haloterrigena), the 16S rRNA genes of mesophilic archaea generally had low G+C contents, and the 16S rRNA genes of thermophilic and hyperthermophilic archaea had much higher G+C contents than the 16S rRNA genes of mesophilic archaea.
FIG. 1.
Correlations between the Topt values of archaeal strains and the G+C contents of their 16S rRNA genes. The archaeal strains were randomly chosen from various publications in which the phylogeny and taxonomy of microorganisms were reviewed (2, 32). The 16S rRNA gene sequences of the strains were obtained from the DDBJ/EMBL/GenBank database. The G+C contents of the 16S rRNA gene sequences were calculated by using Genetyx-Mac.
As we reported previously (24), there is a high correlation between the Topt values and the G+C contents of the 16S rRNA genes for all prokaryotes (R2 = 0.828; n = 406), and PCR-amplified 16S rRNA gene fragments from mesophilic prokaryotes have lower Tm values than PCR-amplified 16S rRNA gene fragments from thermophiles and hyperthermophiles. Since the correlation is obviously high (R2 = 0.903 in archaea), SePA should be more effective for eliminating 16S rRNA gene fragments originating from mesophiles in the archaeal community analysis than for eliminating 16S rRNA gene fragments originating from bacteria.
Melting curve analysis of archaeal 16S rRNA genes.
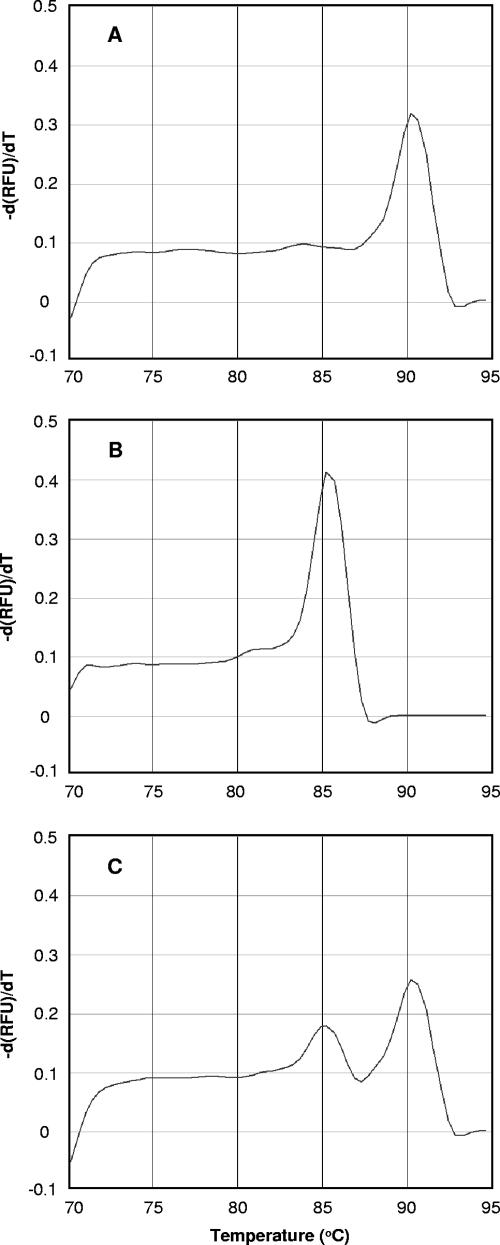
The archaeal 16S rRNA genes obtained from the bulk DNAs in deep-sea hydrothermal fluid (117°C) and surface seawater (29.9°C) were PCR amplified, and their Tm values were determined by melting curve analysis (Fig. 2A and B). The average Tm values of the 16S rRNA gene fragments from the deep-sea hydrothermal fluid were 90.2 ± 0.1°C, whereas the average Tm values of the 16S rRNA gene fragments from the surface seawater were much lower, 85.2 ± 0.2°C. The two peaks of the melting curves were completely separate, which showed that there was a great difference between the G+C contents of the archaeal 16S rRNA genes derived from the deep-sea hydrothermal fluid and the G+C contents of the archaeal 16S rRNA genes derived from the surface seawater.
