Abstract
Soda lakes are naturally occurring highly alkaline and saline environments. Although the sulfur cycle is one of the most active element cycles in these lakes, little is known about the sulfate-reducing bacteria (SRB). In this study we investigated the diversity, activity, and abundance of SRB in sediment samples and enrichment cultures from a range of (hyper)saline soda lakes of the Kulunda Steppe in southeastern Siberia in Russia. For this purpose, a polyphasic approach was used, including denaturing gradient gel electrophoresis of dsr gene fragments, sulfate reduction rate measurements, serial dilutions, and quantitative real-time PCR (qPCR). Comparative sequence analysis revealed the presence of several novel clusters of SRB, mostly affiliated with members of the order Desulfovibrionales and family Desulfobacteraceae. We detected sulfate reducers and observed substantial sulfate reducing rates (between 12 and 423 μmol/dm3 day−1) for most lakes, even at a salinity of 475 g/liter. Enrichments were obtained at salt saturating conditions (4 M Na+), using H2 or volatile fatty acids as electron donors, and an extremely halophilic SRB, strain ASO3-1, was isolated. Furthermore, a high dsr gene copy number of 108 cells per ml was detected in a hypersaline lake by qPCR. Our results indicate the presence of diverse and active SRB communities in these extreme ecosystems.
Soda lakes are extreme environments with pH values up to 11 and salt concentrations up to saturation. These high pH values are maintained due to the high buffering capacity of sodium carbonate/bicarbonate, which are among the major anions in solution. The salinity can vary from 5 to 30 g/liter for hyposaline lakes to up to 500 g/liter for hypersaline lakes. Despite these extreme conditions, soda lakes are highly productive and harbor diverse microbial communities (6, 11, 25, 35) responsible for the element cycling.
The sulfur cycle, driven by haloalkaliphilic sulfur-oxidizing bacteria (SOB) and sulfate-reducing bacteria (SRB), is one of the most active element cycles in soda lakes. SOB are represented by members of the class Gammaproteobacteria belonging to the genera Ectothiorodospira, Thioalkalivibrio, Thioalkalimicrobium, and Thioalkalispira. Bacteria belonging to the first genus are phototrophic sulfur purple bacteria, while the other three genera are obligate chemolithoautotrophs (30), utilizing various reduced inorganic sulfur compounds as electron donors. Members of the Ectothiorodospira and Thioalkalivibrio genera are particularly remarkable in their potential to grow in saturated alkaline brines.
Although more than 100 SOB strains have been isolated from soda lakes (30), so far only four species of SRB, i.e., Desulfonatronovibrio hydrogenovorans (37), Desulfonatronum lacustre (22), Desulfonatronum thiodismutans (23), and Desulfonatronum cooperativum (38) have been isolated, and they are all low-salt-tolerant alkaliphiles. However, although Desulfonatronovibrio hydrogenovorans is a low-salt organism, it was isolated from Lake Magadi, a hypersaline soda lake in Kenya (Africa). According to Oren (21), the upper limit of salt concentration for sulfate reducers appears to be 250 g/liter for incomplete oxidizers and the upper limit for complete oxidizers is ca. 130 g/liter, which might be due to the high energy requirements for the synthesis of compatible solutes. So far, the incomplete oxidizers are mainly represented by members of the order Desulfovibrionales and family Desulfobulbaceae, while the complete oxidizers are mainly represented by SRB belonging to the family Desulfobacteraceae. Interestingly, recent in situ measurements (28) demonstrated sulfate reducing activity in Siberian soda lakes with saturating salinity, indicating the presence of extremely salt-tolerant alkaliphilic SRB species with possible new bioenergetic pathways. The importance of haloalkaliphilic SRB for element cycling in soda lakes can be illustrated by the fact that anaerobic cellulose degradation in soda lake sediments is possible only in the presence of sulfate (10).
Apart from the isolation of pure cultures, culture-independent methods have been used to study SRB community structure in soda lakes. For instance, Scholten and coworkers (25) studied sulfate-reducing bacterial communities from Mono Lake, a meromictic moderate saline soda lake in California in the United States by targeting 16S rRNA, apsA, and dsrAB genes.
Here we describe, for the first time, the diversity, activity, and abundance of SRB in sediment samples from saline and hypersaline soda lakes located in the Kulunda Steppe in southeastern Siberia (Russia). Denaturing gradient gel electrophoresis (DGGE) of dsr gene fragments (4, 8) was used to investigate the diversity of SRB in sediment samples and enrichment cultures, while serial dilutions and quantitative real-time PCR (qPCR) (36) were used to estimate the abundance of SRB. The activity of SRB was investigated by measuring sulfate reduction rates (SRR).
Our results showed the presence and activity of several novel lineages of SRB in the soda lakes, mostly affiliated to the Desulfovibrionales and Desulfobacteraceae. The presence of members of the last group is particularly unexpected, as it was hypothesized that complete oxiders cannot grow at salt concentrations greater than 130 g/liter (21). In this paper we provide novel insight into the diversity of SRB in soda lakes, which is essential for a comprehensive understanding of the sulfur cycle in these extreme ecosystems.
MATERIALS AND METHODS
Site description.
The Kulunda Steppe in Siberia, Russia, stretches from the southeastern part of the Novosibirsk region to the southwestern part of the Altai region and along the northeastern border of Kazah. It harbors numerous saline lakes that differ in size and chemistry. Small and shallow lakes in the southern part of the Kulunda Steppe are characterized by high pH values, high to extremely high carbonate alkalinity, and a total salt content from 50 to 500 g liter−1. The characteristics of the lakes from which sediment samples were obtained and lake numbers are presented in Table 1.
TABLE 1.
