Abstract
Corynebacterium glutamicum was engineered for the production of l-valine from glucose by deletion of the aceE gene encoding the E1p enzyme of the pyruvate dehydrogenase complex and additional overexpression of the ilvBNCE genes encoding the l-valine biosynthetic enzymes acetohydroxyacid synthase, isomeroreductase, and transaminase B. In the absence of cellular growth, C. glutamicum ΔaceE showed a relatively high intracellular concentration of pyruvate (25.9 mM) and produced significant amounts of pyruvate, l-alanine, and l-valine from glucose as the sole carbon source. Lactate or acetate was not formed. Plasmid-bound overexpression of ilvBNCE in C. glutamicum ΔaceE resulted in an approximately 10-fold-lower intracellular pyruvate concentration (2.3 mM) and a shift of the extracellular product pattern from pyruvate and l-alanine towards l-valine. In fed-batch fermentations at high cell densities and an excess of glucose, C. glutamicum ΔaceE(pJC4ilvBNCE) produced up to 210 mM l-valine with a volumetric productivity of 10.0 mM h−1 (1.17 g l−1 h−1) and a maximum yield of about 0.6 mol per mol (0.4 g per g) of glucose.
Corynebacterium glutamicum is an aerobic, gram-positive organism that grows on a variety of sugars and organic acids and is used for the production of amino acids, e.g., l-glutamate, l-lysine, and l-valine (15, 17). The latter amino acid is essential for vertebrates and is used for infusion solutions and as a precursor for the chemical synthesis of herbicides (6, 14). Approximately 500 tons of l-valine is produced annually by fermentation or by extraction from acidic hydrolysates of proteins (6). Due to the growing world market for amino acids, there is an increasing interest in the further development and optimization of efficient fermentative l-valine production processes.
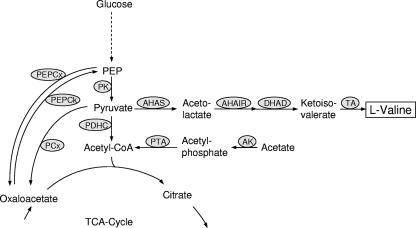
As shown in Fig. 1, l-valine is synthesized from pyruvate in a pathway comprising four reactions, catalyzed by acetohydroxyacid synthase (AHAS; ilvBN gene product), acetohydroxyacid isomeroreductase (AHAIR; ilvC gene product), dihydroxyacid dehydratase (ilvD gene product), and transaminase B (TA; ilvE gene product). In most organisms, these enzymes also catalyze the biosynthesis of l-isoleucine from pyruvate and 2-ketobutyrate. The latter is formed from l-threonine by threonine dehydratase (TD; ilvA gene product). Ketoisovalerate, the last intermediate of l-valine synthesis, is also the precursor for l-leucine and d-pantothenate biosynthesis.
FIG. 1.
The enzymes at the phosphoenolpyruvate-pyruvate-oxaloacetate node with the biosynthetic pathway of l-valine in C. glutamicum. Abbreviations: PK, pyruvate kinase; PCx, pyruvate carboxylase; PEP, phosphoenolpyruvate; PEPCx, phosphoenolpyruvate carboxylase; PEPCk, phosphoenolpyruvate carboxykinase; AK, acetate kinase; PTA phosphotransacetylase; DHAD, dihydroxyacid dehydratase; CoA, coenzyme A; TCA, tricarboxylic acid.
AHAS is the key enzyme of branched-chain amino acid biosynthesis, and the regulation of the C. glutamicum enzyme as well as the regulation of the AHAS genes has been studied in detail (5, 9, 13, 16, 19). Only one AHAS was found in C. glutamicum (13), and this enzyme was shown to be subject to feedback inhibition by l-valine, l-isoleucine, and l-leucine (5, 9, 19). Expression of the ilvBN genes, which form an operon with ilvC, is controlled by an attenuation mechanism, and it increases about twofold in response to shortage of branched-chain amino acids (19). On the other hand, expression of the operon is induced by 2-ketobutyrate (5, 19).
