Abstract
The contamination of polar regions due to the global distribution of anthropogenic pollutants is of great concern because it leads to the bioaccumulation of toxic substances, methylmercury among them, in Arctic food chains. Here we present the first evidence that microbes in the high Arctic possess and express diverse merA genes, which specify the reduction of ionic mercury [Hg(II)] to the volatile elemental form [Hg(0)]. The sampled microbial biomass, collected from microbial mats in a coastal lagoon and from the surface of marine macroalgae, was comprised of bacteria that were most closely related to psychrophiles that had previously been described in polar environments. We used a kinetic redox model, taking into consideration photoredox reactions as well as mer-mediated reduction, to assess if the potential for Hg(II) reduction by Arctic microbes can affect the toxicity and environmental mobility of mercury in the high Arctic. Results suggested that mer-mediated Hg(II) reduction could account for most of the Hg(0) that is produced in high Arctic waters. At the surface, with only 5% metabolically active cells, up to 68% of the mercury pool was resolved by the model as biogenic Hg(0). At a greater depth, because of incident light attenuation, the significance of photoredox transformations declined and merA-mediated activity could account for up to 90% of Hg(0) production. These findings highlight the importance of microbial redox transformations in the biogeochemical cycling, and thus the toxicity and mobility, of mercury in polar regions.
The contamination of polar regions by pollutants that are formed at lower latitudes and are subject to global transport is an important issue. In temperate zones, microbial activities impact the toxicity and mobility of these environmental contaminants (8, 37), but their role in the transformations of pollutants at high latitudes remains largely unexplored. Research to date has focused on the biodegradation of petroleum products (20, 45), with little attention to the importance of polar microbes in the degradation of other types of organic contaminants (39, 62) or the transformations of metals. This lack of information may become critical since high-latitude ecosystems are currently undergoing major alteration due to environmental changes, which in turn may greatly affect the cycling of contaminants (38).
A growing body of literature documents increases in mercury contamination of the Arctic food chain (42) and increased levels of mercury exposure in indigenous populations (59). The accumulation of mercury in Arctic biota, as in temperate zones, is mostly in the form of the potent neurotoxic substance methylmercury (16). Because mercury enters the Arctic biosphere in its inorganic form, bioaccumulation depends on in situ synthesis of methylmercury and its subsequent uptake by the microbes and phytoplankton that occupy the base of the Arctic food web. Hence, processes that either directly or indirectly affect methylmercury production modulate the impact of mercury contamination in the Arctic. The source of this mercury is atmospheric deposition. Modeling studies have estimated that 325 tons of mercury are deposited throughout the Arctic over a 1-year cycle (3), with much of it occurring during polar sunrise (24, 36, 55), leading in some cases to 1,000-fold increases in mercury levels relative to that of background concentration (23). Mercury deposition is thought to be due to the oxidation of atmospheric elemental mercury, Hg(0), the form in which mercury is globally distributed, by reactive halogen radicals from sea salt aerosols (4, 36). Thus, marine and coastal environments in polar regions are more susceptible to mercury deposition than inland areas, and there is a need to better understand the postdepositional fate of mercury in these environments.
Along with photochemical processes (2, 56), the bacterial mercuric reductase enzyme (MerA) affects mercury mobility and bioavailability by converting water-soluble inorganic mercury and methylmercury to the volatile elemental form. This is a detoxification process as evidenced by the resumption of microbial growth after the removal of the gaseous form of Hg(0) (8). Mercury resistance is widespread among microorganisms from diverse environments (44), including extreme environments such as hydrothermal vents (60) and permafrost (41). In temperate regions, microbial reduction of mercury affects the production of dissolved gaseous mercury (DGM) in pristine and in contaminated aquatic systems (6, 7, 9, 57) as well as the degradation of methylmercury in contaminated aquatic systems (52). To date, the role of mercury-resistant microbes in redox cycling of mercury in polar regions has not been examined. Here, we report that merA genes were present and expressed by microbes from remote polar areas. Furthermore, we used a kinetic redox model to assess the importance of microbial mercury reduction in the production of DGM and, thus, its role in mercury biogeochemistry in Arctic coastal waters.
MATERIALS AND METHODS
Sampling sites and sample collection.
Samples were taken from the western shoreline of Cornwallis Island, Nunavut, Canada (75°N, 95°W), during the summer of 2005. Two different types of samples were collected: (i) water and macroalgae, identified as Fucus sp. and Desmarestia sp., present in seawater in gaps between melting sea ice; and (ii) thick photosynthetic microbial mats from coastal lagoons on the seashore, which are fed daily by tides. In both cases, solid (biomass) and liquid (water) samples were collected and immediately frozen until further processing of the biomass. All containers used for sampling were cleaned and rinsed following trace metal protocols and were sterile. Nonpowdered gloves were worn at all times during sampling and further handling of the samples.
Mercury analysis.
Total mercury (THg) concentrations in water were quantified using U.S. EPA method 1631 and an automated mercury fluorescence detector (model 2600; Tekran). DGM in water was measured following the protocol described in reference 48. THg concentration in the sampled biomass was measured by thermal decomposition at 750°C using a direct mercury analyzer (DMA 80; Milestone, MLS). Briefly, prior to analyses, samples were freeze-dried. Prior to combustion, 0.05-g samples were further dried in an oxygen stream passing through a quartz tube located inside a controlled heating coil. The combustion gases were further decomposed on a catalytic column at 750°C. Mercury vapor was collected on a gold amalgamation trap and subsequently desorbed for quantification by atomic absorption spectrometry at 254 nm. The working detection limit of this method was previously calculated to be 0.01 ng of mercury or three times the standard deviation of 10 procedural blanks.
Modeling of mercury redox transformations in coastal Arctic waters.
The relative importance of Hg(0) production by photochemical or biological reactions as well as its destruction by photochemical reactions was modeled using ACUCHEM modeling software (12) and a custom-designed kinetic code. Model parameters are presented in Table 1. We performed sensitivity studies at the initial concentration ranges of THg and DGM as measured in the field in the Cornwallis Island area. We also performed sensitivity studies of the numbers of metabolically active bacteria capable of mercury reduction. The model runs were performed over a period of 10 days to ensure that the concentrations of mercury species, i.e., Hg(II), Hg(0) from the photoreduction of Hg(II), and Hg(0) from the bioreduction of Hg(II), became independent of the initial concentrations used. Additional, shorter-run models were performed and they did not alter the general conclusions.
TABLE 1.