Chemical parameters of soda lake brines from the Kulunda Steppe, southeastern Siberia, Russia, measured in 2005
| Lake no. | Lake name | Salinity (g/liter) | pH | Alkalinity (M)
|
SRR (μmol/dm3 day−1) | Depth (cm) | SO42− concn (mM) | |
|---|---|---|---|---|---|---|---|---|
| CO32− | HCO3− | |||||||
| 1KL | Lake Tanatar III | 93 | 10.08 | 0.84 | 0.18 | NDb | ND | ND |
| 2KL | Lake Tanatar II | 250 | 9.78 | 2.06 | 0.77 | ND | ND | ND |
| 3KL | Lake Tanatar I (east) | 475 (520)a | 10.65 (11.05)a | 5.0 (4.0)a | 0 | 12.7 (100)c | wd | 6.7-8.5 |
| 113 (93) | 0-3 | |||||||
| 44 (78) | 3-6 | |||||||
| 12.6 (27) | 6-10 | |||||||
| 4KL | Lake Tanatar I (south) | 350 | 10.0 | 5.0 | 0.2 | ND | ND | ND |
| 5KL | Lake Tanatar V | 50 | 10.1 | 0.42 | 0.12 | ND | ND | ND |
| 6KL | Lake Tanatar VI | 55 | 10.1 | 0.52 | 0.06 | ND | ND | ND |
| 9KL | Elongated Lake | 320 | 9.15 | 0.32 | 0.14 | ND | ND | ND |
| 10KL | Top Karagay | 58 | 9.7 | 0.14 | 0.1 | ND | ND | ND |
| 11KL | Anonymous | 55 | 9.76 | 0.32 | 0.16 | ND | ND | ND |
| 16KL | Stamp Lake | 380 | 9.27 | 0.38 | 0.18 | ND | ND | ND |
| 18KL | Cock Lake | 59 (74) | 10.1 (10.13) | 0.66 (0.31) | 0.14 (0.13)a | 61 (13) | w | 6.7-8.5 |
| 326 (14) | 0-4 | |||||||
| 3.7 (3) | 4-12 | |||||||
| 10KL-05 | Karacul Lake | 30 | 9.76 | 0.05 | 0.04 | ND | ND | ND |
| 18KL-05 | Picturesque Lake | 405 | 9.95 | 1.3 | 0.7 | 2.8 (0) | w | 278-584 |
| 47 (43) | 0-2 | |||||||
| 423 (0) | 2-6 | |||||||
| 12 (0) | 6-18 | |||||||
| 22KL-05 | Anonymous 2 | 360 | 9.46 | 0.7 | 0.7 | ND | ND | ND |
| 23KL-05 | Narrow Lake | 110 | 9.61 | 0.23 | 0.26 | 1.1 (63) | w | 235-278 |
| 62 (22) | 0-2 | |||||||
| 162 (0) | 2-17 | |||||||
| 52 (5) | 17-32 | |||||||
| 25KL-05 | Bitter Lake 1 | 120 | 10.3 | 0.65 | 0.49 | 2.6 (0) | w | 0.6-2.5 |
| 14.7 (19) | 0-1 | |||||||
| 2.6 (11) | 1-9 | |||||||
| 6 (10) | 9-20 | |||||||
Values in parentheses are values for water near the bottom.
ND, not determined.
Values in parentheses are values for the acid-resistant fraction, as a percentage of the total (sulfur plus pyrite).
w, water near the bottom.
Chemical analyses and sampling.
Field measurements included pH, conductivity, and carbonate alkalinity (titration with 1 M HCl using phenolphthalein and methyl orange as indicators). The salt content, inferred from conductivity measurements, was verified by gravimetry in the laboratory. For DNA extraction, the top 10-cm sediment was sampled from three different places along the littoral and combined in one 50-ml Falcon tube. The tubes were stored at 4°C during transportation to the laboratory and stored at −20°C until further analysis. For the rate measurements, 20-cm cores were taken and then divided in different layers on the basis of color. SRR were measured in 5-ml syringes capped with butyl rubber stoppers using the [35S]SO42− methodology (15). After 1 day of incubation at ambient temperature, the sediments were fixed with 10 M KOH and further processed in the laboratory. Additional analysis of total sulfate content in the sediment pore water was performed in the laboratory after centrifugation of 2-cm3 sample using ion-exchange chromatography (24). Samples for PCR-DGGE, qPCR, and enrichments were taken in the summer of 2003, while SRR measurements and serial dilutions were performed in the summer of 2005. General field measurements were performed during both visits to the sites.
Serial dilutions.
Enumeration of SRB was performed in duplicate by decimal serial dilutions in Hungate tubes with the top 5-cm sediment layer as inoculum. Dilutions were done in low-salt (0.6 M total Na+) and high-salt (4 M total Na+) carbonate media, pH 10 (29) with H2 or lactate and butyrate as an energy source. The inoculation was done in the field. Growth was monitored within 4 to 8 weeks of incubation at room temperature by estimating sulfide production (32).
Enrichment of haloalkaliphilic SRB.
Enrichments of SRB were obtained by inoculating 30-ml vials filled with 10 ml of carbonate media (1, 2, or 4 M total Na+, N2 atmosphere) and 1 ml of sediment slurry. Medium composition was similar as described by Sorokin (29) with minor modifications. Ammonium chloride (5 mM) was used as a nitrogen source, and trace elements (31), vitamins, and a mixture of sodium sulfide (1.2 mM) and sodium dithionite (0.5 mM) were added as reducing agents to the media (17). The slurries for inoculation were prepared by mixing sediment samples from three lakes (i.e., 1KL, 6KL and 18KL [Table 1]) with an equal volume of the appropriate (1, 2, or 4 M) carbonate medium. N2-H2 (50:50 [vol/vol]), ethanol (30 mM), or a mixture of volatile fatty acids (VFAs) (formate [30 mM], lactate [30 mM], and acetate [30 mM]) were added as electron donors. Sulfate (30 mM) was added as an electron acceptor. Bottles were incubated at 37°C in the dark. After several transfers, dilution series of some of the enrichments were prepared as a first step to obtain pure cultures. Growth of SRB was monitored by determining the concentrations of electron donor and acceptor over time (26).