l-Valine-excreting strains of C. glutamicum have been generated by plasmid-bound overexpression of the l-valine biosynthesis genes ilvBNCD or ilvBNCE in combination with a deletion of the ilvA gene encoding TD (23, 24). Improved production of l-valine was achieved by the introduction of the genes encoding a feedback-resistant AHAS enzyme into l-valine-producing C. glutamicum strains (9). Moreover, Radmacher et al. (23) showed that inactivation of the d-pantothenate biosynthesis by deleting the panBC genes led to increased l-valine production of C. glutamicum cultivated under d-pantothenate-limiting conditions. The authors speculated that d-pantothenate limitation leads to an increased drain-off of pyruvate towards the l-valine biosynthetic pathway due to reduced coenzyme A availability for the reaction of the pyruvate dehydrogenase complex (PDHC) (23). In fact, Escherichia coli pyruvate-overproducing strains have been obtained by inactivation of the PDHC (27, 31). Increasing the intracellular availability of the precursor pyruvate by decreasing the PDHC activity thus represents an attractive approach for the optimization of l-valine production by C. glutamicum.
Recently, we identified and functionally characterized the E1p subunit of the PDHC in C. glutamicum and showed that this complex is essential for the growth of this organism on glucose, pyruvate, or lactate (26). In contrast, the PDHC activity was found to be dispensable for the growth of C. glutamicum in medium containing acetate. This growth phenotype opened the possibility to study the effect of PDHC deficiency on metabolite formation from glucose (and acetate) by growing and by nongrowing cells of C. glutamicum. In the present study, we investigate the effect of PDHC inactivation in C. glutamicum on the formation of organic acids and amino acids. We found that resting cells of the PDHC-deficient mutant of C. glutamicum wild type (WT) produce significant amounts of pyruvate, l-valine, and l-alanine. This prompted us to direct the carbon flux more efficiently from glucose to l-valine by additional overexpression of l-valine biosynthetic genes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains used in this study were E. coli DH5α (10), C. glutamicum WT (i.e., ATCC 13032 from the American Type Culture Collection), and its derivative, C. glutamicum ΔaceE (26). Plasmid pJC4ilvBNCE (23) was obtained from L. Eggeling (FZ Jülich).
DNA preparation and transformation.
The isolation of plasmid pJC4ilvBNCE from E. coli and C. glutamicum was performed as described before (8). Plasmid DNA transfer into C. glutamicum was carried out by electroporation, and the recombinant strains were selected on LB-brain heart infusion (BHI) agar plates containing 0.5 M sorbitol and kanamycin (50 μg ml−1) (29).
Culture conditions.
E. coli was grown aerobically in 2× TY complex medium (25) at 37°C as 50-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm. Precultures of C. glutamicum WT, C. glutamicum ΔaceE, and C. glutamicum ΔaceE(pJC4ilvBNCE) were grown in 2× TY medium containing 0.5% (wt/vol) potassium acetate. For amino acid fermentations in shake flasks, the cells of an overnight preculture were washed with 0.9% (wt/vol) NaCl and inoculated into CGXII minimal medium (7) to give an initial optical density at 600 nm (OD600) of about 1. The medium contained 4% (wt/vol) glucose and 1% or 1.5% (wt/vol) potassium acetate. Where indicated in Results, 0.5% (wt/vol) BHI powder (Merck) was added to the medium. The plasmid-carrying strains were grown in the presence of kanamycin (50 μg ml−1). C. glutamicum was grown aerobically at 30°C as 50-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm or as 300-ml cultures in a glass bioreactor (see below).
The batch and fed-batch fermentations were performed at 30°C in a 1-liter glass bioreactor (Biostat B; Braun, Melsungen, Germany). The pH was maintained at 7.0 by online measurement using a standard pH electrode (Mettler Toledo, Giessen, Germany) and the addition of 4 M KOH and 4 M HCl. Foam development was prohibited by manual injection of small amounts (about 20 μl) of silicon antifoam (Roth, Karlsruhe, Germany). The dissolved oxygen was measured online by use of a polarimetric oxygen electrode (Mettler Toledo, Giessen, Germany), and it was adjusted to 30% of saturation in a cascade by stirring at 100 to 1,200 rpm and aerating with 1 to 3 liters of air per min. During the fed-batch processes, adequate amounts of 50% (wt/vol) glucose and 50% (wt/vol) potassium acetate were injected (at the time points indicated below [see Fig. 4]).