Parameters used in simulations of mercury redox state in high Arctic seawater
| Model parameters | Valuea | Reference(s) |
|---|---|---|
| Bacterial cell count (cells ml−1) at a depth ofa: | ||
| Surface | 9.49 × 105 ± 1.75 × 105 | This study |
| 5 m | 5.53 × 105 ± 1.15 × 105 | |
| 10 m | 4.93 × 105 ± 1.7 × 104 | |
| MerA-mediated reduction rates [nmol Hg(II) min−1 mg protein−1] | 10 | 46 |
| Protein content of bacteria (mg protein · cell−1) | 24 | 65 |
| Correction factor for cells containing merA in a given environment | 1% | 35 |
| Correction factor for the effect of cold temperatures on bacterial activity | 1% | 49 |
| Photoreduction rate (h−1) | 0.43-1.58b | This study; 1 |
| Photooxidation rate (h−1) | 0.1-1.4b | 34, 63 |
| UVA attenuation (m−1) | 0.24-4.02c | 10, 25 |
| UVB attenuation (m−1) | 0.05-2.7c | |
| Total mercury (ng · liter−1) | 0.5 | This study |
| Elemental mercury (ng · liter−1) | 0.03c | This study; 1, 19 |
Sensitivity studies were performed on 100%, 50%, 10%, 5%, and 1% cell viability.
The values of 0.5 and 0.6 h−1 were used in the base run. Additional sensitivity studies were performed for lower and upper estimations.
Mean values were employed in the base run.
(i) Biological reduction.
Biological mercury reduction in Arctic seawater was estimated using a merA-mediated reduction rate of 10 nmol Hg(II) min−1 mg protein−1 (±10%) (46), corrected for the protein content of bacterial cells (65) and calculated at the surface and at depths of 5 and 10 m. To take into consideration the harsh arctic conditions, several scenarios were tested. Based on observations indicating that 0.5 to 5% of cells in melted sea ice respire (30), we considered that only 1 and 5% of the cells were active at the surface and 10% at 5- and 10-m depths. Since the reported Hg(II) reduction rate was obtained at an optimal growth temperature (46), we reduced this rate by 99% to account for the effect of temperature on microbial activities as observed for many reactions in cold environments (49). Finally, not all cells in a given environment carry mer operon genes. Using direct cell counts, Liebert and Barkay (35) found that between 1 and 10% of all cells in temperate coastal or marine environments were mercury resistant; to be conservative, we estimated that only 1% of the viable cells expressed mercury resistance in our system.
(ii) Photochemical reactions.
Photochemical rates of reduction and oxidation were based on previously published data for temperate coastal marine systems and ranged from 0.1 h−1 to 1.4 h−1 for photooxidation (33, 34, 63) and from 0.4 h−1 to 1.58 h−1 for photoreduction (1, 63). UV energy attenuation was calculated using an empirical model based on dissolved organic carbon concentrations used for Arctic ocean waters (25). Dissolved organic carbon concentrations for Arctic water ranged from 43 to 225 μmol liter−1, as observed for the central Arctic Ocean (10).
DNA extraction and PCR amplification of merA sequences.
Total DNA was extracted from biomass samples and from the concentrated water wash of these biomass samples, using a PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) according to the manufacturer's guidelines. The water used for the wash was collected with the solid samples at the sampling sites. Biomass in the washed suspensions was concentrated by centrifugation (10,000 rpm for 10 min at 4°C) prior to DNA extraction as described above. The presence of merA genes in DNA extracts was detected using a nested PCR approach using primers specific for merA genes, among all gram-negative bacteria (43). First, a 1,250-bp fragment was amplified using the forward primer A2-n.F (5′-CCA TCG GCG GCW CYT GCG TSA A-3′) and the reverse primer A5-n.R (5′-ACC ATC GTC AGR TAR GGR AAVA-3′) and the following reaction conditions: final volume of 15 to 50 μl containing 1× buffer, 1.5 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate (dNTP), 0.4 μM of each forward and reverse primer, and 1 U Taq polymerase (Fisher). Amplification conditions were as follows: 45 cycles of 10 s at 94°C, 30 to 60 s at 54°C, and 30 s at 72°C. The products of these reactions were separated by electrophoresis on a 1% agarose gel. Numerous bands were observed for most Arctic biomass samples. Amplicons of the predicted size (1,250 bp) were excised from the gel and purified using a QIAGEN DNA gel extraction kit (QIAGEN, Valencia, CA) and used as templates for a second PCR, which amplified a 291-bp fragment internal to the 1,250-bp template. PCR conditions were as described above, with the following changes: a concentration of 0.8 μM was used for both forward primer A7s-n.F (5′-CGA TCC GCA AGT GGC IAC BGT-3′) and reverse primer A5-n.R (5′-ACC ATC GTC AGR TAR GGR AAVA-3′). Attempts to directly amplify the 291-bp fragment from Arctic biomass DNA extracts were unsuccessful.
In order to assess the diversity of microbes present, the 16S rRNA gene was amplified using the forward primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and the reverse primer 907R (5′-CCGTCAATTCATTTGAG-3′) in 25-μl reaction mixtures containing 1× buffer, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.4 μM of each primer, and 1 U Taq polymerase (Fisher). Amplification conditions were as follows: 5 min at 95°C, followed by 35 cycles of 10 s at 94°C, 30 s at 55°C, and 90 s at 72°C, followed by a final extension of 12 min at 72°C.
RNA extraction and cDNA synthesis.
Total RNA was extracted from microbial biomass using an UltraClean microbial RNA kit (MoBio, Carlsbad, CA), following the manufacturer's guidelines. To eliminate contaminating DNA, extracts were treated with RQ1 RNase-free DNase (Promega, Madison, WI), according to the manufacturer's instructions. Reverse transcription was carried out using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions, and random hexamers as primers. cDNA was then used as the template in PCR to detect merA transcripts as described above.
Cloning and sequencing of merA and 16S rRNA gene PCR products.
PCR products used for cloning were separated by gel electrophoresis (2% agarose). The 291-bp merA amplicon was extracted and purified from the gel, as described above, and cloned into the vector pCR4-TOPO, using a TOPO-TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Four libraries were created: (i) sea ice merA genes (SId), (ii) sea ice merA transcripts (SIr), (iii) lagoon merA genes (LGd), and (iv) lagoon merA transcripts (LGr). Plasmid DNA was isolated from 96 clones from each library using a PureLink96 plasmid kit (Invitrogen) and the plasmids screened for inserts of the correct size by restriction digestion with PvuII or by amplification of the cloned region following the PCR protocol described above. Clones containing the 291-bp insert were sequenced using the M13r primer and ABI dye terminator chemistry (BigDye, v.3.1; Applied Biosystems, Foster City, CA) on an ABI 3100 genetic analyzer. The same protocol was used to clone 16S rRNA gene PCR products from sea ice and lagoon DNA.
ARDRA.