FIG. 2.
Melting curve profiles for the PCR amplicons amplified from bulk DNAs that were extracted from deep-sea hydrothermal fluid (A), surface seawater (B), and a mixture of equal amounts of bulk DNAs extracted from surface seawater and deep-sea hydrothermal fluid (C). The profiles are plots of the negative first derivatives of relative fluorescence units (RFU) [−d(RFU)/dT] versus temperature.
The archaeal 16S rRNA genes obtained from a DNA mixture that contained equal amounts of the bulk DNAs extracted from the two samples were PCR amplified. The melting curve for the PCR amplicons is shown in Fig. 2C. Two separate peaks were detected in the melting curve analysis and were completely consistent with the Tm values for the PCR amplicons from the surface seawater (85.2°C) and the deep-sea hydrothermal fluid (90.2°C). These results clearly indicated that the PCR performed with the DNA mixture as the template amplified archaeal 16S rRNA genes both in surface seawater and deep-sea hydrothermal fluid.
Archaeal community in deep-sea hydrothermal fluid.
Approximately 800-bp archaeal 16S rRNA gene fragments obtained from the bulk DNAs in the deep-sea hydrothermal fluid were amplified. A clone library was constructed, and then 53 clones were randomly selected and sequenced (Table 1). The clones were divided into six OTUs based on an alignment analysis of the sequences (ARCS-01 to ARCS-06). The phylogenetic analysis revealed that these OTUs belonged to the genera Vulcanisaeta, Archaeoglobus, Methanotorris, and Thermococcus. ARCS-01 and ARCS-02 were closely related to Vulcanisaeta distributa and were members of the Crenarchaeota, which is a group of heterotrophic, anaerobic, hyperthermophilic archaea commonly isolated from terrestrial hot springs (19). The ARCS-01 and ARCS-02 clones accounted for 79.2% of all the clones. ARCS-03 and ARCS-04 exhibited the highest level of homology to Archaeoglobus profundus, a thermophilic sulfur-reducing archaeon belonging to the Euryarchaeota (20), and accounted for 15.1% of all the clones. The closest match for ARCS-05 was Methanotorris sp. strain Mc-I-70, a thermophilic methanogen isolated from a deep-sea hydrothermal vent field (40). The ARCS-05 clones accounted for 3.8% of all the clones. ARCS-06 was closely related to Thermococcus kodakaraensis, which is often found in terrestrial geothermal areas (29), and it was a minor component of the clones examined. The phylogenetic analysis indicated that all of the clones belonged to clusters composed primarily of hyperthermophilic archaeal strains or environmental clones that were collected from geothermal and hydrothermal hot environments.
TABLE 1.