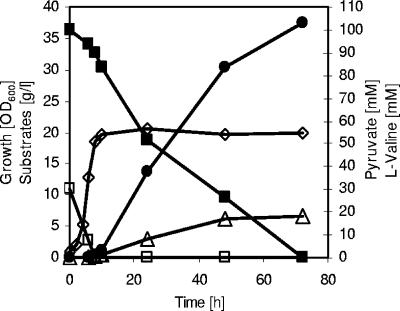
FIG. 4.
l-Valine accumulation during a representative fed-batch fermentation of C. glutamicum ΔaceE(pJC4ilvBNCE) on CGXII medium initially containing glucose (4%), acetate (1%), and BHI (0.5%). ⋄, growth; ▪, glucose; □, acetate; ▵, pyruvate; •, l-valine. Three independent fed-batch fermentations were performed, with all three showing comparable results.
Analytics.
One milliliter of the culture was harvested by centrifugation (13,000 rpm, 10 min, room temperature), and the supernatant was used for determination of amino acid, glucose, and/or organic acid concentrations in the culture fluid. The amino acid concentration was determined by reversed-phase high-pressure liquid chromatography (on an HP 1100 instrument; Hewlett-Packard, Waldbronn, Germany) with fluorimetric detection (excitation at 230 nm and emission at 450 nm) after automatic precolumn derivatization with ortho-phthaldialdehyde (18). Separation was carried out at 40°C on a Multohyp octyldecyl silane column (particle size, 5 μm; 125 by 4 mm; CS-Chromatographie, Langerwehe, Germany). The elution buffer consisted of a polar phase (0.1 M sodium acetate, pH 7.2) and a nonpolar phase (methanol). Quantification was done by calculation of the concentration by use of an internal standard (l-ornithine at 100 μM) and by a 10-point calibration curve for each amino acid. Glucose, acetate, and lactate concentrations were determined by enzymatic tests (Roche Diagnostics, Penzberg, Germany). The pyruvate concentrations were determined enzymatically according to the method of Bergmeyer (1).
For the measurement of intracellular pyruvate concentrations, the cells were cultivated in 50 ml CGXII medium. Samples of 5 ml were taken for measurements during growth and production phases, at about 9 h and 31 h after inoculation, respectively. All following steps were performed below −10°C to prevent changes in the metabolome (4, 12). After centrifugation of the samples at 10,400 × g for 5 min at −20°C, the cell pellets were resuspended in 5 ml ice-cold NaCl (0.9% [wt/vol]) and centrifuged again. The supernatants were discarded and the cells resuspended in 1 ml methanol (−80°C) and 1 ml Tris-EDTA buffer (pH 7.0). For cell extraction, 2 ml chloroform was added, and the suspension was shaken for 2 h at −20°C. After extraction, the pyruvate in the methanol phase was determined by liquid chromatography-tandem mass spectrometry (Finnigan TSQ Quantum Ultra; Thermo) according to the method of Luo et al. (18a). Quantification was done by external calibration due to the absence of matrix effects in the appropriate dilutions. For the calculation of the intracellular pyruvate concentration, a specific cell volume of 1.95 μl/mg was assumed (21). All intracellular concentrations are based on data from three independent experiments with two replicates of each analytical measurement.