For amplified ribosomal DNA restriction analysis (ARDRA), 50 randomly picked colonies per 16S rRNA gene library were each grown overnight at 37°C in LB containing antibiotics. Plasmid DNA extraction was performed using a PureLink 96-well kit (Invitrogen) and the insert amplified using primers 27F and 907R. Ten microliters of each amplicon was digested for 3 h at 37°C with 2.5 U of HaeIII and 2.5 U of EcoRI in 20-μl reaction mixtures. Restriction patterns were separated on 3% NuSieve agarose gels and stained with SYBR green I DNA stain. Each different restriction pattern was defined as an operational taxonomic unit (OTU). The distribution of OTUs in each library was determined. Two representatives of each OTU were sequenced using primers M13f and M13r and compared to those in the GenBank database, using BlastN. The efficiency of our sampling effort was assessed using EstimateS (version 7.5; http://purl.oclc.org/estimates). Sample-based rarefaction curves and richness estimations and the coverage percentage of clone libraries were computed.
merA clone library diversity analysis.
A distance matrix of aligned merA sequences was generated with model F84 for nucleotide substitution using the DNADIST program in PHYLIP (version 3.6; Department of Genomic Sciences, University of Washington, Seattle, WA; http://evolution.genetics.washington.edu/phylip.html). This distance matrix was then used to calculate sample coverage and richness estimates with DOTUR (54). Phylotypes were assigned using the furthest-neighbor-clustering algorithms. merA phylotypes were assigned based on a nucleotide sequence identity of 97%. Several tests were then applied to the merA clone libraries to estimate sampling efficiency by rarefaction analysis (29) and to calculate nonparametric richness estimates with abundance-based coverage estimator (ACE) (18) and Chao1 (17) as a function of sampling effort from rarefaction curves following 1,000 randomizations.
Phylogenetic analysis.
The merA nucleotide sequences were trimmed and assembled using SeqMan (DNAStar, Madison, WI). DNA sequences were then translated into amino acids and aligned with reference MerA sequences using the ClustalW function in MegAlign (DNAStar). The alignment was exported to MEGA 3.1, and a neighbor-joining tree was built with 1,000 bootstrap replicates. 16S rRNA nucleotide sequences were trimmed and assembled in a similar manner and the nearest relatives to each OTU determined using BlastN.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study were deposited in the GenBank database with the following accession numbers: DQ408728 to DQ408744 and EF379214 to EF379240.
RESULTS
Amplification and cloning of merA from Arctic DNA and RNA samples.
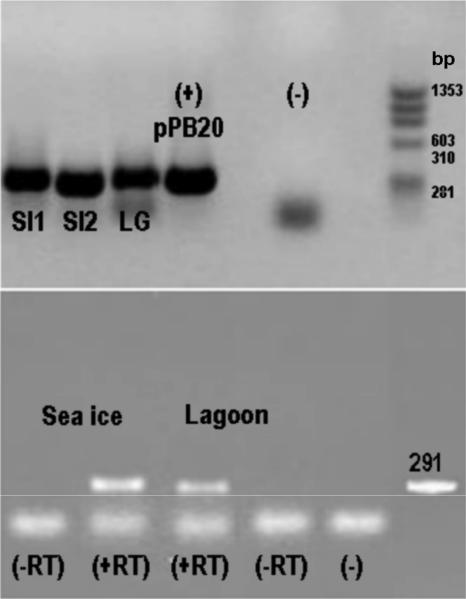
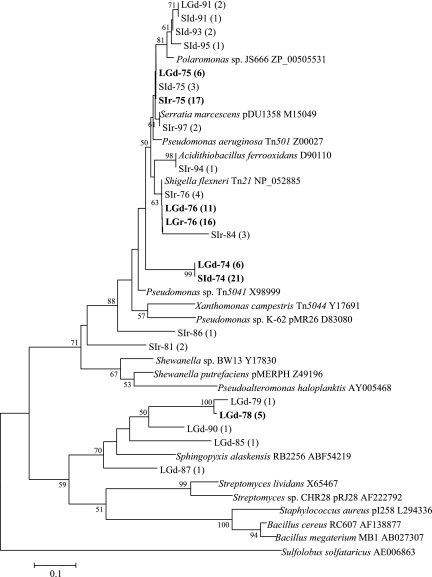
Previously described primers for merA (43) were used to amplify the merA gene from DNA and RNA extracts of microbial biomass samples collected from a benthic biofilm in a coastal lagoon and from a biofilm associated with macroalgae that were retrieved from seawater between blocks of coastal sea ice (Fig. 1). Four merA clone libraries were constructed using the PCR products of the lagoon and sea ice communities. Seventeen unique MerA phylotypes were obtained which encompassed the known diversity of MerA among bacteria (Fig. 2). Over 29% of these phylotypes, all from the lagoon gene library, were related to a putative locus in Sphingopyxis alaskensis, a psychrophilic bacterium that had been isolated from Alaskan waters. These MerA phylotypes formed a distinct cluster most closely related to the MerA phylotypes from gram-positive bacteria (Fig. 2). A further 18% of the phylotypes observed clustered with a putative MerA from the genome sequence of Polaromonas sp. strain JS666, an organism that is closely related to the Antarctic bacterium Polaromonas vacuolata. The remaining merA phylotypes, which included the majority of the clones, were closely related to the genetically and biochemically characterized MerA phylotypes from Tn21, Tn501, and pDU1358 (Fig. 2).
FIG. 1.
merA genes and transcripts in microbial biomass. (Top) Gel shows 291-bp merA PCR products obtained with sea ice (SI) and lagoon (LG) DNA extracts as templates. SI1 and LG, DNA obtained by extracting biomass with its associated microbes; SI2, extract of a fraction obtained by washing alga with site water; (+) pPB20, positive control consisting of merA from Pseudomonas stutzeri OX (50); PCR blank lane (−) and DNA size markers (in bp). (Bottom) Gel shows merA PCR products following reverse transcriptase reactions (+RT) of RNA extracted from sea ice and lagoon biomass. (−RT), no RT controls; (−) PCR blank lane; a 291-bp PCR fragment is shown.
FIG. 2.
Phylogenetic distribution of merA phylotypes. The dendrogram was constructed from a ClustalW alignment of the trimmed amino acid sequences by neighbor-joining analysis using MEGA 3.1. Bootstrap values greater than 50 are indicated. The number of clones in a particular phylotype is indicated in parentheses. Phylotypes representing >25% of the clones in a library are highlighted in bold type. Phylotype designations that contain the letters LG originated in the lagoon biomass and those that contain SI in the macroalgae; d and r indicate that the phylotype originated in DNA and RNA clone libraries, respectively. Reference sequences from GenBank include the accession numbers.
Diversity analysis of merA.
