Abstract
Escherichia coli serotype O157:H7 was detected among bacteria collected from the Ganges River. O157:H7 isolates tested positive for stx1, stx2, and eae gene sequences. Identification of potentially pathogenic isolates from extensively used source water indicates that O157:H7 may be a significant but as yet underacknowledged public health concern in India.
Escherichia coli serotype O157:H7 is an important pathogen of humans, causing hemorrhagic colitis and hemolytic-uremic syndrome (HUS) (15). Disease progression is characterized by bloody diarrhea and can lead to HUS with a high associated mortality in susceptible individuals, including the elderly and very young. O157:H7 isolates are grouped with other Shiga toxin-producing E. coli (STEC) isolates in recognition of the major virulence factors responsible for pathogenicity (16). O157:H7 isolates are also grouped with enterohemorrhagic E. coli, with enterohemorrhagic E. coli considered to be a subset of STEC (5). Other virulence factors, such as intimin, encoded by the eae gene, are also expressed by O157:H7.
O157:H7 was first associated with disease outbreaks in the United States in 1982 (19). The virulence properties and genetic diversity of O157:H7 isolates have been widely studied in the United States and other developed countries (8). Far less is known about O157 prevalence in developing countries, where diarrheal disease and associated mortality are much more pervasive. The first major outbreak of bloody diarrhea in the developing world associated with O157 occurred in Swaziland in 1992 (6). O157 infection may have accounted for tens of thousands of cases during this epidemic.
In India, the status of STEC and O157 prevalence and contribution to disease is uncertain (22). In 2002, researchers in Calcutta, India, reported finding non-O157 STEC isolates in 1.4% of stool samples from humans suffering from bloody diarrhea (12, 13). They concluded that STEC was not an important cause of diarrhea in India.
Study in Varanasi, India.
The Swatcha Ganga Research Laboratory (SGRL) has monitored Ganges River water quality in Varanasi, India, since 1993. Data collected between 1993 and 2004 demonstrate the seriously polluted nature of the Ganges in Varanasi caused by release of raw sewage into the river (11). In the most polluted part of the river, the average biological oxygen demand (BOD) level exceeds 40 mg/ml and the average fecal coliform count (FCC) is greater than 107 CFU per 100 ml. Residents who live near the Ganges suffer from a high incidence of waterborne diseases, including cholera and dysentery (11). Risk factors for disease include poor sanitation and regular use of the river for personal hygiene, laundry, and utensil washing.
While conducting our health survey in 2004, river samples were collected from five sites (Fig. 1). BOD and FCC were measured by the SGRL by following standard procedures (1). Samples were also processed as follows. Water samples were filtered under vacuum through a Whatman no. 1 prefilter layered on top of a 0.45-μm-pore-size membrane filter, both of 47 mm in diameter (Whatman Corp., Florham Park, NJ). Samples were also filtered through 25-mm-diameter (0.2-μm-pore-size) polycarbonate membranes (Millipore, Billerica, MA). Membranes were sealed in plastic bags, packaged, and shipped to the microbiology lab at Montana State University (MSU). Import permits were obtained from the Centers for Disease Control and Prevention, Atlanta, GA.
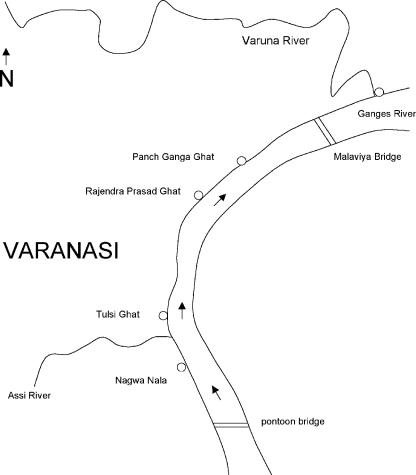
FIG. 1.
Map of the Ganges River in Varanasi, showing the locations of the water sampling sites of Nagwa Nala, Tulsi Ghat, Rajendra Prasad Ghat, Panch Ganga Ghat, and the Varuna River's confluence with the Ganges. The distance between Nagwa Nala and the Varuna-Ganges confluence is about 7 km.
Screening for O157:H7.