RESULTS
C. glutamicum ΔaceE forms pyruvate, l-alanine, and l-valine.
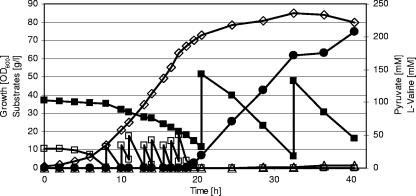
Previously, we analyzed the C. glutamicum aceE gene, encoding the E1p subunit of the PDHC, and constructed and characterized an E1p-deficient mutant, C. glutamicum ΔaceE (26). This mutant carries a deletion in the chromosomal aceE locus, shows no PDHC activity, and is unable to grow on minimal medium containing glucose as the carbon and energy source unless supplemented with acetate. To test C. glutamicum ΔaceE for its ability to form organic acids and/or amino acids from glucose and acetate and to compare its metabolite formation with that of the parental C. glutamicum WT, both strains were grown in a 1-liter bioreactor in CGXII medium containing 4% (wt/vol) glucose and 1.5% (wt/vol) potassium acetate, and growth, glucose, and acetate utilization and pyruvate, lactate, and amino acid accumulation were monitored over the course of the experiment. C. glutamicum WT grew with maximal growth rates of 0.36 h−1 to a final OD600 of about 65, metabolized glucose and acetate simultaneously, and did not form any of the tested metabolites to concentrations higher than 1 mM (not shown). For the first 6 to 8 hours, C. glutamicum ΔaceE also grew exponentially, with a maximal growth rate of 0.36 h−1, and simultaneously used acetate and glucose. During this growth phase, the strain also did not excrete any of the metabolites tested into the culture broth (Fig. 2). As expected, the PDHC-deficient mutant stopped growth as soon as the acetate concentration dropped to <1 mM and the culture reached a maximal OD600 of about 19. However, the mutant continued to metabolize glucose and, in contrast to C. glutamicum WT, started to produce significant amounts of pyruvate (30 to 35 mM), l-alanine (25 to 30 mM), l-threonine (2 to 5 mM), and l-valine (30 to 35 mM) (Fig. 2). Lactate, acetate, and amino acids other than l-alanine, l-threonine, and l-valine were not formed in the course of the cultivation of C. glutamicum ΔaceE. These results show that PDHC deficiency is an ideal condition to breed C. glutamicum strains producing either pyruvate, l-alanine, or l-valine. Furthermore, the results indicate that inactivation of the competing pathways of l-isoleucine biosynthesis and of d-pantothenate synthesis, e.g., by deleting the ilvA and/or panB gene(s), is not necessary for efficient l-valine formation.
FIG. 2.
Representative batch fermentation of C. glutamicum ΔaceE on CGXII medium initially containing glucose (4%), acetate (1.5%). (A) ⋄, growth; ▪, glucose; □, acetate. (B) ▵, pyruvate; ○, l-alanine; •, l-valine; ×, l-threonine. Three independent fermentations were performed, with all three showing comparable results.
l-Valine production by C. glutamicum ΔaceE strains overexpressing ilv genes.
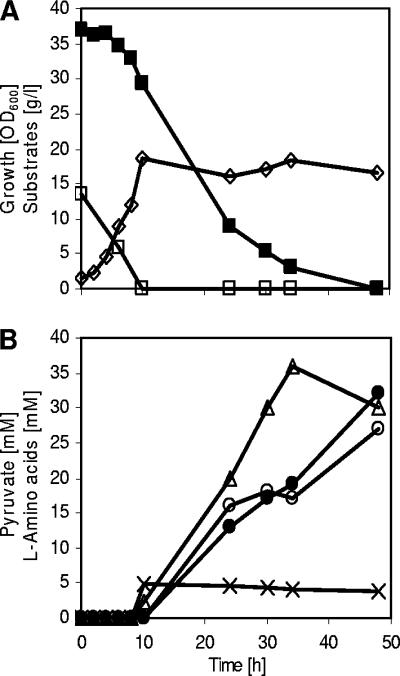
In order to force the carbon flux in C. glutamicum ΔaceE from pyruvate towards l-valine production, we transformed this strain with plasmid pJC4ilvBNCE. This plasmid carries the genes encoding three of the four enzymes involved in l-valine (and l-isoleucine) biosynthesis (see the introduction) and has previously been used to construct l-valine-producing derivatives of C. glutamicum WT, C. glutamicum ΔilvA, and C. glutamicum ΔilvA ΔpanBC (reference 23; also see the introduction). l-Valine fermentations with C. glutamicum ΔaceE(pJC4ilvBNCE) were carried out in CGXII medium with 0.5% (wt/vol) BHI, 4% (wt/vol) glucose, and 1% (wt/vol) potassium acetate. Figure 3 shows the course of a typical fermentation. The culture grew within 8 h to an OD600 of about 20, consuming acetate completely (1%) and glucose to a minor part (0.5%). The cells then stopped growth, and they further consumed glucose and accumulated up to about 105 mM l-valine within 64 h. Although C. glutamicum ΔaceE(pJC4ilvBNCE) produced relatively high amounts of l-valine, it nevertheless excreted nearly 20 mM pyruvate into the medium, indicating that there is still precursor available for l-valine production. No l-alanine was formed in C. glutamicum carrying ΔaceE in combination with plasmid pJC4ilvBNCE. These results show that pyruvate supply on one hand and the enzymes for the l-valine biosynthetic pathway on the other are major bottlenecks for l-valine overproduction by C. glutamicum.