Special enrichments were made to look at the presence of SRB capable of sulfite fermentation. For this, anaerobic sodium carbonate medium containing 0.6, 2, and 4 M total Na+, pH 10, was supplemented with 1 to 2 mM sulfide as a reductant and with 5 to 10 mM sodium sulfite as the only energy source. The culture was inoculated with 10% (vol/vol) of sediment sample composed of 10 individual samples from Kulunda soda lakes. The development was monitored by microscopy and increase in sulfide concentration and decrease in sulfite concentration.
DNA extraction.
Prior to DNA extraction, the sediments were washed three times with 1 M NaCl and 10 mM Tris (pH 7.5) to lower the pH and salt concentration. The genomic DNA from 11 sediment samples was extracted using the UltraClean soil DNA extraction kit (MoBio Laboratories), following the manufacturer's instructions with minor modifications. Briefly, 200 μl of 0.1 M AlNH4(SO4)2 was added to the sediment to remove potential PCR inhibitors, such as humic acids (2). Subsequently, the cells were lysed by a combination of detergents and mechanical disruption. The released DNA was bound to a silica spin filter. The filter was washed, and the DNA was recovered in Milli-Q water. The quality of the extracted DNA was examined on 1% (wt/vol) agarose gels in 1× TBE buffer (1× TBE buffer is 90 mM Tris-borate and 2 mM EDTA, pH 8) after staining with ethidium bromide. Images were obtained using the Gel Doc 2000 system (Bio-Rad, Hercules, CA).
PCR of dsr gene fragments.
In order to perform DGGE based on the dsrB gene, a nested PCR approach was used to increase the sensitivity of the amplification. First, the entire dsrAB gene of ca. 1,900 bp was amplified. Amplification of the extracted DNA was performed in a 25-μl final volume with 12.5 μl Master Mix QIAGEN (QIAGEN, Hilden, Germany), MgCl2 (final concentration of 1.75 mM), and 0.5 μl (final concentration of 0.4 μM) of primer DSR1F (5′-ACSCACTGGAAGCACG-3′) and DSR4R (5′-GTGTAGCAGTTACCGCA-3′) (33). The following PCR conditions were used: (i) 5 min at 94°C; (ii) 30 cycles, with 1 cycle consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s; and (iii) a final extension at 72°C for 10 min. Subsequently, a 350-bp fragment portion of the dsrB gene was amplified using primers DSRp2060F (5′-GC clamp-CAACATCGTYCAYACCCAGGG-3′) (8) and DSR4R. Two PCRs were performed per each sample in a final volume of 25 μl, with 12.5 μl Master Mix QIAGEN (QIAGEN, Hilden, Germany), MgCl2 (final concentration of 1.75 mM), and 1 μl of each primer (final concentration of 0.2 μM). The reaction was carried out using a so-called touchdown protocol: (i) 5 min at 95°C; (ii) 20 cycles, with 1 cycle consisting of 40 s at 95°C, decreasing of the annealing temperature from 65°C to 55°C, and 1 min at 72°C. In addition, another 20 cycles (1 cycle consisting of 40 s at 94°C, 40 s at 55°C, and 1 min at 72°C) were performed, followed by a final extension at 72°C for 10 min. The yield and quality of the PCR products were examined on 1% (wt/vol) agarose gels stained with ethidium bromide.
DGGE of dsrB gene fragments.
The dsrB amplicons were analyzed by DGGE as described by Dar et al. (4) using a gradient of 30 to 65% urea-formamide in 6% polyacrylamide gels. The electrophoresis was performed in 1× TAE buffer (1× TAE buffer is 40 mM Tris, 20 mM acetic acid, and 1 mM EDTA, pH 8) at 60°C for 5 h at a constant voltage of 150 V. After electrophoresis, the gels were stained with ethidium bromide and destained in Milli-Q water. Images were obtained using the Gel Doc 2000 system (Bio-Rad, Hercules, CA). Bands of interest were excised, reamplified, and run again on denaturing gels to check their purity. PCR products for sequencing were reamplified using primers without GC clamp, run on a 1.5% (wt/vol) agarose gel, and purified with the QIAquick gel extraction kit (QIAGEN, Hilden, Germany). The ∼350-bp purified PCR products were then sent to a company (BaseClear, Leiden, The Netherlands) for sequencing.
Comparative sequence analysis.
Partial dsrB sequences were compared with sequences in the GenBank database using the BLAST search tool for the identification of closely related sequences. Nucleic acid sequences were aligned with the most similar sequences using the ClustalW and BioEdit Sequence Alignment Editor software, and subsequently translated into amino acid sequences. Deduced aligned amino acid sequences were then imported into the ARB software package (19) for phylogenetic analysis.
A consensus tree was calculated from nearly complete sequences using neighbor-joining, maximum-parsimony, and maximum-likelihood algorithms and different filters. Subsequently, the partial sequences were added to the tree using the QUICK_ADD parsimony tool and individually removed to avoid long branch attraction. Deletions and insertions were not included in the calculation. In addition, only the positions encoded on the added sequences were considered for the phylogenetic reconstruction. Finally, a consensus tree was generated.
qPCR.
Quantification of the dsrB copy number in the extracted DNA was performed using the I-Cycler (Bio-Rad Laboratories, Veenendaal, The Netherlands). SYBR green (Bio-Rad Laboratories, Veenendaal, The Netherlands) was used as a double-stranded DNA (dsDNA) binding dye, whereby the fluorescence intensity of the dye increases with the amount of amplified dsDNA. Baseline and threshold calculations were performed with the I-Cycler software (version 4).
Amplification was done with SYBR green Super Mix (Bio-Rad Laboratories, Veenendaal, The Netherlands) and the same set of primers used for the dsrB gene at a final concentration of 0.2 μM. The amplification consists of 35 cycles, with 1 cycle consisting of denaturation (40 s at 95°C), annealing (40 s at 55°C), and elongation (1 min at 72°C).