16S rRNA gene sequences obtained from deep-sea hydrothermal fluid (117°C) and surface seawater (29.9oC) in the south Mariana area
| Library | OTU | No. of clones | 16S rRNA gene
|
Archaeal division | Closest database match (% homology) | |
|---|---|---|---|---|---|---|
| Length (bp) | G+C content (%) | |||||
| Hydrothermal fluid | ARCS-01 | 24 | 810 | 68.0 | Crenarchaeota | Vulcanisaeta distributa IC-065 (95.2) |
| ARCS-02 | 18 | 810 | 67.2 | Crenarchaeota | Vulcanisaeta distributa IC-065 (95.3) | |
| ARCS-03 | 6 | 814 | 66.2 | Euryarchaeota | Archaeoglobus profundus (98.5) | |
| ARCS-04 | 2 | 814 | 66.0 | Euryarchaeota | Archaeoglobus profundus (97.5) | |
| ARCS-05 | 2 | 794 | 65.5 | Euryarchaeota | Methanotorris sp. strain Mc-I-70 (97.3) | |
| ARCS-06 | 1 | 797 | 65.6 | Euryarchaeota | Thermococcus kodakaraensis (99.8) | |
| Total | 53 | |||||
| Surface seawater | SURF-01 | 18 | 789 | 54.0 | Crenarchaeota | SRI-298 (81.1) |
| SURF-02 | 11 | 791 | 54.4 | Crenarchaeota | SRI-298 (78.7) | |
| SURF-03 | 8 | 791 | 54.7 | Euryarchaeota | DJ3.25-13 (82.1) | |
| SURF-04 | 7 | 778 | 54.9 | Crenarchaeota | SRI-298 (81.4) | |
| SURF-05 | 4 | 791 | 54.2 | Euryarchaeota | DJ3.25-13 (81.4) | |
| SURF-06 | 2 | 792 | 56.4 | Crenarchaeota | SRI-298 (78.0) | |
| SURF-07 | 2 | 792 | 56.1 | Crenarchaeota | SRI-298 (78.5) | |
| Total | 52 | |||||
The G+C contents of the 16S rRNA gene sequences ranged from 65.5% (ARCS-05) to 68.0% (ARCS-01), and the overall average G+C content was 67.3% (Table 1). These extremely high G+C contents were consistent with the high Tm values determined by the melting curve analysis (Fig. 2A).
Archaeal community in surface seawater.
A 16S rRNA gene clone library for the archaeal community in surface seawater was constructed in the same manner as the 16S rRNA gene clone library for the deep-sea hydrothermal fluid sample, and 52 clones in the library were sequenced (Table 1). The clones were divided into seven OTUs based on an alignment analysis of their sequences (SURF-01 to SURF-07). The phylogenetic analysis indicated that none of these OTUs was related to any known archaeal taxonomic group. Although none of the database similarity values exceeded 82%, all of the OTUs were related to environmental clone sequences found in marine or hypersaline environments, including SRI-298 (34) and DJ3.25-13 (7). The G+C contents of the 16S rRNA gene sequences ranged from 54.0% (SURF-01) to 56.4% (SURF-06), and the overall average was 54.5% (Table 1). The overall average G+C content was clearly lower than that of archaeal 16S rRNA genes that originated from the deep-sea hydrothermal fluid. The low G+C contents resulted in the low Tm values for the 16S rRNA gene fragments in the melting curve analysis (Fig. 2B).
Heat treatment, digestion, and cloning of archaeal 16S rRNA genes.
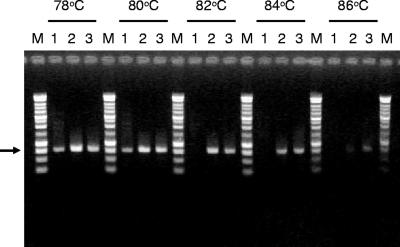
We examined the abilities to survive heat denaturation and digestion with Exo I of archaeal 16S rRNA gene fragments amplified from three samples: (i) hydrothermal fluid, (ii) surface seawater, and (iii) a mixture of equal amounts of bulk DNA extracted from the two environmental samples. The extent of survival of each 16S rRNA gene fragment was visualized by electrophoresis (Fig. 3). The PCR amplicons amplified from the surface seawater were denatured by heat treatment at 82, 84, and 86°C and were completely removed by subsequent digestion with Exo I. In contrast, the 16S rRNA gene fragments from the deep-sea hydrothermal fluid survived heat treatment even at 84°C, as well as subsequent digestion. Furthermore, some PCR amplicons from the deep-sea hydrothermal fluid survived the heat treatment at 86°C and digestion with Exo I.
FIG. 3.
Heat denaturation and digestion with Exo I of archaeal 16S rRNA genes, as visualized in a 1.0% agarose gel. Lanes 1, archaeal 16S rRNA gene fragments obtained from surface seawater; lanes 2, archaeal 16S rRNA genes obtained from a mixture of equal amounts of bulk DNAs obtained from deep-sea hydrothermal fluid and surface seawater; lanes 3, archaeal 16S rRNA gene fragments obtained from deep-sea hydrothermal fluid; lanes M, DNA marker (1-kb DNA ladder; Promega, Madison, WI). The heat treatment temperatures are indicated at the top. Each 16S rRNA gene fragment was approximately 800 bp long (arrow).