FIG. 3.
l-Valine accumulation during a representative batch cultivation of C. glutamicum ΔaceE(pJC4ilvBNCE) on CGXII medium initially containing glucose (4%), acetate (1%), and BHI (0.5%). ⋄, growth; ▪, glucose; □, acetate; ▵, pyruvate; •, l-valine. Three independent fermentations were performed, with all three showing comparable results.
Intracellular pyruvate concentrations in C. glutamicum and its derivatives.
The production pattern of C. glutamicum ΔaceE and C. glutamicum ΔaceE(pJC4ilvBNCE) suggested that the PDHC deficiency brought about by deletion of the aceE gene might have led to a high intracellular accumulation of pyruvate. To test for this hypothesis, we determined the intracellular pyruvate concentrations in C. glutamicum WT and its derivatives, the ΔaceE and ΔaceE(pJC4ilvBNCE) strains, during exponential growth and at the stationary phase, the latter being the production phase for C. glutamicum ΔaceE and C. glutamicum ΔaceE(pJC4ilvBNCE). All three strains showed very low intracellular pyruvate concentrations (<0.05 mM) in the exponential growth phase. In the stationary/production phase, however, C. glutamicum WT, ΔaceE, and ΔaceE(pJC4ilvBNCE) showed intracellular pyruvate concentrations of about 1.9 mM, 25.9 mM, and 2.3 mM, respectively. These results indicate that in the absence of acetate as the carbon and energy source, the aceE deletion in C. glutamicum leads to an intracellular accumulation of pyruvate. Furthermore, these results indicate that overexpression of the genes encoding l-valine biosynthetic enzymes leads to a drain-off of the pyruvate towards l-valine.
Fed-batch fermentations with C. glutamicum ΔaceE(pJC4ilvBNCE).
In order to test the suitability of C. glutamicum ΔaceE(pJC4ilvBNCE) for an improved l-valine production process, we established a fed-batch fermentation based on mixed substrate (Fig. 4). These fermentations were carried out in CGXII medium containing 0.5% (wt/vol) BHI, 4% glucose, and an initial potassium acetate concentration of 1% (wt/vol). To allow growth to a high cell density, adequate amounts of a 50% (wt/vol) acetate stock solution were repeatedly added (eight times) to the growing cells (Fig. 4). Using this technique, an OD600 of about 70 was obtained after 19 h (Fig. 4). During this growth period, about 20 g glucose/liter was consumed in addition to acetate; however, neither l-valine nor pyruvate was excreted into the medium. After having consumed the last pulse of acetate, the cells started to excrete l-valine, and at time points 20 h and 33 h, we added pulses of 4% (wt/vol) glucose. As shown in Fig. 4, the cells accumulated about 210 mM l-valine within 21 h with a volumetric productivity of 10.0 mM h−1 (1.17 g liter−1 h−1) and a maximal yield of 0.6 mol l-valine per mol of glucose (between 19 h and 33 h in Fig. 4). The overall yield in the production phase (between 19 h and 40 h) was about 0.5 mol per mol of glucose. In addition to l-valine, the cells also still excreted small amounts of pyruvate (5 mM) into the medium, indicating that l-valine production by C. glutamicum ΔaceE(pJC4ilvBNCE) can be further increased. Taken together, all these results demonstrate that C. glutamicum ΔaceE with plasmid-bound overexpression of l-valine biosynthetic genes is a very useful platform for optimizing l-valine production with C. glutamicum.