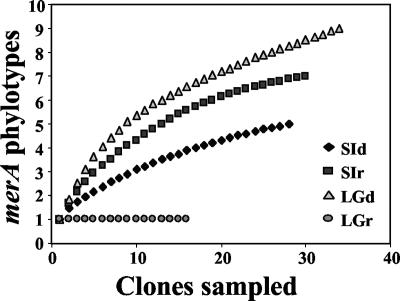
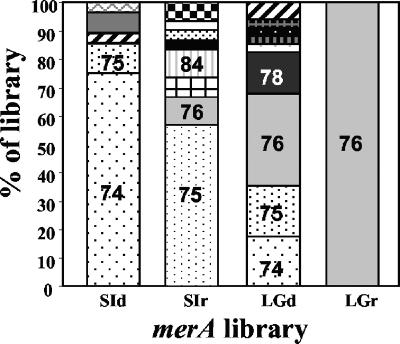
Rarefaction curves were constructed for each merA library to assess our sampling effort (Fig. 3). Both sea ice merA libraries were sampled adequately as the rarefaction curves approached a plateau. Furthermore, ACE and Chao1 estimates indicated that in both libraries, we captured approximately 80% of the predicted phylotypes (Table 2). While the distribution of phylotypes in the sea ice RNA library was more even than that in the sea ice DNA library (Fig. 3), the richness (Chao1 and ACE) and diversity (H′) estimates were the same within the sampling error (Table 2). The sea ice DNA library was dominated by Arct74, which was most closely related to the MerA of Tn21 (Fig. 2 and 4). Arct74 was not detected in the sea ice RNA library. This library was dominated by the second most abundant clone from the DNA library, Arct75 (Fig. 4). With the exception of Arct75, there was no overlap between the sea ice DNA and RNA merA libraries.
FIG. 3.
merA phylotype accumulation curves showing the number of clones sampled versus the number of merA phylotypes observed.
TABLE 2.
Diversity analysis of the merA gene fragment librariesa
| Library | N | n | Sampling efficiency tests
|
||
|---|---|---|---|---|---|
| Chao1 | ACE | H′ | |||
| SId | 28 | 5 | 5.5 (5.0-13.2) | 6.7 (5.2-21.5) | 0.88 (0.47-1.28) |
| Sir | 30 | 6 | 7.3 (7.0-13) | 8.1 (7.3-11.5) | 1.41 (1.03-1.79) |
| LGd | 34 | 9 | 12 (9.4-32) | 13.4 (9.7-35.7) | 1.84 (1.56-2.12) |
| LGr | 16 | 1 | 1 (1-1.1) | ND | 0 (−0.087-0.087) |
N, number of clones sequenced; n, number of merA phylotypes observed; Chao1, Chao1 nonparametric richness estimate; ACE, abundance-based coverage estimator; H′, Shannon-Weaver diversity index. Numbers in parentheses represent 95% confidence intervals; ND, not determined.
FIG. 4.
Relative abundance of merA phylotypes within the four clone libraries. A threshold of 97% nucleotide identity was used to define each merA phylotype. SI, sea ice; LG, lagoon. Different shading patterns indicate different phylotypes, and sections with similar patterns in each bar denote similar phylotypes that were present in more than one library. Phylotype numbers, provided for those phylotypes that represent at least 10% of a library, correspond to those used as phylotype designations shown in Fig. 2.
The lagoon DNA library was the most diverse, with an H′ of 1.84, while the lagoon RNA library was the least diverse, with an H′ of 0, corresponding to the presence of a single phylotype, Arct76 (Table 2). ACE- and Chao1-based coverage estimates indicated that our sequencing effort captured 75% of the predicted phylotypes from the lagoon DNA library. The single phylotype in the lagoon RNA library was most closely related to MerA of Tn21. Arct76 comprised about one-third of the lagoon DNA library (Fig. 4). A Tn21-like MerA locus was previously described in a bacterium obtained from 8,000-year-old permafrost (31), and thus, this MerA phylotype may be common in cold environments.
Bacterial community analysis.
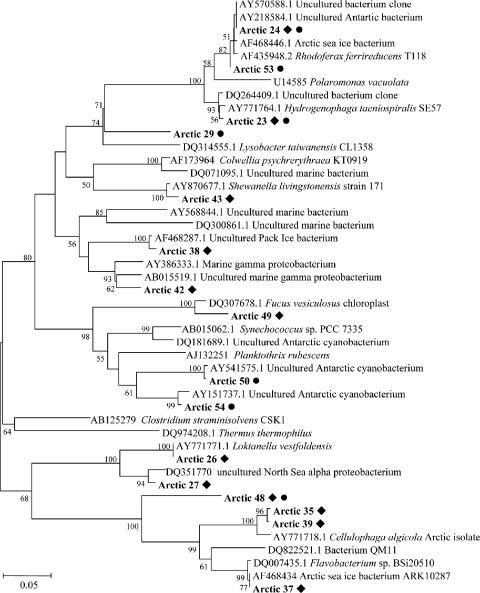
We constructed and analyzed 16S rRNA gene clone libraries to examine whether microbes that are commonly found in polar regions were present in the sampled lagoon and sea ice biomass. This was confirmed by sequence analysis (Fig. 5) showing that Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, the Cytophaga-Flavobacterium-Bacteroides (CFB) group, and Cyanobacteria dominated the cloned 16S rRNA gene libraries. The sea ice community was dominated by Proteobacteria (55% of sequenced clones), while the lagoon was dominated by Cyanobacteria (45% of sequenced clones). The second most abundant phyla were Proteobacteria (36%) and the CFB group (30%) for the lagoon and the sea ice, respectively; BlastN analysis revealed that the closest relatives to these dominant 16S phylotypes were bacteria that are often found in polar coastal, marine, and sympagic environments (14), such as Loktanella vestfoldensis or uncultured Antarctic and Arctic Proteobacteria (Fig. 5). The 16S libraries were not sampled to exhaustion (data not shown).
FIG. 5.
16S rRNA-based phylogeny of Arctic microbial biomass. The dendrogram was constructed from a ClustalW alignment of the trimmed nucleotide sequences by neighbor-joining analysis using MEgA 3.1. Bootstrap values greater than 50 are indicated. ⧫, phylotypes represented in the sea ice library; •, lagoon library.
Total mercury levels in water and microbial mats.
Total mercury concentrations from coastal waters at both the lagoon and sea ice sites were between 9.4 and 11.5 pmol · liter−1, corresponding to previously observed concentrations in Arctic seawater (23). Modeling the inorganic speciation of mercury in these samples using MINEQL+ v.4.5 (53) showed the dominance of negatively charged chlorocomplexes (e.g., HgCl42−). To further assess the mercury levels to which the sampled microbial communities were exposed, we measured THg in Desmarestia sp. (sea ice sample) and in the microbial mats (lagoon sample). The data showed the former and latter to have concentrations of 52.5 ± 0.9 ng · g−1 (dry weight) and 27.3 ± 2.9 ng · g−1 (dry weight), respectively, which were 2 to 5 times lower than those reported at lower latitudes for periphyton in boreal lakes (22).