Despite a high incidence of waterborne disease among Varanasi residents living near the Ganges, it is unlikely that specific diagnoses of STEC morbidity and mortality would be reported, particularly in poorer neighborhoods. The high incidence of dysentery provided a rationale for testing river water for the presence of O157:H7. In the MSU lab, the 25-mm polycarbonate membranes were stained with fluorescein-labeled, goat anti-E. coli O157:H7 antibody (Kirkegaard & Perry, Gaithersburg, MD) (17) and mounted on slides. Stained cells were counted using a Zeiss Axioskop epifluorescence microscope. Samples from all five sampling locations were positive for anti-O157:H7 antibody-reactive bacteria (Table 1). An estimate of over 103 cells (presumed to be O157:H7) per ml of river water at each site suggested the presence of the bacteria in high numbers throughout the Ganges in Varanasi. We noted that this estimate of the O157:H7 cell number from direct cell counts was in excess of the corresponding FCC. Bacteria immobilized on the polycarbonate filters in the Varanasi lab were likely to be a collection of both culturable and nonculturable cells. It was also possible that the antibody-reactive cells included bacteria of other species sharing O157:H7 epitopes that might cross-react with the antibody. Accordingly, we resuscitated cells for further characterization to confirm the presence of E. coli O157:H7 in the Ganges.
TABLE 1.
Direct cell counts for bacterial staining with anti-O157:H7 fluorescent antibody, compared with FCC and BODa
| Site | O157:H7 direct cell count estimate (cells/ml) | FCCb (CFU/100 ml) | BODb (mg/ml) |
|---|---|---|---|
| Nagwa Nala | 4.21 × 103 | NT | NT |
| Tulsi Ghat | 1.80 × 103 | 4.5 × 104 | 6.9 |
| Rajendra Prasad Ghat | 2.61 × 103 | 3.7 × 104 | 7.0 |
| Panch Ganga Ghat | 1.50 × 103 | 2.3 × 104 | 2.9 |
| Varuna confluence | 3.91 × 103 | 2.0 × 107 | 53.0 |
Measured at the SGRL in Varanasi.
Results of testing water samples collected 26 February 2004 at Ganges River sites. NT, not tested.
Recovery and testing of culturable bacteria.
Each 47-mm membrane was cut into eight pieces. Enrichment for pathogenic E. coli followed methods described in the FDA Bacteriological Analytical Manual Online (9). Membrane pieces were vortexed in brain heart infusion (Difco, Detroit, MI) to release bacteria into the medium for incubation to resuscitate cells. Selective growth for coliform cells included incubation using double-strength tryptone phosphate broth and growth on MacConkey agar (Difco).
After growth on MacConkey agar, subcultures from 150 colonies presumed to be coliform bacteria were transferred to nutrient agar plates and incubated overnight. Colonies containing only gram-negative rods were screened by growth on CHROMagar O157 chromogenic medium (CHROMagar, Paris, France). This medium has been found to be highly sensitive for identifying strains of E. coli O157 and to a lesser degree E. coli O111 (2). Based on color, colonies presumed to be O157 were transferred from CHROMagar O157 to fresh nutrient agar plates, incubated, and analyzed using both API 20E (bioMerieux, Hazelwood, MO) and BBL crystal enteric/nonfermenter (Becton Dickinson, Sparks, MD) identification kits. Bacteria from these isolates were also immobilized on polycarbonate filters and tested with anti-O157:H7 fluorescein-labeled antibody.
Testing with the biochemical identification kits indicated that these isolates were E. coli. Antibody screening indicated that four isolates were of E. coli serotype O157:H7. The manufacturer claims that the anti-O157:H7 antibody is highly specific for E. coli O157:H7 and provided data showing the antibody to be nonreactive or only weakly reactive with other species that might possess O157 epitopes (H. M. M. Webster, personal communication).
DNA was prepared from O157:H7 isolates by use of a FastDNA SPIN kit for soil (Qbiogene, Solon, OH) and tested for the presence of Shiga toxin (stx1 and stx2) and eae gene sequences by use of PCR protocols (4, 24). A TaKaRa LA Taq polymerase kit (TaKaRa Bio, Otsu, Japan) was used. PCR product sizes were analyzed using agarose gel electrophoresis and ethidium bromide staining. Three isolates tested positive for stx1, stx2, and eae sequences (Table 2) . While screening for virulence genes is not a definitive test of pathogenicity, the combined presence of stx2 and eae in STEC has been identified as a risk factor for HUS (7).
TABLE 2.