For PCR-amplified 16S rRNA genes from the DNA mixture, the survival pattern agreed almost entirely with that of PCR products derived from the deep-sea hydrothermal fluid (Fig. 3). A clone library was constructed from 16S rRNA gene fragments that originated from the DNA mixture and survived heat treatment at 84°C and digestion with Exo I. Seventy clones in the library were analyzed and divided into six OTUs (MIX-01 to MIX-06) based on an alignment analysis of the 16S rRNA gene sequences (Table 2). The phylogenetic analysis suggested that these OTUs belonged to the genera Vulcanisaeta, Archaeoglobus, Methanotorris, and Thermococcus. The compositions of these OTUs were very similar to those of the OTUs containing hyperthermophilic archaea obtained from the deep-sea hydrothermal fluid. The results led to the conclusion that the 16S rRNA genes of mesophiles in the surface seawater were completely eliminated by heat treatment at 84°C and subsequent digestion with Exo I.
TABLE 2.
Frequencies and levels of homology of 16S rRNA genes that were amplified from a DNA mixture (bulk DNAs extracted from deep-sea hydrothermal fluid and surface seawater) and that survived heat treatment at 84°C and subsequent digestion with Exo I
| OTU | No. of clones | Archaeal division | Closest database match (% homology) |
|---|---|---|---|
| MIX-01 | 23 | Crenarchaeota | ARCS-02 (100) |
| MIX-02 | 29 | Crenarchaeota | ARCS-01 (99.9) |
| MIX-03 | 8 | Euryarchaeota | ARCS-03 (99.8) |
| MIX-04 | 7 | Euryarchaeota | ARCS-04 (100) |
| MIX-05 | 2 | Euryarchaeota | ARCS-05 (99.8) |
| MIX-06 | 1 | Euryarchaeota | ARCS-06 (99.8) |
| Total | 70 |
Efficiency of elimination of 16S rRNA genes of mesophilic archaea.
In the previous study, we reported the SePA using bacterial 16S rRNA gene fragments amplified from terrestrial hot spring and river water samples (24). In the SePA using these terrestrial samples, 16S rRNA genes obtained from mesophilic bacteria in the river water were almost, but not completely, eliminated. In this study, the SePA using the archaeal 16S rRNA gene fragments completely eliminated 16S rRNA genes of mesophilic archaea derived from the surface seawater (Table 2). One of the reasons for the improved efficiency of elimination of the fragments from mesophiles in this study is that archaeal rRNA gene fragments were used for the analysis. Compared with the correlation between the Topt values and the G+C contents of the 16S rRNA genes in bacteria, the correlation in archaea is obviously high (R2 = 0.903) (Fig. 1). Therefore, SePA is more effective for elimination of 16S rRNA gene fragments derived from mesophilic archaea than for elimination of 16S rRNA gene fragments derived mesophilic bacteria.
Another reason is that the difference between the environmental temperatures affected the efficiency with which the 16S rRNA genes of the mesophiles were eliminated. The previously described imperfect elimination by SePA when the terrestrial hot spring and river water samples were used appeared to result from insufficient differences between the environmental temperatures of the samples (Table 3). On the other hand, the complete elimination by SePA when deep-sea hydrothermal fluid and surface seawater were used was due to a sufficient difference in the environmental temperatures between deep-sea hydrothermal fluid and surface seawater and to the clearly different Tm values for 16S rRNA genes of the archaeal communities in the two samples (Table 3).
TABLE 3.