Modeling of mercury speciation in seawater.
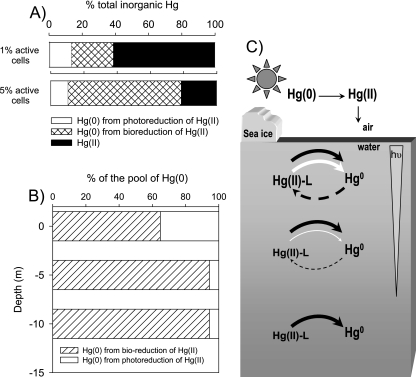
To assess if microbial reduction can constitute a major pathway for mercury cycling in polar regions, we used a kinetic redox model (13) in which photooxidation, photoreduction, and microbial mercury reduction were considered. Mercury speciation was modeled at the surface and at depths of 5 and 10 m (Fig. 6). It should be noted that microbially mediated reduction rates were corrected for Arctic conditions that accounted for cell viability and metabolism alteration because of low temperatures (see Materials and Methods). At the sea surface, the steady-state pool of elemental mercury could represent 40 to 80% of the inorganic mercury pool, depending on the assumed proportion of viable bacterial cells. When the model considered 1% of the cells active, >20% of the total pool of inorganic mercury was biogenic Hg(0); and when 5% of the cells were considered active, >55% of the inorganic mercury pool was biogenic Hg(0) (Fig. 6A). At depths of 5 and 10 m, due to UVA and UVB energy attenuation, the relative importance of photochemical processes decreased, and microbial reduction could account for up to 94% of the pool of elemental mercury (Fig. 6B and C).
FIG. 6.
Results of modeling of the relative importance of photochemical reactions versus biologically mediated reduction in mercury redox cycling in seawater in Arctic near-coastal environments. (A) Relative distribution of inorganic mercury species and proportion of Hg(0) formed by photochemical and microbial processes at the sea surface. In this example, photoreduction and photooxidation rates were considered to be 0.5 and 0.6 h−1, respectively, and the proportion of active bacterial cells was assumed to be 1 or 5%. (B) Relative importance of photochemical versus microbial contributions to the pool of Hg(0) at the surface and at depths of 5 and 10 m. (C) A proposed model for redox cycling of mercury in the water column of Arctic near-coastal marine environments. Arrow width correlates with the relative quantitative contribution of the depicted process to the total activity at each depth. Black, white, and dashed-line arrows represent microbially mediated reduction, photoreduction triggered mostly by UVB, and photooxidation triggered mostly by UVA, respectively. The gray inverted triangle depicts the gradient of light penetration with depth.
DISCUSSION
Here we report that merA genes and transcripts were detected in high-Arctic microbial biomass that contained microbes previously described as inhabiting polar environments. These results suggest that mercury-resistant organisms are present and active in Arctic coastal environments where critical redox transformations of mercury occur and where methylmercury is accumulated in the marine food chain. Furthermore, modeling efforts suggested an important role for the prokaryotic MerA in the production of Hg(0) in the high Arctic.
The concentration of Hg(0) in natural waters results from the balance between its production (reduction) and its destruction (oxidation). Because of the attenuation of UV radiation with depth in the water column, both photoreduction and photooxidation decline with increasing depth. Moreover, in coastal and marine systems, because of the presence of chloride, photoreduction is hampered and, in the absence of an alternative pathway for Hg(0) production, oxidation reactions may be prevalent and likely more significant than evasion in controlling the fate of Hg(0) (1, 3, 63). Microbial reduction can account for a significant component of the mercury redox cycling in coastal marine systems from temperate zones [up to 20% of the pool of Hg(0)] (51), although the mechanisms involved remain unknown. Our model (Fig. 6) suggests that in the high-Arctic coastal marine environment, the activity of the prokaryotic MerA is an important source of Hg(0) and, as it is not dependent on light, can occur at any depth or under sea ice during the winter. These predictions for the role of MerA in Hg(0) production are supported by recent data showing that during wintertime, the pool of total mercury in surface seawater beneath sea ice is comprised of up to 50% elemental mercury and can reach a concentration of ca. 0.6 pmol · liter−1 (V. St. Louis, personal communication), 5- to 10-fold higher than that observed in coastal seawater during the ice-free season. Although very few data exist as to the metabolic activity of microbes under sea ice during polar winter, it is possible that they further increase the pool of reduced mercury at a time of the year when photooxidation processes are greatly reduced. Moreover, because mer operon functions also degrade MeHg, bacteria may decrease the pool of methylmercury available for uptake by Arctic food webs. In this regard, it is important to note that macroalgae such as Desmarestia sp. were reported to produce both monomethylmercury and dimethylmercury in polar seawater (47). The importance of mercury-resistant microbes that are associated with this macroalgae-to-mercury cycling is critical to the understanding of contamination pathways in the Arctic.
The discovery of merA transcripts in Arctic microbial biomass was unexpected. The expression of merA is tightly controlled by intracellular concentrations of divalent mercury (15), and merA induction requires exposure concentrations in the nmol · liter−1 range in both pure cultures (64) and aquatic microbial communities (52). The mercury concentrations we present here and those reported for late spring and summer in meltwater, lakes, and seawater are in the pmol · liter−1 range (36, 58), typical of pristine environments. Furthermore, this mercury was present mostly as negatively charged chlorocomplexes, which are poorly bioavailable to microbes (5, 27). Therefore, not only was mercury concentration low in the coastal environments sampled here, but its bioavailability was likely also limited by high concentrations of Cl−. The induction of merA observed in our study may reflect (i) the existence of environmental conditions that enhance mercury bioavailability or (ii) the presence of microniches with high mercury concentrations within the sampled biomass. Our analysis of mercury concentrations involved homogenization steps and would miss such niches.
To understand why merA was expressed in the high Arctic, an understanding of both the processes that control mercury bioavailability and how these processes are impacted by the uniqueness of the microbial habitats in high latitudes is needed. These are not well understood beyond the passive diffusion of Hg-sulfide complexes (11) and the facilitated transport of inorganic and organic mercury complexes (26). Niches with high concentrations of mercury can occur in the high Arctic, possibly in association with the complex and heterogeneous nature of sea ice (21). One of the biomass samples described here (SI) was obtained from waters with sea ice, and considering the slow degradation of RNA in frozen environments (61), a sea ice origin for the merA transcripts detected in this sample may be plausible. This proposition is supported by the dominance of bacteria endemic to sympagic environments in the sampled communities (Fig. 5). The conditions which control the uptake of mercury are still unclear and may well be as diverse as the environments studied. The occurrence of merA expression in Arctic coastal and marine aquatic environments exhibiting low mercury levels underscores the gap in our current knowledge of Hg-microbe interactions, and further investigation of the biogeochemical cycling of mercury in polar environments should elucidate some key aspects of the Arctic mercury cycle.