The standard curve was calculated on the basis of a partially dsrB purified PCR product of known copy number. Using the standard curve, we determined the dsr copy number per milliliter of sediment. The qPCR measurements were done in triplicate. The optimal DNA dilution was first tested and then used for the final qPCR. The amplification efficiency (E) was calculated from the slope of the standard curve using the formula: percent E = 10−1/slope × 100. Since SYBR green might also bind to nonspecific dsDNA, a melting curve was performed to assure the specificity of the PCR.
Nucleotide sequence accession numbers.
The sequences determined in this study were deposited in GenBank under the following accession numbers: EF055361 to EF055374 for the sediment samples and EF055375 to EF055384 for the enrichment samples.
RESULTS
Sulfate reduction rates in soda lake sediments.
SRR were measured in sediments of five lakes with increasing salinity. Sulfate was present at very high concentrations, up to 584 mM, in sediment pore waters of lakes with a salinity of more than 150 g/liter, while in less saline lakes, such as Cock Lake and Bitter Lake 1, it was present at lower concentrations, 8.5 and 2.5 mM, respectively (Table 1). The maximal sulfate reduction rates were measured for lakes Picturesque and Cock, 423 and 326 μmol/dm3 day−1, respectively. Up to 35% of the 35S label from sulfate was found in the acid-resistant fraction (i.e., sulfur plus pyrite). Substantial SRR (44 μmol/dm3 day−1) were even found for Lake Tanatar I, which has an extremely high salinity of 475 g/liter.
Serial dilutions and enrichments.
Samples from the same sediments were also used for enumeration and enrichments. Enumeration of culturable haloalkaliphilic SRB was done by serial dilutions using 0.6 and 4 M sodium carbonate-based medium, pH 10, with either H2 or VFA (lactate and butyrate) as an electron donor (Table 2). The results demonstrated the presence of relatively dense populations (up to 106 cells per ml sediment) of haloalkaliphilic SRB in the sediments of the investigated lakes (Table 2). Further attempts to grow SRB from primary dilutions were successful for only some of the samples, especially those grown in 4 M sodium carbonate-based medium. The dominant morphotype in all these enrichments was a motile, vibrio-shaped cell.
TABLE 2.
Enumeration of SRB in sediments of Kulunda soda lakes in southeastern Siberia, Russia
| Lake no. | Depth (cm) | Total [Na+] (M) in medium | No. of SRB
|
|
|---|---|---|---|---|
| Heterotrophica | Autotrophicb | |||
| 10KL-05 | 0-5 | 0.6 | 105 | 105 |
| 6-15 | 0.6 | 105 | 104 | |
| 16-25 | 0.6 | 102 | 102 | |
| 25KL-05 | 0-1 | 0.6 | 106 | 104 |
| 1-9 | 0.6 | NDc | 103 | |
| 18KL | 0-4 | 0.6 | 106 | ND |
| 4-12 | 0.6 | 105 | ND | |
| 18KL-05 | 0-2 | 4.0 | 103 | 104 |
| 2-6 | 4.0 | 102 | 102 | |
| 6-18 | 4.0 | 102 | ND | |
| 22KL-05 | 0-10 | 4.0 | 105 | 105 |
| 23KL-05 | 0-2 | 4.0 | 106 | ND |
Grown on a mixture of lactate and butyrate.
Grown on a mixture of H2 and CO2.
ND, not determined.
Incubation in a mineral medium of a mixture of three sediments from lakes of the Kulunda region in southeastern Siberia in Russia revealed that sulfate reduction activity was found on all of substrates tested, i.e., H2, a mixture of VFAs (formate, lactate, and acetate), and ethanol. Sulfate-reducing activity was found up to 4 M sodium for H2 and ethanol and was highest at 2 M sodium for all substrates tested. In all enrichments, vibrio-shaped cells were dominant.
Enrichments for sulfite fermentation gave a positive enrichment culture with 2 M Na+ and 10 mM sulfite. From this enrichment, a pure culture, strain ASO3-1, was isolated using the dilution to extinction method.
Diversity analysis using DGGE of dsrB gene fragments.
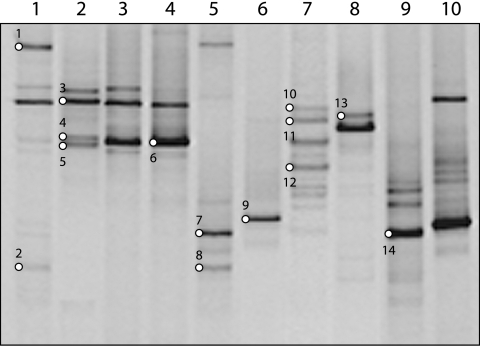
The SRB diversity in the sediment samples and enrichment cultures was analyzed by DGGE using the dsrB subunit as a molecular marker (8). For the sediment samples, as many as seven distinct bands could be observed (Fig. 1, lane 7). Samples 2KL, 3KL, and 4KL show patterns similar to each other (Fig. 1, lanes 2 to 4). The other samples showed different profiles, indicating a moderate diversity of SRB in this habitat.
FIG. 1.
DGGE analysis of the partial dsrB gene fragments from sediment samples of different soda lakes from the Kulunda Steppe (southwestern Siberia, Russia). Lane 1, 1KL; lane 2, 2KL; lane 3, 3KL; lane 4, 4KL; lane 5, 5KL; lane 6, 6KL; lane 7, 9KL; lane 8, 10KL; lane 9, 11KL; lane 10, 16KL (see Table 1 for more information on the soda lakes). Bands indicated by a white circle and number were excised from the gel and sequenced.
Regarding the enrichments, the same sediment samples analyzed in this study were used as inoculum. Eleven out of 21 enrichments (results not shown) revealed an identical DGGE profile with a single dominant band, whose sequence (i.e., E_H2_1_E [sequence names explained in the legend to Fig. 2) corresponds to a novel cluster affiliated to Desulfonatronovibrio hydrogenovorans (Fig. 2). Sequence E_H2_1_E was present in all enrichments except for two, in which VFA were used as electron donor at a salinity of 2 M.