Comparison of environmental temperatures and prokaryotic 16S rRNA genes of microbial communities in terrestrial hot spring, river water, deep-sea hydrothermal fluid, and surface seawater samples
| Samplesa | Environmental temp (°C) | Tm (°C) | G+C content (%) | Reference |
|---|---|---|---|---|
| THS | 76.0 | 86.8b | 58.6c | 24 |
| RIV | 14.0 | 84.5b | 52.9c | 24 |
| Difference | 62.0 | 2.3 | 5.7 | |
| DHF | 117 | 90.2d | 67.3e | This study |
| SSW | 29.9 | 85.2d | 54.5e | This study |
| Difference | 87.1 | 5.0 | 12.8 |
THS, terrestrial hot spring; RIV, river water; DHF, deep-sea hydrothermal fluid; SSW, surface seawater.
Determined by a melting curve analysis of bacterial 16S rRNA genes (ca. 1,500 bp).
Calculated from bacterial 16S rRNA gene sequences (ca. 1,500 bp).
Determined by a melting curve analysis of archaeal 16S rRNA genes (ca. 800 bp).
Calculated from archaeal 16S rRNA gene sequences (ca. 800 bp).
In our studies, we have shown that the efficiency of elimination of 16S rRNA genes of mesophilic prokaryotes increases as the difference in the environmental temperatures between the samples increases. Since the subsurface environmental temperature increases with depth, the difference between the temperature of the core sample and the temperature of the drilling fluid is likely to increase with drilling depth. Therefore, SePA has a great advantage in phylogenetic analysis targeting hyperthermophilic prokaryotes in core samples obtained by deep drilling into deeper and hotter crusts.
Utility of SePA in deep drilling.
In seafloor drilling, surface seawater is generally used as the drilling fluid. Surface seawater commonly contains mesophilic prokaryotes at concentrations of about 105 to 106 cells ml−1 (42). On the other hand, the cell density of hyperthermophilic prokaryotes in the deep-subsurface hot biosphere is lower than that of mesophiles in the drilling fluid. Cragg et al. (6) actually counted microbial cells in subsurface sediments (at 90 to 110°C) in a deep-sea hydrothermal vent field in the Juan de Fuca Ridge by using the acridine orange staining method. The cell densities obtained were 104 to 105 cells cm−3 on average. In the subsurface hot rocks of a deep-sea hydrothermal area, the Manus Basin, the microbial cell densities were clearly lower and were estimated to be less 104 cells cm−3 (23). Given that these cell densities are adequate, core samples recovered from deep-subsurface hot environments are likely to be exposed to drilling fluid that contains mesophilic organisms at concentrations that are 10 to 1,000 times higher than the concentrations of the hyperthermophiles in core samples. This would be a serious problem for culture-independent community analysis of hyperthermophiles in deep crust.
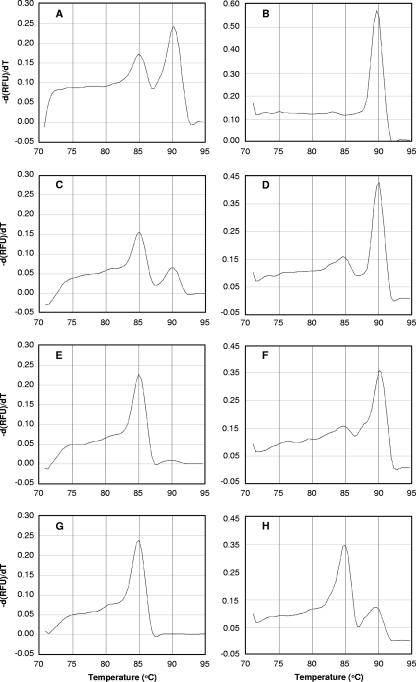
To determine whether the SePA method is effective even with a rock sample excessively contaminated with mesophile-containing drilling fluid, an additional investigation was performed using four mixtures of bulk DNAs extracted from two environmental samples, deep-sea hydrothermal fluid and surface seawater, at ratios of 1:1, 1:10, 1:100, and 1:1,000. The archaeal 16S rRNA genes in these four DNA mixtures were PCR amplified, and then the Tm of each PCR amplicon of an archaeal 16S rRNA gene was determined by real-time PCR (Fig. 4). While PCR amplicons from the 1:1 and 1:10 DNA mixtures clearly produced two peaks of Tm values at around 85 and 90°C, which originated from mesophiles and hyperthermophiles, respectively (Fig. 4A and C), the number of amplicons from hyperthermophiles in the latter sample (1:10) was obviously lower. The difference was also conspicuous in more highly diluted samples (1:100 and 1:1,000). There was a slight peak of PCR amplicons from hyperthermophiles in the 1:100 DNA mixture, and there was only a trace in the 1:1,000 mixture (Fig. 4E and G). The tests using DNA samples excessively contaminated with mesophilic archaea indicated that 16S rRNA genes of hyperthermophilic archaea in the deep-sea hydrothermal fluid were almost entirely overwhelmed by amplicons from mesophiles and could not be detected by the conventional culture-independent community analysis.