Many of the MerA phylotypes reported here from Arctic biomass are related to those from microbes that thrive at high latitudes. Forty-eight percent were most closely related to putative mer loci from the complete genomes of Sphingopyxis alaskensis and Polaromonas vacuolata, two microbes first isolated from polar environments. This finding attests to the authenticity of the MerA loci that were isolated here as representative of those loci that are present and expressed in microbial communities from polar regions. The remaining MerA phylotypes, which constitute the majority of the clones sequenced, were closely related to MerA phylotypes from Tn21, Tn501, and pDU1358. A Tn21-related mercury resistance transposon, Tn5060, has previously been described in an 8,000- to 10,000-year-old Siberian permafrost isolate of Pseudomonas sp. (31).
Phylotypes related to Tn21 and Tn501 merA phylotypes dominated in an anaerobic enrichment of mercury-contaminated sediment (43) as well as in the microbial biomass of an acidic and sulfidic mercury-rich spring in Yellowstone National Park (T. Barkay, unpublished data). These data suggest that this merA locus, which represents the most recently evolved MerA lineage (Fig. 2), is broadly distributed in many environments. The total number of merA phylotypes observed here (Fig. 2) was low compared to that in the anaerobic enrichment with its 39 merA phylotypes. Others have shown that selection for the target genes may increase phylotype diversity (28), while enhanced activities in environmental incubations were associated with a reduced phylotype diversity (40). A more rigorous examination of the relationship of merA diversity to mercury resistance and reduction in environmental samples is needed for a clear understanding of how phylotype diversities is related to the potential for mercury transformations in various environments.
Two contrasting patterns were observed when the diversities of the DNA and RNA clone libraries that were obtained from the same community were examined: (i) a high diversity of merA genes and a low diversity of transcripts in the lagoon and (ii) a high diversity of both genes and transcripts in the sea ice samples. At this point, we can only speculate on why the lagoon and the sea ice communities showed such different patterns. One possibility is that the high diversity of merA transcripts in the sea ice reflects the heterogeneous nature of microbial niches within sea ice (see above). Alternatively, it may be related to the in situ temperature at the time of sampling. While Arctic coastal seawater temperatures are usually slightly below 0°C, the temperatures in coastal lagoons may reach a mesophilic range (58), and the temperature at the lagoon at sampling time was 7°C. Higher temperatures in the lagoon may have accelerated the turnover rate of transcripts, while colder conditions in open waters may have slowed mRNA degradation to the point that we detected a greater diversity of expressed merA genes in sea ice.
The diversity of expressed genes, not that of the genes that are present in a community, is at the heart of what determines ecosystem function as related to environmental conditions. While experimental approaches to studying the expression of biogeochemical functions have been widely used in the last decade, few studies have compared the diversities of functional genes and mRNA transcripts in microbial biomass. Knauth et al. (32) used terminal restriction fragment length polymorphism analysis of both DNA and RNA extracted from microbial biomass of the rice rhizosphere to examine how nitrogen availability and plant host species affected the diversity of the nitrogenase gene nifH. As in our study of merA, their results showed significant differences between nifH DNA and mRNA profiles, underscoring the importance of mRNA-based approaches to understanding functional gene diversity. The observation of different merA gene and transcript profiles presented here highlights the need to further develop an understanding of how patterns of microbial gene expression are affected by environmental parameters in heterogeneous environments such as the Arctic.
Our discovery of merA expression and the results of the model calculations that simulated redox transformations suggest an active role for microbial activities in the cycling of mercury in Arctic coastal and marine environments. Clearly, microbial redox transformations have the potential to drive mercury dynamics in cold environments, and Arctic microbes that possess and express merA genes stand to be key players in determining the environmental mobility and toxicity of mercury in high-latitude polar regions.
Acknowledgments
We thank H. Vandermeulen for his help with alga identification, Paul del Giorgio for access to the flow cytometer, J. Brenchley and V. Miteva for stimulating discussions, and B. E. Keatley for comments on the manuscript.
Support by the U.S. National Science Foundation (CHE-0221978 and ATM-0322022), the U.S. National Institute of Environmental Health and Safety (P42 ES004911), and the Fond Québecois de la Recherche sur la Nature et les Technologies for funding is acknowledged.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Amyot, M., G. A. Gill, and F. M. M. Morel. 1997. Production and loss of dissolved gaseous mercury in coastal seawater. Environ. Sci. Technol. 31:3606-3611. [Google Scholar]
- 2.Amyot, M., G. Mierle, D. R. S. Lean, and D. J. McQueen. 1994. Sunlight-induced formation of dissolved gaseous mercury in lake waters. Environ. Sci. Technol. 28:2366-2371. [DOI] [PubMed] [Google Scholar]
- 3.Ariya, P. A., A. P. Dastoor, M. Amyot, W. H. Schroeder, L. Barrie, K. Anlauf, F. Raofie, A. Ryzhkov, D. Davignon, J. Lalonde, and A. Steffen. 2004. The Arctic: a sink for mercury. Tellus. Ser. B 56:397-403. [Google Scholar]
- 4.Ariya, P. A., A. Khalizov, and A. Gidas. 2002. Reactions of gaseous mercury with atomic and molecular halogens: kinetics, product studies, and atmospheric implications. J. Phys. Chem. A 106:7310-7320. [Google Scholar]