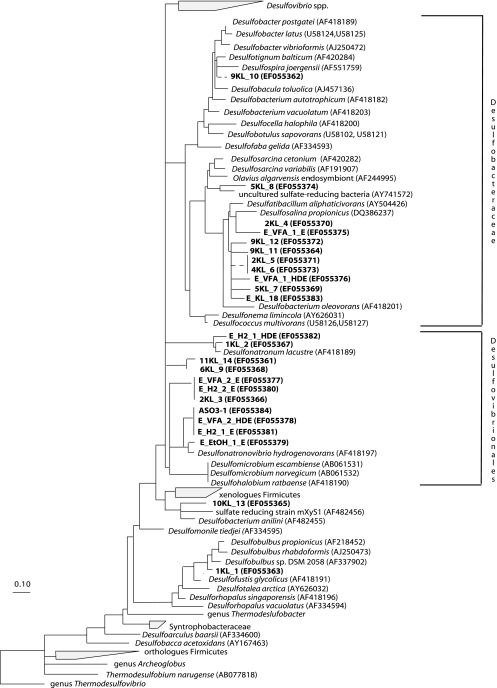
FIG. 2.
Consensus phylogenetic tree based on amino acid sequences of the dsrAB gene. The partial sequences determined in this study are in bold type. They were added to the tree using the QUICK_ADD parsimony tool and individually removed to avoid long branch attraction. Deletions and insertions were not included in the calculation. Branching orders that were not supported by all tree construction methods are shown as multiforcations. The band number (Fig. 1) is preceded by the lake number (Table 1) for sediment samples. For enrichment samples, the following code was used: E_H2/VFA/EtOH (ethanol [EtOH]) (electron donor)_1/2 (total Na+ [in molar concentration])_E/HDE/HDO/LDO (type of enrichment). E stands for enrichment obtained by regular transfers of an incubation that was originally inoculated with the sediment mixture sample. HDE are enrichments that were obtained by regular transfer of the highest positive dilution of the initial sediment incubation at that specific condition, whereas HDO and LDO are the first enrichments with the highest and the lowest positive dilutions, respectively, in which the original sediment sample was used as inoculum.
Identification of SRB.
Altogether, 24 representative dsrB fragments were sequenced from sediment samples and enrichment cultures. Comparative sequence analysis was conducted on both nucleotide and deduced amino acid translated sequences. The resulting topology from DNA (not shown) and protein (Fig. 2) sequences was largely congruent, differing only in the exact placement of individual sequences within low branching clusters. All of our sequences fell in the Deltaproteobacteria class of the phylum Proteobacteria, with a majority in the Desulfovibrionales (11 sequences) and the Desulfobacteriaceae (11 sequences) (Fig. 2). One sequence (KL1_1) fell within the Desulfobulbaceae and showed a similarity of 93% at the protein level with the dsr sequence of the glycolate-oxidizing SRB Desulfofustis glycolicus. Another sequence (KL10_13), from the low-salt lake 10KL, clustered together with sequences of Desulfobacter anilinii and the sulfate-reducing strain mXyS1 (77% similarity), both belonging to the order Desulfobacterales.
Four sequences from sediment samples and seven from enrichment samples were affiliated with members of the Desulfovibrionales. Nine sequences formed two novel clusters associated with Desulfonatronovibrio hydrogenovorans (with similarities between 68% and 86%), and two other sequences were related to Desulfonatronum lacustre (with 91% and 94% similarity, respectively). One sequence obtained from the high-saline Lake Tanatar II (i.e., KL2_3), and two sequences (E_H2_2_E and E_VFA_2_E) from samples both enriched at 2 M Na+, but with different electron donors (i.e., H2 and VFA), were identical and affiliated with Desulfonatronovibrio hydrogenovorans (Fig. 2).
Only sequences from low-saline lakes, i.e., KL1_2 from Lake Tanatar III, and E_H2_1_HDE, obtained from samples enriched at 1 M Na+ and H2 as electron donor, were affiliated with Desulfonatronum lacustre.
Eleven sequences clustered within the Desulfobacteraceae. Nine sequences were associated with Desulfosalina propionicus, a so far nondescribed halophilic SRB isolated from the Great Salt Lake (Utah), and seven of them formed a new lineage. Two sequences (KL2_5 and KL4_6) obtained from two different lakes, i.e., Lake Tanatar I and Lake Tanatar II (Table 1), had identical sequences. All sequences from sediment samples were obtained from extremely saline lakes (from 250 to 350 g/liter Na+). Another sequence, KL5_8 from Lake Tanatar V, clustered together with Desulfosarcina variabilis (91% similarity), and another one, KL9_10, was related to Desulfotignum balticum (96% similarity), a facultative halophilic organism.
Quantification of SRB by qPCR.
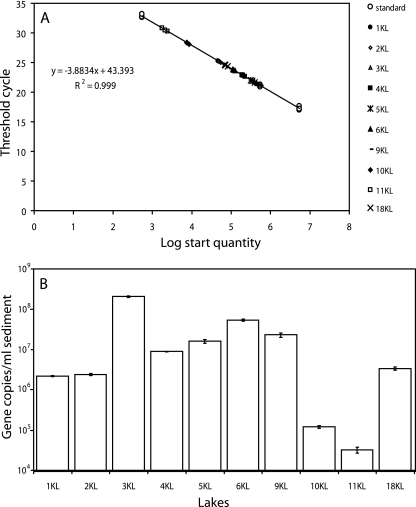
We applied qPCR to estimate the abundance of SRB in the sediment samples of different soda lakes. The standard curve was based on a partially dsrB purified PCR product, in which the dsr copy numbers were known (Fig. 3A). All the sediment samples analyzed fit exactly the standard curve.
FIG. 3.
Abundance of the SRB in sediment samples by qPCR. (A) Standard curve for qPCR measurements of the dsr gene. The standard curve was calculated on the basis of a partially dsrB purified PCR product of known copy number. (B) SRB abundance (logarithmic scale) in the different soda lakes. Error bars represent standard deviations.