FIG. 4.
Melting curve profiles for the archaeal 16S rRNA genes amplified from bulk DNAs extracted from deep-sea hydrothermal fluid and surface seawater mixed at ratios of 1:1 (A), 1:10 (C), 1:100 (E), and 1:1,000 (G) and of second-PCR amplicons from archaeal 16S rRNA genes amplified from the 1:1 (B), 1:10 (D), 1:100 (F), and 1:1,000 (H) DNA mixtures after heat denaturation at 84°C and digestion with Exo I. The profiles are plots of the negative first derivatives of relative fluorescence units (RFU) [−d(RFU)/dT] versus temperature.
Even in such an undesirable case, the SePA method effectively eliminated 16S rRNA genes of contaminating mesophilic archaea. Each of the four PCR amplicons was heat denatured at 84°C and digested with Exo I, and this was followed by further PCR amplification and a melting curve analysis. Figures 4B, D, F, and H show the melting curves for the second-PCR amplicons. For the second-PCR amplicons derived from the 1:1 DNA mixture, the peak at 85°C completely disappeared, indicating that all of the PCR amplicons that originated from mesophilic archaea were eliminated (Fig. 4B). The second-PCR amplicons derived from the 1:10 DNA mixture produced a predominant peak at approximately 90°C, and most of the amplicons derived from mesophiles were eliminated by denaturation and digestion (Fig. 4D). This remarkable effect was also observed for the second-PCR amplicons derived from the 1:100 DNA mixture. Whereas in the melting curve for the first-PCR amplicons there was a large peak at 85°C and just a faint peak at around 90°C, in the melting curve for the second-PCR amplicons the peak that originated from mesophiles (at 85°C) was considerably lower and the peak that originated from hyperthermophiles was prominently higher (Fig. 4F). A similar increase for the peak that originated from hyperthermophiles was observed even when the second-PCR amplicon from the 1:1,000 DNA mixture was used. The trace peak at 90°C for the first-PCR amplicons (Fig. 4G) was conspicuously higher in the second-PCR curve after denaturation and digestion (Fig. 4H). These results show that SePA can be used to detect 16S rRNA genes of hyperthermophilic archaea in deep-subsurface hot environments, even if the core samples are excessively contaminated with mesophiles. SePA is a simple but exceedingly useful method for culture-independent community analysis of hyperthermophilic archaea in the deep-subsurface biosphere.
Acknowledgments
We are grateful to the captains and crews of the R/V Yokosuka and Shinkai 6500 and to the operation teams of Shinkai 6500 for helping us collect the deep-sea hydrothermal fluid samples.
This work was supported in part by a Grant-in-Aid for Scientific Research (grant 16310024) and by a Grant-in-Aid for Young Scientists (grant 18710007) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Blöchl, E., R. Rachel, S. Burggraf, D. Hafenbradl, H. W. Jannasch, and K. O. Stetter. 1997. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113°C. Extremophiles 1:14-21. [DOI] [PubMed] [Google Scholar]
- 2.Boone. D. R., and R. W. Castenholz. 2001. Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, NY.