DISCUSSION
l-Valine-producing strains of C. glutamicum have previously been obtained by undirected mutagenesis (20, 28) and by overexpression of genes encoding (deregulated) l-valine biosynthetic enzymes, in combination with the deletion of ilvA and/or of the d-pantothenate biosynthetic genes panBC (9, 23, 24). The knockout of the ilvA gene eliminates TD activity and thus the formation of 2-ketobutyrate, which together with pyruvate is a substrate for l-isoleucine biosynthesis via AHAS, AHAIR, dihydroxyacid dehydratase, and TA. This leads to an isoleucine auxotrophy on the one hand and to an enhanced pyruvate availability on the other. Both these effects have been proposed to be responsible for enhanced l-valine production, since the former should lead to a suspension of the allosteric AHAS inhibition and of the attenuation control of the ilvBNC genes by l-isoleucine (5, 19) and the latter to an enhanced pyruvate flux into the l-valine biosynthetic pathway. As shown here, ilvA deletion (or TD inactivation) is not a prerequisite for effective l-valine production with C. glutamicum, indicating that the allosteric regulation of AHAS and the ilvBNC expression control by intracellular l-isoleucine are not very critical for the l-valine production process. Deletion of the panBC genes in C. glutamicum has been shown to result in pantothenate auxotrophy (23). Due to the finding that a panC mutant excreted pyruvate and alanine into the growth medium and due to the hypothesis that reduced coenzyme A availability in pantothenate-auxotrophic C. glutamicum strains should lead to reduced PDHC activity, Radmacher et al. (23) speculated that in this mutant increased quantities of precursor are directly available for l-valine synthesis. In accordance with this hypothesis, we here show that a PDHC-deficient mutant of C. glutamicum WT without any further genetic engineering is able to produce significant amounts of l-valine from glucose. In addition, C. glutamicum ΔaceE excreted significant amounts of pyruvate and l-alanine, indicating that l-valine production in this strain is limited by the reactions from pyruvate to l-valine and not by pyruvate availability. This hypothesis is corroborated by our results obtained with C. glutamicum ΔaceE carrying the ilvBNCE genes on plasmid pJC4. In batch fermentations, this strain produced much more l-valine, less pyruvate, and no l-alanine, and in fed-batch fermentations, the product spectrum was almost completely shifted towards l-valine. Since C. glutamicum ΔaceE(pJC4ilvBNCE) in shake flask batch fermentation produced as much l-valine as or even more l-valine than did previously constructed l-valine-producing strains (references 3, 9, and 23; also see below), our results indicate that effective l-valine production with C. glutamicum can easily be attained by optimization of the pyruvate availability and increasing the intracellular content of l-valine biosynthetic enzymes AHAS, AHAIR, and TA. Furthermore, the results show that effective l-valine production neither relies on isoleucine and/or pantothenate auxotrophy nor requires the presence of deregulated l-valine biosynthetic enzymes.
Independently of the presence or absence of plasmid pJC4ilvBNCE and independently of the fermentation process, l-valine production in C. glutamicum ΔaceE was decoupled from growth, i.e., took place only after cessation of the growth, or in other words, after depletion of the acetate required for growth of the mutant. This phenotype is surprising in view of the fact that l-valine (and also l-lysine, l-leucine, l-alanine, and l-threonine) production with C. glutamicum generally occurs in the course of the growth (2, 9, 11). One explanation for the production phenotype of C. glutamicum ΔaceE might be that the presence of acetate may have a negative effect on glucose uptake and/or on one or several enzymes involved in glycolysis; thus, the pyruvate availability in the growth phase might be restricted to the requirements for cell mass biosynthesis. In favor of this argument, we previously showed that glucose consumption and the carbon flux from external glucose to pyruvate was reduced when C. glutamicum grows on glucose plus acetate instead of on glucose alone (30). Moreover, we here show that in fact the intracellular pyruvate concentrations in C. glutamicum WT as well as in the aceE deletion mutants were very low in the course of the growth and much higher in the stationary/production phase. Here it is interesting to note that AHAS, the first enzyme in l-valine synthesis from pyruvate, has a relatively weak affinity for pyruvate (Km = 8.3 mM) (5, 16) compared to those of enzymes competing with AHAS for pyruvate, e.g., that of pyruvate carboxylase (Km = 1.3 mM) (22).