In four lakes (i.e., lakes 1KL, 2KL, 4KL, and 18KL), the copy numbers of the dsr gene have been found to be in the order of 106 per ml sediment, while in lakes 5KL, 6KL, and 9KL, they were found to be 1 order of magnitude higher (Fig. 3B). In lakes 10KL and 11KL, two low-salt lakes which have similar conditions, the lowest copy number was found, i.e., 105 and 104, respectively (Fig. 3B). In contrast, the highest dsr gene copy numbers (108) per ml sediment were detected in lake 3KL, the most saline lake investigated in this study.
DISCUSSION
Here we describe the diversity, activity, and abundance of SRB in sediment samples from Siberian soda lakes in Russia. For this purpose, we used a polyphasic approach, which included both culture-dependent (i.e., enrichment and serial dilutions) and independent (i.e., PCR-DGGE, comparative sequence analysis, and qPCR) techniques.
SRB activity.
SRB activity was detected by SRR measurements. In sediments of soda lakes from the Kulunda Steppe in southeastern Siberia in Russia, we observed SRR that are significantly higher than the values measured for less alkaline and saline habitats. For instance, SRR were observed in the range of 0.5 to 13 μmol/dm3 day−1 for freshwater sediments (16), 5.6 to 104 μmol/dm3 day−1 for a sediment of a Yellowstone hot spring, and from 15 to 20 μmol/dm3 day−1 for marine sediments (12). The highest SRR reported to the best of our knowledge, 5 × 103 μmol/dm3 day−1, was measured in a marine sediment covered by a Beggiatoa mat (1).
There are few data available on SRR in soda lakes; in Big Soda Lake (Nevada) and in Mono Lake (California), the SRR in the anoxic water were significantly lower than in the above-mentioned habitats (20, 27): 0.74 × 10−3 to 3.2 × 10−3 μmol/dm3 day−1 for Big Soda Lake and 2.3 × 10−3 μmol/dm3 day−1 for Mono Lake. In the Mongolian soda lake sediments (28), and especially in hyposaline and less alkaline (pH 9.4 to 9.5) lakes in the southeastern Transbaikal area in Russia, the SRR were considerably higher, i.e., 3 × 103 μmol/dm3 day−1 (9). For Cock and Picturesque Lakes, we measured SRR that were only slightly lower (Table 1). Especially unexpected was the very high SRR measured in Picturesque Lake, since it has extremely high salinity (405 g/liter).
Even at a salt concentrations of 520 g/liter (Lake Tanatar I), substantial SRR were observed (Table 1). This indicates again that a yet undiscovered group of extremely haloalkaliphilic SRB might be active in these hypersaline lakes.
SRB quantification.
Estimation of the number of sulfate reducers was done by both culture-independent, i.e., quantitative real-time PCR on genomic DNA, as well as culture-dependent techniques, i.e., serial dilution analysis. The highest dsr gene copy number was detected in Lake Tanatar I, with 108 cells per ml sediment, assuming that SRB possess only one dsr gene copy (14). More than one dsr gene copy has been detected in some Desulfovibrio species (14); therefore, our qPCR analysis might give an overestimation of the SRB population. However, our results are in accordance (Fig. 3) with data observed in Mono Lake, a moderate-salt soda lake, in which SRB have been found in the order of 0.5 × 107 to 6 × 107 cells per ml. Enumerations obtained from serial dilutions were lower than the ones detected by qPCR (Table 2), probably due to cultivation biases (13).
SRB diversity in soda lakes.
Most of the SRB fell into two major groups, i.e., the Desulfovibrionales and Desulfobacteraceae. Concerning the first group, sequences were affiliated with Desulfonatronovibrio hydrogenovorans, which belongs to the Desulfohalobiaceae family and to Desulfonatronum lacustre, the only member of the Desulfonatronumaceae family (18). Both these strains are low-salt-tolerant alkaliphiles isolated from soda lakes. They are incomplete oxidizers and are able to use hydrogen and a few organic compounds as an electron donor, which is a common feature of the members of the Desulfovibrionales order. In this study, two novel clusters related to Desulfonatronum hydrogenovorans were found, revealing that the diversity within this group is higher than anticipated. The identified SRB belonging to this group originated from both high- and low-salinity samples. Since we found that two DGGE bands at the same position but from different enrichment samples resulted from the same sequence (data not shown), we assume this is generally valid. Band E_H2_1_E was excised from a sample enriched at 1 M Na+ but was also present at higher salt concentrations. Therefore, we can conclude that this group included SRB, such as E_H2_1_E, having a broad range of salt tolerance. On the other hand, the group apparently includes highly specialized organisms, such as the extremely haloalkaliphilic isolate ASO3-1, which is unique not only for its sodium requirement (2 M Na+) and tolerance (4 M Na+), but also for its ability to grow without any organic substrate by sulfite fermentation. This isolate is, to the best of our knowledge, the only sulfate reducer known to be able to grow at this high salt concentration. Therefore, it would be interesting to detect this new SRB ecotype with specific probes in other hypersaline environments. The use of probes may, however, not be specific, since two sequences originating from low-salt enrichments, i.e., E_VFA_2_HDE and E_H2_1_E, showed 100% identity with the sequence of the obligate extremely high-salt isolate ASO3-1 (Fig. 2). Clearly, this indicates that physiological differences may not be observable at the dsr gene level.
All the sequences within the family Desulfobacteraceae were related to halophilic SRB, such as Desulfotignum balticum, Desulfosarcina variabilis, and Desulfosalina propionicus, and almost all of them were detected in hypersaline lakes. Desulfobacter halotolerans, isolated from the sediments of Great Salt Lake in Utah (3), can tolerate up to 130 g/liter NaCl. We detected sequences related to this family in sediments from lakes with salinity up to 475 g/liter. The occurrence of sequences within this group is interesting, because so far, all known bacteria belonging to this family are complete oxidizers and little is known about their salinity tolerance.