- 3.Brody, R., K. Doherty, and P. Zimmerman. 1986. Processivity and kinetics of the reaction of exonuclease-I from Escherichia coli with polydeoxyribonucleotides. J. Biol. Chem. 261:7136-7143. [PubMed] [Google Scholar]
- 4.Burggraf, S., K. O. Stetter, P. Rouviere, and C. R. Woese. 1991. Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst. Appl. Microbiol. 14:346-351. [DOI] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cragg, B. A., M. Summit, and R. J. Parkes. 2000. Bacterial profiles in a sulfide mound (Site 1035) and an area of active fluid venting (Site 1036) in hot hydrothermal sediments from Middle Valley (northwest Pacific), p. 1-18. In R. A. Zierenberg, Y. Fouquet, D. J. Miller, and W. R. Normark (ed.), Proceedings of the ODP, scientific results, vol. 169. Ocean Drilling Program, College Station, TX. [Google Scholar]
- 7.Cytryn, E., D. Minz, R. S. Oremland, and Y. Cohen. 2000. Distribution and diversity of archaea corresponding to the limnological cycle of a hypersaline stratified lake (Solar Lake, Sinai, Egypt). Appl. Environ. Microbiol. 66:3269-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drescher, J., T. Kirsten, and K. Schafer. 1998. The rate gas inventory of the continental crust, recovered by the KTB continental deep drilling project. Earth Planet. Sci. Lett. 154:247-263. [Google Scholar]
- 9.Eder, W., W. Ludwig, and R. Huber. 1999. Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch. Microbiol. 172:213-218. [DOI] [PubMed] [Google Scholar]
- 10.Emmermann, R. 1995. Abenteuer Tiefbohrung. Geowissenschaften 13:114-128. [Google Scholar]
- 11.Galtier, N., and J. R. Lobry. 1997. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 44:632-636. [DOI] [PubMed] [Google Scholar]
- 12.Galtier, N., N. Tourasse, and M. Gouy. 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283:220-221. [DOI] [PubMed] [Google Scholar]
- 13.Gold, T. 1992. The deep, hot biosphere. Proc. Natl. Acad. Sci. USA 89:6045-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold, T. 1999. Life at borders, p. 11-36. In T. Gold (ed.), The deep hot biosphere: the myth of fossil fuels. Springer-Verlag, New York, NY.
- 15.Großkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanada, S. 2003. Filamentous anoxygenic phototrophs in hot springs. Microbes Environ. 18:51-61. [Google Scholar]
- 17.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 18.Hugenholtzt, P., and T. Huber. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 53:289-293. [DOI] [PubMed] [Google Scholar]
- 19.Itoh, T., K. Suzuki, and T. Nakase. 2002. Vulcanisaeta distributa gen. nov., sp. nov., and Vulcanisaeta souniana sp. nov., novel hyperthermophilic, rod-shaped crenarchaeotes isolated from hot springs in Japan. Int. J. Syst. Evol. Microbiol. 52:1097-1104. [DOI] [PubMed] [Google Scholar]
- 20.Kashefi, K., J. M. Tor, D. E. Holmes, C. V. Gaw Van Praagh, A. L. Reysenbach, and D. R. Lovley. 2002. Geoglobus ahangari gen. nov., sp. nov., a novel hyperthermophilic archaeon capable of oxidizing organic acids and growing autotrophically on hydrogen with Fe(III) serving as the sole electron acceptor. Int. J. Syst. Evol. Microbiol. 52:719-728. [DOI] [PubMed] [Google Scholar]
- 21.Kashefi, K., and D. R. Lovley. 2003. Extending the upper temperature limit for life. Science 301:934. [DOI] [PubMed] [Google Scholar]