Brik Ternbach et al. (2) recently used fed-batch experiments with C. glutamicum ΔilvA ΔpanBC(pJC4ilvBNCD) for model development and improvement of the l-valine fermentation process. In an experiment designed to maximize the total volumetric productivity, the final product concentration was almost 200 mM, and the maximal volumetric productivity was 6.2 mM l-valine per h. However, the cells produced several by-products, such as l-alanine, l-glycine, and 2-ketoglutarate (2). Leuchtenberger et al. (15) reported on a C. glutamicum mutant (VR3) which was obtained by classical mutagenesis and which produced up to 850 mM l-valine with a yield of 0.3 to 0.4 g per g of sucrose, which is equivalent to 0.45 to 0.6 mol per mol of glucose. Comparing these numbers with the data presented here for the fed-batch fermentation with C. glutamicum ΔaceE(pJC4ilvBNCE), i.e., a final l-valine concentration of 210 mM, a volumetric productivity of 10.0 mM l-valine per h and a yield of 0.5 to 0.6 mol l-valine per mol of glucose within 21 h of production, the performance of our simple model strain becomes strikingly evident. However, the maximal theoretical yield amounts to 1 mol l-valine per mol of glucose (i.e., 0.65 g per g), indicating that l-valine production with C. glutamicum can be further optimized. Moreover, taking the numbers given above in account, we neglect that C. glutamicum ΔaceE(pJC4ilvBNCE) does not produce l-valine in the growth phase. With respect to l-valine process development, the opportunity for a simple growth-decoupled production is a significant advantage allowing for more efficient use of the biocatalyst.
Further approaches to improve l-valine production with C. glutamicum ΔaceE(pJC4ilvBNCE) will aim at unraveling and overcoming the metabolic or regulatory reason for the nonproduction phenotype during the growth phase to allow both growth-coupled and -decoupled product formation. However, although not yet maximized as l-valine production strains, derivatives of C. glutamicum ΔaceE may prove useful as cost-effective biocatalysts for the production of pyruvate itself and/or of products derived from this intermediate.
Acknowledgments
We thank Lothar Eggeling for providing plasmid pJC4ilvBNCE and Joy Schreiner for critically reading the manuscript. We thank Andreas Karau and Robert Gerstmeir (Degussa AG) for valuable discussions.
The support of the Fachagentur Nachwachsende Rohstoffe of the BMVEL (grant 04NR004/22000404) is gratefully acknowledged.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Bergmeyer, H. U. 1983. Methods of enzymatic analysis, 3rd ed., vol. 6. Verlag Chemie, Weinheim, Germany.
- 2.Brik Ternbach, M., C. Bollmann, C. Wandrey, and R. Takors. 2005. Application of model discriminating experimental design for modeling and development of a fermentative fed-batch l-valine production process. Biotechnol. Bioeng. 91:356-368. [DOI] [PubMed] [Google Scholar]
- 3.Chassagnole, C., A. Diano, F. Létisse, and N. D. Lindley. 2003. Metabolic network analysis during fed-batch cultivation of Corynebacterium glutamicum for pantothenic acid production: first quantitative data and analysis of by-product formation. J. Biotechnol. 104:261-272. [DOI] [PubMed] [Google Scholar]
- 4.de Koning, W., and K. van Dam. 1992. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal. Biochem. 204:118-123. [DOI] [PubMed] [Google Scholar]
- 5.Eggeling, I., C. Cordes, L. Eggeling, and H. Sahm. 1987. Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of alfa-ketobutyrate to l-isoleucine. Appl. Microbiol. Biotechnol. 25:346-351. [Google Scholar]
- 6.Eggeling, L. 2001. Amino acids, p. 281-303. In C. Ratledge and B. Kristiansen (ed.), Basic bio/technology. Cambridge University Press, London, United Kingdom.