Characterization of O157:H7 isolates from the Ganges River, Varanasi
| Isolate | Sampling site | Resulta of:
|
|||
|---|---|---|---|---|---|
| PCR for stx1 | PCR for stx2 | PCR for eae | Sorbitol fermentation test | ||
| Referenceb | NAc | + | + | + | − |
| No. 1 | Nagwa Nala | + | + | + | + |
| No. 2 | Nagwa Nala | − | − | − | + |
| No. 3 | Tulsi Ghat | + | + | + | + |
| No. 4 | Varuna River | + | + | + | + |
Profiles for O157:H7 strains were determined using PCR analysis with an array of virulence gene primers. +, positive; −, negative; stx1, Shiga toxin 1 gene; stx2, Shiga toxin 2 gene; eae, enterocyte attachment and effacement (intimin) gene. The ability to ferment sorbitol was tested using API 20E and BBL crystal enteric/nonfermenter identification kits.
The O157:H7 reference strain is from the E. coli Reference Center, Penn State University (identification no. 3A-3299-85).
NA, not applicable.
Two of our E. coli O157:H7 isolates were from the Nagwa Nala confluence with the Ganges, the southernmost site routinely monitored by the SGRL. Given that one isolate lacked the stx and eae genes, these two Nagwa Nala isolates are unlikely to be clonally related. A third isolate was from Tulsi Ghat, only a few hundred meters upstream of a pump station that collects river water for the city's main water treatment plant. The fourth isolate was from the Varuna River's confluence with the Ganges, about 7 km north of Nagwa Nala.
Sorbitol phenotype.
In the 1980s, O157:H7 isolates were originally described as being sorbitol negative. This led to use of selective media, such as sorbitol MacConkey agar (SMAC), to identify sorbitol-negative strains. Since then, sorbitol-positive strains of O157:H7 have been isolated frequently (3). Sorbitol utilization is unrelated to pathogenicity of O157:H7 (10).
In contrast to the sorbitol-negative O157:H7 control, the four Ganges isolates were all sorbitol positive (Table 2). This finding supports the need to consider selection methods for O157:H7 that are independent of the sorbitol phenotype. In conducting an Internet literature search, we uncovered only one report of an attempt to identify O157:H7 in water sources in India. Sharma et al. (20) tested wastewater as well as surface water, groundwater, and drinking water samples in Delhi and reported no detection of O157:H7. Their methods relied on screening for sorbitol-negative bacteria using SMAC. A Government of India website accessed in January 2007 (14) recommends that laboratories test for O157:H7 by using SMAC. Based on our O157:H7 isolates being sorbitol positive, attempts to detect O157:H7 using SMAC may underestimate the prevalence of these bacteria in India.
Public health significance.
The government of Uttar Pradesh, India, estimates that 309 million liters of sewage is generated daily in Varanasi (21). Due to frequent power outages and shortcomings of sewage collection and treatment, most of the city's sewage is released untreated into the Ganges River. Detection of potentially pathogenic O157:H7 bacteria in the river is alarming. Visiting pilgrims routinely use the river for religious bathing. Many poorer residents of Varanasi use the river daily for bathing and washing laundry and as a source of water for cooking. These people may be at great risk for contracting O157:H7 infections.
Outbreaks of O157:H7 infection involving recreational water use have been documented in the United States (18). An infectious dose of O157:H7 bacteria is estimated to be quite low, in the range of 10 to 100 cells (9, 16). Given the presence of O157:H7 bacteria in the Ganges River, screening of patients presenting with bloody diarrhea at hospitals and clinics would be warranted to determine whether O157:H7 is associated with disease in Varanasi. Clinical screening for O157:H7 may not be considered feasible in a resource-limited, developing country such as India. However, if O157:H7 is a significant cause of disease in India, this has important implications for case management of severe diarrheal disease, given the controversial nature of treating O157:H7 infections and the risk of adverse sequelae (23).
Acknowledgments
This publication was made possible by a travel grant award from the NSF EPSCoR program of MSU and by NIH grant no. P20 RR-16455-03/5 from the INBRE-BRIN program of the National Center for Research Resources.
This publication's contents are solely the responsibility of the authors and do not necessarily represent the official views of the NSF or NIH.
The SGRL of the Sankat Mochan Foundation is operated with funding from Oz GREEN, Australia.
Steve Hamner thanks members of the Sankat Mochan Foundation and the San Francisco-based Friends of the Ganges for their encouragement and support for completion of this work. Steve Hamner dedicates the publication of this study to Kirti Tsenshab Rinpoche.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.American Public Health Association. 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, DC.