- 5.Barkay, T., M. Gillman, and R. R. Turner. 1997. Effects of dissolved organic carbon and salinity on bioavailability of mercury. Appl. Environ. Microbiol. 63:4267-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barkay, T., C. Liebert, and M. Gillman. 1989. Environmental significance of the potential for mer(Tn21)-mediated reduction of Hg2+ to Hg0 in natural waters. Appl. Environ. Microbiol. 55:1196-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barkay, T., C. Liebert, and M. Gillman. 1989. Hybridization of DNA probes with whole-community genome for detection of genes that encode microbial responses to pollutants: mer genes and Hg2+ resistance. Appl. Environ. Microbiol. 55:1574-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355-384. [DOI] [PubMed] [Google Scholar]
- 9.Barkay, T., R. R. Turner, A. Vandenbrook, and C. Liebert. 1991. The relationships of Hg(II) volatilization from a fresh-water pond to the abundance of mer-genes in the gene pool of the indigenous microbial community. Microb. Ecol. 21:151-161. [DOI] [PubMed] [Google Scholar]
- 10.Benner, R., P. Louchouarn, and R. M. W. Amon. 2005. Terrigenous dissolved organic matter in the Arctic Ocean and its transport to surface and deep waters of the North Atlantic. Global Biogeochem. Cycles 19:GB2025. [Google Scholar]
- 11.Benoit, J. M., C. C. Gilmour, R. P. Mason, and A. Heyes. 1999. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters. Environ. Sci. Technol. 33:951-957. [Google Scholar]
- 12.Braun, W., J. T. Herron, and D. K. Kahaner. 1988. ACUCHEM: a computer-program for modeling complex chemical-reaction systems. Int. J. Chem. Kinet. 20:51-62. [Google Scholar]
- 13.Braune, B., D. Muir, B. DeMarch, M. Gamberg, K. Poole, R. Currie, M. Dodd, W. Duschenko, J. Eamer, B. Elkin, M. Evans, S. Grundy, C. Hebert, R. Johnstone, K. Kidd, B. Koenig, L. Lockhart, H. Marshall, K. Reimer, J. Sanderson, and L. Shutt. 1999. Spatial and temporal trends of contaminants in Canadian Arctic freshwater and terrestrial ecosystems: a review. Sci. Total Environ. 230:145-207. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmeyer, R., K. Knittel, J. Jürgens, H. Weyland, R. Amann, and E. Helmke. 2003. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl. Environ. Microbiol. 69:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 16.Campbell, L. M., R. J. Norstrom, K. A. Hobson, D. C. G. Muir, S. Backus, and A. T. Fisk. 2005. Mercury and other trace elements in a pelagic Arctic marine food web (Northwater Polynya, Baffin Bay). Sci. Total Environ. 351:247-263. [DOI] [PubMed] [Google Scholar]
- 17.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 18.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 19.Costa, M., and P. S. Liss. 1999. Photoreduction of mercury in sea water and its possible implications for Hg(0) air-sea fluxes. Mar. Chem. 68:87-95. [Google Scholar]
- 20.De Domenico, M., A. Lo Giudice, L. Michaud, M. Saitta, and V. Bruni. 2004. Diesel oil and PCB-degrading psychrotrophic bacteria isolated from antarctic seawaters (Terra Nova Bay, Ross Sea). Polar Res. 23:141-146. [Google Scholar]
- 21.Deming, J. W. 2002. Psychrophiles and polar regions. Curr. Opin. Microbiol. 5:301-309. [DOI] [PubMed] [Google Scholar]
- 22.Desrosiers, M., D. Planas, and A. Mucci. 2006. Mercury methylation in the epilithon of boreal shield aquatic ecosystems. Environ. Sci. Technol. 40:1540-1546. [DOI] [PubMed] [Google Scholar]
- 23.Douglas, T. A., M. Sturm, W. R. Simpson, S. Brooks, S. E. Lindberg, and D. K. Perovich. 2005. Elevated mercury measured in snow and frost flowers near Arctic sea ice leads. Geophys. Res. Lett. 32:L04502. [Google Scholar]
- 24.Ebinghaus, R., H. H. Kock, C. Temme, J. W. Einax, A. G. Lowe, A. Richter, J. P. Burrows, and W. H. Schroeder. 2002. Antarctic springtime depletion of atmospheric mercury. Environ. Sci. Technol. 36:1238-1244. [DOI] [PubMed] [Google Scholar]
- 25.Gibson, J. A. E., W. F. Vincent, B. Nieke, and R. Pienitz. 2000. Control of biological exposure to UV radiation in the Arctic Ocean: comparison of the roles of ozone and riverine dissolved organic matter. Arctic 53:372-382. [Google Scholar]
- 26.Golding, G. R., C. A. Kelly, R. Sparling, P. C. Loewen, J. W. M. Rudd, and T. Barkay. 2002. Evidence for facilitated uptake of Hg(II) by Vibrio anguillarum and Escherichia coli under anaerobic and aerobic conditions. Limnol. Oceanogr. 47:967-975. [Google Scholar]
- 27.Gutknecht, J. 1981. Inorganic mercury (Hg2+) transport through lipid bilayer membranes. J. Membr. Biol. 61:61-66. [Google Scholar]
- 28.Hobel, C. F. V., V. T. Marteinsson, G. Ó. Hreggvidsson, and J. K. Kristjánsson. 2005. Investigation of the microbial ecology of intertidal hot springs by using diversity analysis of 16S rRNA and chitinase genes. Appl. Environ. Microbiol. 71:2771-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Junge, K., H. Eicken, and J. W. Deming. 2004. Bacterial activity at −2 to −20°C in Arctic wintertime sea ice. Appl. Environ. Microbiol. 70:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kholodii, G., S. Mindlin, M. Petrova, and S. Minakhina. 2003. Tn5060 from the Siberian permafrost is most closely related to the ancestor of Tn21 prior to integron acquisition. FEMS Microbiol. Lett. 226:251-255. [DOI] [PubMed] [Google Scholar]
- 32.Knauth, S., T. Hurek, D. Brar, and B. Reinhold-Hurek. 2005. Influence of different Oryza cultivars on expression of nifH gene pools in roots of rice. Environ. Microbiol. 7:1725-1733. [DOI] [PubMed] [Google Scholar]
- 33.Lalonde, J. D., M. Amyot, A. M. L. Kraepiel, and F. M. M. Morel. 2001. Photooxidation of Hg(0) in artificial and natural waters. Environ. Sci. Technol. 35:1367-1372. [DOI] [PubMed] [Google Scholar]
- 34.Lalonde, J. D., M. Amyot, J. Orvoine, F. M. M. Morel, J. C. Auclair, and P. A. Ariya. 2004. Photoinduced oxidation of Hg0(aq) in the waters from the St. Lawrence estuary. Environ. Sci. Technol. 38:508-514. [DOI] [PubMed] [Google Scholar]
- 35.Liebert, C., and T. Barkay. 1988. A direct viable counting method for measuring tolerance of aquatic microbial communities to Hg2+. Can. J. Microbiol. 34:1090-1095. [Google Scholar]
- 36.Lindberg, S. E., S. Brooks, C. J. Lin, K. J. Scott, M. S. Landis, R. K. Stevens, M. Goodsite, and A. Richter. 2002. Dynamic oxidation of gaseous mercury in the Arctic troposphere at polar sunrise. Environ. Sci. Technol. 36:1245-1256. [DOI] [PubMed] [Google Scholar]
- 37.Lovley, D. R. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 1:35-44. [DOI] [PubMed] [Google Scholar]