- 22.Khachane, A. N., K. N. Timmis, and V. A. dos Santos. 2005. Uracil content of 16S rRNA of thermophilic and psychrophilic prokaryotes correlates inversely with their optimal growth temperatures. Nucleic Acids Res. 33:4016-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura, H., R. Asada, A. Masta, and T. Naganuma. 2003. Distribution of microorganisms in the subsurface of the Manus Basin hydrothermal vent field in Papua New Guinea. Appl. Environ. Microbial. 69:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura, H., M. Sugihara, K. Kato, and S. Hanada. 2006. Selective phylogenetic analysis targeted at 16S rRNA genes of thermophiles and hyperthermophiles in deep-subsurface geothermal environments. Appl. Environ. Microbial. 72:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lackschewitz, K. S., C. W. Devey, P. Stoffers, R. Botz, A. Eisenhauer, M. Kummetz, M. Schmidt, and A. Singer. 2004. Mineralogical, geochemical and isotopic characteristics of hydrothermal alteration processes in the active, submarine, felsic-hosted PACMANUS field, Manus Basin, Papua New Guinea. Geochim. Cosmochim. Acta 68:4405-4427. [Google Scholar]
- 26.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 27.Lin, L.-H., P.-L. Wang, D. Rumble, J. Lippmann-Pipke, E. Boice, L. M. Pratt, B. S. Lollar, E. L. Brodie, T. C. Hazen, G. L. Andersen, T. Z. DeSantis, D. P. Moser, D. Kershaw, and T. C. Onstott. 2006. Long-term sustainability of a high-energy, low diversity crustal biome. Science 314:479-482. [DOI] [PubMed] [Google Scholar]
- 28.Lipman, D. J., and W. R. Person. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 29.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkes, R. J., B. A. Cragg, S. J. Bale, J. M. Getliff, K. Goodman, P. A. Rochelle, J. C. Fry, A. J. Weightman, and S. M. Harvey. 1994. Deep bacterial biosphere in Pacific Ocean sediments. Nature 371:410-413. [Google Scholar]
- 31.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reysenbach, A.-L., D. Götz, and D. Yernool. 2002. Microbial diversity of marine and terrestrial thermal springs, p. 345-421. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life. Foundation of Earth's biosphere. Wiley-Liss, Inc., New York, NY.
- 33.Shipboard Scientific Party. 2002. Leg 193 summary, p. 1-84. In R. A. Binns, F. J. A. S. Barriga, D. J. Miller, et al (ed.), Proceedings of the ODP, initial reports, vol. 193. Ocean Drilling Program, College Station, TX. [Google Scholar]
- 34.Skirnisdottir, S., G. O. Hreggvidsson, S. Hjörleifsdottir, V. T. Marteinsson, S. K. Petursdottir, O. Holst, and J. K. Kristjansson. 2000. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, D. C., A. J. Spivack, M. R. Fisk, S. A. Haveman, H. Staudigel, and the Leg 185 Shipboard Scientific Party. 2000. Methods for quantifying potential microbial contamination during deep ocean coring. ODP technical note 28. http://www-odp.tamu.edu/publications/tnotes/tn28/INDEX.HTM.
- 36.Smith, D. C., A. J. Spivack, M. R. Fisk, S. A. Haveman, H. Staudigel, and Ocean Drilling Program Leg 185 Shipboard Scientific Party. 2000. Tracer based estimates of drilling-induced microbial contamination of deep-sea crust. Geomicrobiol. J. 17:207-219. [Google Scholar]
- 37.Somerville, C. C., I. T. Knight, W. L. Straube, and R. R. Colwell. 1989. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl. Environ. Microbiol. 55:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 39.Stetter, K. O. 1996. Hyperthermophilic prokaryotes. FEMS Microbiol. Rev. 18:149-158. [Google Scholar]
- 40.Takai, K., T. Gamo, U. Tsunogai, N. Nakayama, H. Hirayama, K. N. Nealson, and K. Horikoshi. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269-282. [DOI] [PubMed] [Google Scholar]
- 41.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]