- 2.Bettelheim, K. A. 1998. Reliability of CHROMagar O157 for the detection of enterohaemorrhagic Escherichia coli (EHEC) O157 but not EHEC belonging to other serogroups. J. Appl. Microbiol. 85:425-428. [DOI] [PubMed] [Google Scholar]
- 3.Bopp, C. A., F. W. Brenner, J. G. Wells, and N. A. Stockbine. 1999. Escherichia, Shigella, and Salmonella, p. 459-474. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, DC.
- 4.Chakraborty, S., J. S. Deokule, P. Garg, S. K. Bhattacharya, R. K. Nandy, G. B. Nair, S. Yamasaki, Y. Takeda, and T. Ramamurthy. 2001. Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J. Clin. Microbiol. 39:3241-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Investig. 107:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Effler, P., M. Isaacson, L. Arntzen, R. Heenan, P. Canter, T. Barrett, L. Lee, C. Mambo, W. Levine, A. Zaidi, and P. M. Griffin. 2001. Factors contributing to the emergence of Escherichia coli O157 in Africa. Emerg. Infect. Dis. 7:812-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ethelberg, S., K. E. P. Olsen, F. Scheutz, C. Jensen, P. Schiellerup, J. Engberg, A. M. Petersen, B. Olesen, P. Gerner-Smidt, and K. Molbak. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 10:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, P. 1995. Escherichia coli serotype O157:H7: novel vehicles of infection and emergence of phenotypic variants. Emerg. Infect. Dis. 1:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng, P., and S. D. Weagant. 2002. Diarrheagenic Escherichia coli. U.S. FDA bacteriological analytical manual online 2002. http://www.cfsan.fda.gov/∼ebam/bam-4a.html.
- 10.Fratamico, P. M., R. L. Buchanan, and P. H. Cooke. 1993. Virulence of an Escherichia coli O157:H7 sorbitol-positive mutant. Appl. Environ. Microbiol. 59:4245-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamner, S., A. Tripathi, R. K. Mishra, N. Bouskill, S. C. Broadaway, B. H. Pyle, and T. E. Ford. 2006. The role of water use patterns and sewage pollution in incidence of water-borne/enteric diseases along the Ganges River in Varanasi, India. Int. J. Environ. Health Res. 16:113-132. [DOI] [PubMed] [Google Scholar]
- 12.Khan, A., S. C. Das, T. Ramamurthy, A. Sikdar, J. Khanam, S. Yamasaki, Y. Takeda, and G. B. Nair. 2002. Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources in Calcutta, India. J. Clin. Microbiol. 40:2009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan, A., S. Yamasaki, T. Sato, T. Ramamurthy, A. Pal, S. Datta, N. R. Chowdhury, S. C. Das, A. Sikdar, T. Tsukamoto, S. K. Bhattacharya, Y. Takeda, and G. B. Nair. 2002. Prevalence and genetic profiling of virulence determinants of non-O157 Shiga toxin-producing Escherichia coli isolated from cattle, beef, and humans, Calcutta, India. Emerg. Infect. Dis. 8:54-62. [PubMed] [Google Scholar]
- 14.Ministry of Health and Family Welfare. 2006. Food Safety India: Escherichia coli O157:H7. http://foodsafetyindia.nic.in/ecolifaq.htm.
- 15.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyle, B. H., S. C. Broadaway, and G. A. McFeters. 1999. Sensitive detection of Escherichia coli O157:H7 in food and water by immunomagnetic separation and solid-phase laser cytometry. Appl. Environ. Microbiol. 65:1966-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 20.Sharma, S., I. Singh, and J. S. Virdi. 2003. Microbial contamination of various water sources in Delhi. Curr. Sci. 84:1398-1399. [Google Scholar]
- 21.Uttar Pradesh Jal Nigam. 2006. Ganga action plan (phase II) at Varanasi. http://www.upjn.org/pollution121.htm.
- 22.Wani, S. A., F. Pandit, I. Samanta, M. A. Bhat, and A. S. Buchh. 2004. Molecular epidemiology of Shiga-toxin producing Escherichia coli in India. Curr. Sci. 87:1345-1353. [Google Scholar]
- 23.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. L. Tar. 2000. The risk of hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu, J., and J. B. Kaper. 1992. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 6:411-417. [DOI] [PubMed] [Google Scholar]