Life at such extreme conditions is energetically very costly. Oren (21) described two different microbial strategies that are used by microorganisms to deal with the osmotic stress at this high salinity. One is the so-called salt-in strategy, in which the intracellular salt concentration is kept osmotically equivalent to the external one by using K+. This strategy is energetically relatively less expensive, and so far, it is known to be mostly used by organisms belonging to the orders of the aerobic halophilic archaea Halobacteriales and the anaerobic bacteria Haloanaerobiales. The second strategy is based on organic compatible solutes, such as ectoine, glycerol, and glycine-betaine, which are energetically very expensive to produce. This might be one of the reasons that restrict the presence of microorganisms with a metabolism of low energetic yield in hypersaline habitats. Until recently, it appeared that the upper salt concentration limits for SRB are ca. 250 g/liter for incomplete oxidizers and 130 g/liter for complete oxidizers (21). Assuming that all sulfate reducers belonging to the Desulfobacteraceae are complete oxidizers, the fact that we detected sequences of organisms within this family at a salt concentration of 475 g/liter might indicate either the presence of an unknown type of complete oxidizing SRB with an inorganic osmoadaptation strategy or the presence of spatial-temporal low-salt microniches.
Only one match was found between sequences retrieved from sediment (KL2_3) and enrichment (E_VFA_2_E and E_H2_2_E) samples, within the order Desulfovibrionales. Since the sediment samples investigated in this study were used as inoculum for the enrichments, we expected to find more than one match, although we cannot exclude the use of nonoptimal enrichment conditions (13, 21).
We did not detect sequences related to the gram-positive Desulfotomaculum group, as was found in another study (25). Since these SRB have been described as growing best at low salt concentrations (5, 34), they might be not present at all or only in very low numbers in the soda lakes of the Kulunda steppe in southwestern Siberia in Russia.
Unfortunately, for different logistic reasons, samples were taken in different years, i.e., in 2003, samples were taken for qPCR, for enrichments, and for PCR-DGGE, and in 2005, samples were taken for SRR and serial dilutions. However, both sampling campaigns were done in the same month (July), which is essential, since lake properties (e.g., total salt content) may vary in different seasons. Furthermore, sampling was performed in the same parts of the lakes, which were marked in 2003. The water chemistry of the lakes was highly similar for both years, and SRR measured in sediments of some of the lakes gave very similar values in both years. However, despite these similarities, we cannot exclude the possibility that there might have been changes in the microbial communities in the 2 years.
In summary, we investigated, for the first time, the SRB communities in saline and hypersaline soda lakes using a polyphasic approach. Different novel clusters were detected, including organisms able to grow at salt concentrations close to saturation, with a higher broad range of salt tolerance than thought before. The presence and vitality of such extremely halophilic SRB have been proved by SRR and enrichment analysis. Overall, our results demonstrated the existence of active SRB communities with a high diversity in soda lakes.
Acknowledgments
This work was supported by the Dutch Science Foundation for Applied Research (STW), NWO-RFBR (grant 047.011.2004.010), and by the Program on Molecular and Cell Biology of RAS.
We thank Shabir Dar for help with dsr DGGE and Ann-Charlotte Toes for help with qPCR. Marzia Miletto is thanked for helpful discussions.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 2.Braid, M. D., L. M. Daniels, and C. L. Kitts. 2003. Removal of PCR inhibitors from soil DNA by chemical flocculation. J. Microbiol. Methods 52:389-393. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, K. K., and K. Ingvorsen. 1997. Desulfobacter halotolerans sp. nov., a halotolerant acetate-oxidizing sulfate-reducing bacterium isolated from sediments of Great Salt Lake, Utah. Syst. Appl. Microbiol. 20:366-373. [Google Scholar]
- 4.Dar, S. A., L. Yao, U. van Dongen, J. G. Kuenen, and G. Muyzer. 2007. Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers. Appl. Environ. Microbiol. 73:594-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Detmers, J., H. Strauss, U. Schulte, A. Bergmann, K. Knittel, and J. Kuever. 2004. FISH shows that Desulfotomaculum spp. are the dominating sulfate-reducing bacteria in a pristine aquifer. Microb. Ecol. 47:236-242. [DOI] [PubMed] [Google Scholar]
- 6.Duckworth, A. W., W. D. Grant, B. E. Jones, and R. van Steenbergen. 1996. Phylogenetic diversity of soda lakes. FEMS Microbiol. Ecol. 19:181-191. [Google Scholar]
- 7.Foti, M., S. Ma, D. Y. Sorokin, J. L. W. Rademaker, J. Gijs Kuenen, and G. Muyzer. 2006. Genetic diversity and biogeography of haloalkaliphilic sulphur oxidizing bacteria belonging to the genus Thioalkalivibrio. FEMS Microbiol. Ecol. 55:95-101. [DOI] [PubMed] [Google Scholar]
- 8.Geets, J., B. Borremans, L. Diels, D. Springael, J. Vangronsveld, D. van der Lelie, and K. Vanbroekhoven. 2006. dsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J. Microbiol. Methods 66:194-205. [DOI] [PubMed] [Google Scholar]
- 9.Gorlenko, V. M., B. B. Namsaraev, A. V. Kulyrova, D. G. Zavarzina, and T. N. Zhilina. 1999. Activity of sulfate-reducing bacteria in bottom sediments of soda lakes of the southeastern Transbaikal region. Microbiology 68:580-585. [Google Scholar]