- 7.Eikmanns, B. J., M. Metzger, D. Reinscheid, M. Kircher, and H. Sahm. 1991. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl. Microbiol. Biotechnol. 34:617-622. [DOI] [PubMed] [Google Scholar]
- 8.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K. U. Ludtke, and H. Sahm. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 9.Elišáková, V., M. Pátek, J. Holátko, J., Nešvera., D. Leyval, J. L. Goergen, and S. Delaunay. 2005. Feedback-resistant acetohydroxy acid synthase increases valine production in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71:207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Hüser, A. T., C. Chassagnole, N. D. Lindley, M. Merkamm, A. Guyonvarch, V. Elišáková, M. Pátek, J. Kalinowski, I. Brune, A. Pühler, and A. Tauch. 2005. Rational design of a Corynebacterium glutamicum pantothenate production strain and its characterization by metabolic flux analysis and genome-wide transcriptional profiling. Appl. Environ. Microbiol. 71:3255-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, N. B. S., K. V. Jokumsen, and J. Villadsen. 1999. Determination of the phosphorylated sugars of the Embden-Meyerhoff-Parnas pathway in Lactobacillus lactis using a fast sampling technique and solid phase extraction. Biotechnol. Bioeng. 63:356-362. [PubMed] [Google Scholar]
- 13.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leuchtenberger, W. 1996. Amino acids—technical production and use, p. 465-502. In H.-J. Rehm, G. Reed, A. Pühler, P. Stadler, and M. Roehr (ed.), Biotechnology, vol. 6. VCH, Weinheim, Germany. [Google Scholar]
- 15.Leuchtenberger, W., K. Huthmacher, and K. Drauz. 2005. Biotechnological production of amino acids and derivates: current status and prospects. Appl. Microbiol. Biotechnol. 69:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Leyval, D., D. Uy, S. Delaunay, J. L. Goergen, and J. M. Engasser. 2003. Characterisation of the enzyme activities involved in the valine biosynthetic pathway in a valine-producing strain of Corynebacterium glutamicum. J. Biotechnol. 104:241-252. [DOI] [PubMed] [Google Scholar]
- 17.Liebl, W. 1991. The genus Corynebacterium—nonmedical, p. 1157-1171. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, vol. 2. Springer, New York, NY. [Google Scholar]
- 18.Lindroth, P., and K. Mopper. 1979. High performance liquid chromatographic determination of subpicomole amounts of amino acids by precolumn fluorescence derivatization with o-phthalaldehyde. Anal. Chem. 51:1667-1674. [Google Scholar]
- 18a.Luo, B., K. Grönke, R. Takors, C. Wandrey, and M. A. Oldiges. A simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and TCA cycle by liquid chromatography-mass spectrometry. J. Chromatogr. A, in press. [DOI] [PubMed]
- 19.Morbach, S., C. Junger, H. Sahm, and L. Eggeling. 2000. Attenuation control of ilvBNC in Corynebacterium glutamicum: evidence of leader peptide formation without the presence of a ribosome binding site. J. Biosci. Bioeng. 90:501-507. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama, K., S. Kitada, and S. Kinoshita. 1961. l-Valine production using microbial auxotroph. J. Gen. Appl. Microbiol. 7:52-69. [Google Scholar]
- 21.Petersen, S., C. Mack, A. A. de Graaf, C. Riedel, B. J. Eikmanns, and H. Sahm. 2001. Metabolic consequences of altered phosphoenolpyruvate carboxykinase activity in Corynebacterium glutamicum reveal anaplerotic mechanisms in vivo. Metab. Eng. 3:344-361. [DOI] [PubMed] [Google Scholar]
- 22.Peters-Wendisch, P. G., V. F. Wendisch, S. Paul, B. J. Eikmanns, and H. Sahm. 1997. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology 143:1095-1103. [DOI] [PubMed] [Google Scholar]
- 23.Radmacher, E., A. Vaitsiková, U. Burger, K. Krumbach, H. Sahm, and L. Eggeling. 2002. Linking central metabolism with increased pathway flux: l-valine accumulation by Corynebacterium glutamicum. Appl. Environ. Microbiol. 68:2246-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahm, H., and L. Eggeling. 1999. d-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes encoding l-valine synthesis for d-pantothenate overproduction. Appl. Environ. Microbiol. 65:1973-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., D. W. Russell, N. Irwin, and U. A. Janssen. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Schreiner, M. E., D. Fiur, J. Holátko, M. Pátek, and B. J. Eikmanns. 2005. E1 enzyme of the pyruvate dehydrogenase complex in Corynebacterium glutamicum: molecular analysis of the gene and phylogenetic aspects. J. Bacteriol. 187:6005-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomar, A., M. A. Eitemann, and E. Altmann. 2003. The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli. Appl. Microbiol. Biotechnol. 62:76-82. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchida, T., F. Yoshinaga, and K. Kubota. 1975. Production of l-valine by 2-thiazolealanine resistant mutants derived from glutamic acid bacteria. Agric. Biol. Chem. 39:1319-1322. [Google Scholar]
- 29.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogenic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 30.Wendisch, V., A. A. de Graaf, H. Sahm, and B. J. Eikmanns. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zelic, B., T. Gerharz, M. Bott, D. Vasic-Racki, C. Wandrey, and R. Takors. 2003. Fed-batch process for pyruvate production by recombinant Escherichia coli YYC202 strain. Eng. Life Sci. 3:299-305. [Google Scholar]