- 38.Macdonald, R. W., T. Harner, and J. Fyfe. 2005. Recent climate change in the Arctic and its impact on contaminant pathways and interpretation of temporal trend data. Sci. Total Environ. 342:5-86. [DOI] [PubMed] [Google Scholar]
- 39.Master, E. R., and W. W. Mohn. 1998. Psychrotolerant bacteria isolated from Arctic soil that degrade polychlorinated biphenyls at low temperatures. Appl. Environ. Microbiol. 64:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metcalfe, A. C., M. Krsek, G. W. Gooday, J. I. Prosser, and E. M. H. Wellington. 2002. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl. Environ. Microbiol. 68:5042-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mindlin, S., L. Minakhin, M. Petrova, G. Kholodii, S. Minakhina, Z. Gorlenko, and V. Nikiforov. 2005. Present-day mercury resistance transposons are common in bacteria preserved in permafrost grounds since the Upper Pleistocene. Res. Microbiol. 156:994-1004. [DOI] [PubMed] [Google Scholar]
- 42.Muir, D., B. Braune, B. DeMarch, R. Norstrom, R. Wagemann, L. Lockhart, B. Hargrave, D. Bright, R. Addison, J. Payne, and K. Reimer. 1999. Spatial and temporal trends and effects of contaminants in the Canadian Arctic marine ecosystem: a review. Sci. Total Environ. 230:83-144. [DOI] [PubMed] [Google Scholar]
- 43.Ní Chadhain, S. M., J. K. Schaefer, S. Crane, G. J. Zylstra, and T. Barkay. 2006. Analysis of mercuric reductase (merA) gene diversity in an anaerobic mercury-contaminated sediment enrichment. Environ. Microbiol. 8:1746-1752. [DOI] [PubMed] [Google Scholar]
- 44.Osborn, A. M., K. D. Bruce, P. Strike, and D. A. Ritchie. 1997. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 19:239-262. [DOI] [PubMed] [Google Scholar]
- 45.Pepi, M., A. Cesaro, G. Liut, and F. Baldi. 2005. An antarctic psychrotrophic bacterium Halomonas sp. ANT-3b, growing on n-hexadecane, produces a new emulsifying glycolipid. FEMS Microbiol. Ecol. 53:157-166. [DOI] [PubMed] [Google Scholar]
- 46.Philippidis, G. P., L. H. Malmber, W. S. Hu, and J. L. Schottel. 1991. Effect of gene amplification on mercuric ion reduction activity of Escherichia coli. Appl. Environ. Microbiol. 57:3558-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pongratz, R., and K. G. Heumann. 1998. Production of methylated mercury and lead by polar macroalgae: a significant natural source for atmospheric heavy metals in clean room compartments. Chemosphere 36:1935-1946. [Google Scholar]
- 48.Poulain, A. J., M. Amyot, D. Findlay, S. Telor, T. Barkay, and H. Hintelmann. 2004. Biological and photochemical production of dissolved gaseous mercury in a boreal lake. Limnol. Oceanogr. 49:2265-2275. [Google Scholar]
- 49.Price, P. B., and T. Sowers. 2004. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl. Acad. Sci. USA 101:4631-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reniero, D., E. Mozzon, E. Galli, and P. Barbieri. 1998. Two aberrant mercury resistance transposons in the Pseudomonas stutzeri plasmid pPB. Gene 208:37-42. [DOI] [PubMed] [Google Scholar]
- 51.Rolfhus, K. R., and W. F. Fitzgerald. 2004. Mechanisms and temporal variability of dissolved gaseous mercury production in coastal seawater. Mar. Chem. 90:125-136. [Google Scholar]
- 52.Schaefer, J. K., J. Yagi, J. R. Reinfelder, T. Cardona, K. M. Ellickson, S. Tel-Or, and T. Barkay. 2004. Role of the bacterial organomercury lyase (MerB) in controlling methylmercury accumulation in mercury-contaminated natural waters. Environ. Sci. Technol. 38:4304-4311. [DOI] [PubMed] [Google Scholar]
- 53.Schecher, W. D., and D. C. McAvoy. 1992. Mineql+: a software environment for chemical-equilibrium modeling. Comput. Environ. Urban Syst. 16:65-76. [Google Scholar]
- 54.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schroeder, W. H., K. G. Anlauf, L. A. Barrie, J. Y. Lu, A. Steffen, D. R. Schneeberger, and T. Berg. 1998. Arctic springtime depletion of mercury. Nature 394:331-332. [Google Scholar]
- 56.Sellers, P., C. A. Kelly, J. W. M. Rudd, and A. R. MacHutchon. 1996. Photodegradation of methylmercury in lakes. Nature 380:694-697. [Google Scholar]
- 57.Siciliano, S. D., N. J. O'Driscoll, and D. R. Lean. 2002. Microbial reduction and oxidation of mercury in freshwater lakes. Environ. Sci. Technol. 36:3064-3068. [DOI] [PubMed] [Google Scholar]
- 58.St. Louis, V. L., M. J. Sharp, A. Steffen, A. May, J. Barker, J. L. Kirk, D. J. Kelly, S. E. Arnott, B. Keatley, and J. P. Smol. 2005. Some sources and sinks of monomethyl and inorganic mercury on Ellesmere Island in the Canadian high Arctic. Environ. Sci. Technol. 39:2686-2701. [DOI] [PubMed] [Google Scholar]
- 59.Van Oostdam, J., S. G. Donaldson, M. Feeley, D. Arnold, P. Ayotte, G. Bondy, L. Chan, E. Dewaily, C. M. Furgal, H. Kuhnlein, E. Loring, G. Muckle, E. Myles, O. Receveur, B. Tracy, U. Gill, and S. Kalhok. 2005. Human health implications of environmental contaminants in Arctic Canada: a review. Sci. Total Environ. 351-352:165-246. [DOI] [PubMed] [Google Scholar]
- 60.Vetriani, C., Y. S. Chew, S. M. Miller, J. Yagi, J. Coombs, R. A. Lutz, and T. Barkay. 2005. Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl. Environ. Microbiol. 71:220-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vlassov, A. V., S. A. Kazakov, B. H. Johnston, and L. F. Landweber. 2005. The RNA world on ice: a new scenario for the emergence of RNA information. J. Mol. Evol. 61:264-273. [DOI] [PubMed] [Google Scholar]
- 62.Welander, U. 2005. Microbial degradation of organic pollutants in soil in a cold climate. Soil Sediment Contam. 14:281-291. [Google Scholar]
- 63.Whalin, L. M., and R. P. Mason. 2006. A new method for the investigation of mercury redox chemistry in natural waters utilizing deflatable Teflon bags and additions of isotopically labeled mercury. Anal. Chim. Acta 558:211-221. [Google Scholar]
- 64.Wright, J. G., M. J. Natan, F. M. Macdonnell, D. M. Ralston, and T. V. Ohalloran. 1990. Mercury(II) thiolate chemistry and the mechanism of the heavy-metal biosensor MerR. Prog. Inorg. Chem. 38:323-412. [Google Scholar]
- 65.Zubkov, M. V., B. M. Fuchs, H. Eilers, P. H. Burkill, and R. Amann. 1999. Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl. Environ. Microbiol. 65:3251-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]