- 10.Grant, S., D. Y. Sorokin, W. D. Grant, B. E. Jones, and S. Heaphy. 2004. A phylogenetic analysis of Wadi el Natrun soda lake cellulase enrichment cultures and identification of cellulase genes from these cultures. Extremophiles 8:421-429. [DOI] [PubMed] [Google Scholar]
- 11.Jones, B. E., W. D. Grant, A. W. Duckworth, and G. G. Owenson. 1998. Microbial diversity of soda lakes. Extremophiles 2:191-200. [DOI] [PubMed] [Google Scholar]
- 12.Jørgensen, B. B., and F. Bak. 1991. Pathways and microbiology of thiosulfate transformation and sulfate reduction in a marine sediment (Kattegat, Denmark). Appl. Environ. Microbiol. 57:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 14.Kondo, R., D. B. Nedwell, K. J. Purdy, and S. de Queiroz Silva. 2004. Detection and enumeration of sulfate-reducing bacteria in estuarine marine sediments by competitive PCR. Geomicrobiol. J. 21:145-157. [Google Scholar]
- 15.Lein, A., N. Pimenov, C. Guillou, J.-M. Martin, C. Lancelot, I. Rusanov, S. Yusupov., Y. Miller, and M. Ivanov. 2002. Seasonal dynamics of the sulfate reduction rate on the north-western Black Sea shelf. Estuar. Coast. Shelf Sci. 54:385-401. [Google Scholar]
- 16.Li, J., K. J. Purdy, S. Takii, and H. Hayashi. 1999. Seasonal changes in ribosomal RNA of sulfate-reducing bacteria and sulfate reducing activity in freshwater lake sediment. FEMS Microbiol. Ecol. 28:31-39. [Google Scholar]
- 17.Lomans, B. P., R. Maas, R. Luderer, H. J. M. Op den Camp, A. Pol, C. van der Drift, and G. D. Vogels. 1999. Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl. Environ. Microbiol. 65:3641-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lûmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oremland, R. S., J. F. Stolz, and J. T. Hollibaugh. 2004. Microbial arsenic cycle in Mono Lake, California. FEMS Microbiol. Ecol. 48:15-27. [DOI] [PubMed] [Google Scholar]
- 21.Oren, A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pikuta, E. V., T. N. Zhilina, G. A. Zavarzin, N. A. Kostrikina, G. A. Osipov, and F. A. Rainey. 1998. Desulfonatronum lacustre gen. nov., sp. nov.: a new alkaliphilic sulfate-reducing bacterium utilizing ethanol. Microbiology 67:105-113. [Google Scholar]
- 23.Pikuta, E. V., R. B. Hoover, A. K. Bej, D. Marsic, W. B. Whitman, D. Cleland, and P. Krader. 2003. Desulfonatronum thiodismutans sp. nov., a novel alkaliphilic, sulfate-reducing bacterium capable of lithoautotrophic growth. Int. J. Syst. Evol. Microbiol. 53:1327-1332. [DOI] [PubMed] [Google Scholar]
- 24.Pimenov, N. V., and E. A. Bonch-Osmoloyskaya. 2006. Activity studies in thermal environments. Methods Microbiol. 35:29-53. [Google Scholar]
- 25.Scholten, J. C. M., S. B. Joye, J. T. Hollibaugh, and J. C. Murrell. 2005. Molecular analysis of the sulfate reducing and archaeal community in a meromictic soda lake (Mono Lake, California) by targeting 16S rRNA, mcrA, apsA, and dsrAB genes. Microb. Ecol. 50:29-39. [DOI] [PubMed] [Google Scholar]
- 26.Scholten, J. C. M., and A. J. M. Stams. 1995. The effect of sulfate and nitrate on methane formation in a fresh water sediment. Antonie Leeuwenhoek 68:309-315. [DOI] [PubMed] [Google Scholar]
- 27.Smith, R. L., and R. S. Oremland. 1987. Big Soda Lake (Nevada). Pelagic sulfate reduction. Limnol. Oceanogr. 32:794-803. [Google Scholar]
- 28.Sorokin, D. Y., V. M. Gorlenko, B. B. Namsaraev, Z. B. Namsaraev, A. M. Lysenko, B. T. Eshinimaev, V. N. Khmelenina, Y. A. Trotsenko, and J. G. Kuenen. 2004. Prokaryotic communities of the north-eastern Mongolian soda lakes. Hydrobiologia 522:235-248. [Google Scholar]
- 29.Sorokin, D. Y., and J. G. Kuenen. 2005. Haloalkaliphilic sulfur-oxidizing bacteria in soda lakes. FEMS Microbiol. Rev. 29:685-702. [DOI] [PubMed] [Google Scholar]
- 30.Sorokin, D. Y., H. Banciu, L. A. Robertson, and J. G. Kuenen. 2006. Haloalkaliphilic sulfur-oxidizing bacteria, p. 969-984. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, vol. 2. Ecophysiology and biochemistry. Springer, New York, NY. [Google Scholar]
- 31.Stams, A. J. M., J. B. van Dijk, C. Dijkema, and C. M. Plugge. 1993. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl. Environ. Microbiol. 59:1114-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trüper, H. G., and H. G. Schlegel. 1964. Sulphur metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie Leeuwenhoek 30:225-238. [DOI] [PubMed] [Google Scholar]
- 33.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseay, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widdel, F. 2006. The genus Desulfutomaculum, p. 787-794. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, vol. 4. Firmicutes. Springer, New York, NY. [Google Scholar]
- 35.Zavarzin, G. A., T. N. Zhilina, and V. V. Kevbrin. 1999. The alkaliphilic microbial community and its functional diversity. Microbiology 68:503-521. [Google Scholar]
- 36.Zhang, T., and H. P. Fang. 2006. Application of real-time polymerase chain reaction for quantification of microorganisms in environmental samples. Appl. Microbiol. Biotechnol. 70:281-289. [DOI] [PubMed] [Google Scholar]
- 37.Zhilina, T. N., G. A Zavarzin, F. A. Rainey, E. N. Pikuta, G. A. Osipov, and N. A. Kostrikina. 1997. Desulfonatronovibrio hydrogenovorans gen. nov., sp. nov., an alkaliphilic sulfate-reducing bacterium. Int. J. Syst. Bacteriol. 47:144-149. [DOI] [PubMed] [Google Scholar]
- 38.Zhilina, T. N., D. G. Zavarzina, J. Kuever, A. M. Lysenko, and G. A. Zavarzin. 2005. Desulfonatronum cooperativum sp. nov., a novel hydrogenotrophic, alkaliphilic, sulfate-reducing bacterium, from a syntrophic culture growing on acetate. Int. J. Syst. Evol. Microbiol. 55:1001-1006. [DOI] [PubMed] [Google Scholar]