Abstract
Phylogenetic analysis of bacteria preserved within an ice wedge from the Fox permafrost tunnel was undertaken by cultivation and molecular techniques. The radiocarbon age of the ice wedge was determined. Our results suggest that the bacteria in the ice wedge adapted to the frozen conditions have survived for 25,000 years.
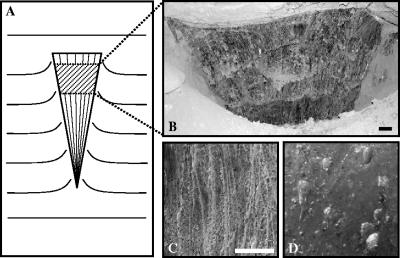
Ice wedges are wedge-shaped ancient ice (Fig. 1A) and are among the most common features in permafrost regions, including northern and central Alaska (7). They grow as a result of repeated cycles of frost cracking followed by the infiltration of snow, meltwater, soil, and other material into open frost cracks (17). Material incorporated into the ice wedge quickly becomes frozen, and the ice and the ice in soil and organic particles are thus preserved in a frozen state. The Fox permafrost tunnel in Alaska (13), where numerous buried ice wedges are exposed in the tunnel wall (Fig. 1B), is preserved at a temperature of roughly −3°C by the U.S. Army's Cold Regions Research and Engineering Laboratory. Ice wedges in the tunnel exhibit numerous thin, vertical bands of sediment and ice veinlets characteristic of undisturbed ice wedges (Fig. 1C), as well as numerous small air bubbles (Fig. 1D), suggesting that their shapes and fabrics exhibit no signs of thawing (7). If they have not been thawed, it is important to know their age. Microorganisms derived from meltwater and soil particles also might have been trapped and preserved in a frozen state since ice wedge formation. Although there are a number of examples of bacteria in frozen environments (1, 3, 8-12, 19, 21, 24, 25, 30-32, 35-37), no systematic analysis of bacteria within a dated ice wedge has ever been done. Therefore, the objectives of this study were to determine the age of the ice wedge sample collected from the Fox tunnel, to isolate living bacteria, to classify both the isolates and bacterial DNA extracted from the melted ice wedge sample on the basis of the partial 16S rRNA gene sequence, and to examine the temperature sensitivity of ice wedge isolates.
FIG. 1.
Fabrics of the ice wedge in the Fox permafrost tunnel. Each scale bar indicates 0.1 m. (A) Schematic pattern of ice wedge in permafrost. (B) Exposed part of the ice wedge. (C) Foliation of ice indicating annual veinlets. (D) Air bubbles in the ice (1 to 2 mm in diameter).
An ice wedge sample was collected from the Fox permafrost tunnel (64°57.084′N, 147°37.250′W) and was kept frozen during transportation to the laboratory. The sample was separated into two portions. The radiocarbon date and δ13C as a carbon isotopic ratio of the methane derived from one portion of the ice sample (approximately 2.5 kg) were measured with a Tandetron accelerator mass spectrometry system at Nagoya University. The second portion of the sample (about 50 g) was surface sterilized by immersing it in a 70% ethanol solution and by burning it to remove the ethanol or contaminated surface ice. We confirmed that the newly exposed surface of ice was not contaminated by stamping it on cultivation agar and incubating it at 15°C. It was then melted and spread on agar media after aseptic dilution. The cultivation media used were Luria broth (LB), LBG (LB plus 10 g/liter glucose), R2B (21), 100-fold-diluted LB and LBG, Hickey-Tresner revised medium with antibiotics (0.4 g/liter peptone, 0.2 g/liter yeast extract and meat extract, 2.0 g/liter soluble starch, 0.05 g/liter nystatin, 0.01 g/liter cycloheximide, 0.005 g/liter nalidixic acid), minimal medium (1.0 g/liter K2HPO4 and NH4Cl, 0.2 g/liter MgSO4·7H2O, 0.01 g/liter FeSO4·7H2O and CaCl2·2H2O, 0.1 mg/liter trace elements), minimal medium plus 5.0 g/liter glucose, and MME-1 and MME-2 (minimal medium containing 1.0% and 10% filter-sterilized ice extract obtained from the supernatant of the melted ice wedge, respectively). All of the media contained 20 g/liter agar, and the pH was adjusted to 7.0 with 1 N HCl or 1 N NaOH. Plates were incubated aerobically at 15°C in the dark for 3 months. Different types of colonies were selected and purified by restreaking on fresh media of the same kind. The partial 16S rRNA genes (Escherichia coli positions 27 to 520) were amplified and sequenced from 270 colonies with an AmpliTaq PCR kit and a Big Dye Terminator cycle sequencing ready reaction kit (Applied Biosystems). Total nucleic acids were extracted from the precipitates of surface-sterilized melted ice sample with ISOIL (NIPPONGENE). The partial 16S rRNA gene clone library was constructed with the pGEM-T Easy vector (Promega). Automatic and manual sequence alignments were performed with the ARB program package (16). A phylogenetic tree was constructed with PHYLIP, version 3.65 (6). The growth of 24 representatives was examined at −5°C, 4°C, 15°C, 27°C, and 37°C by measuring diameters of colonies.
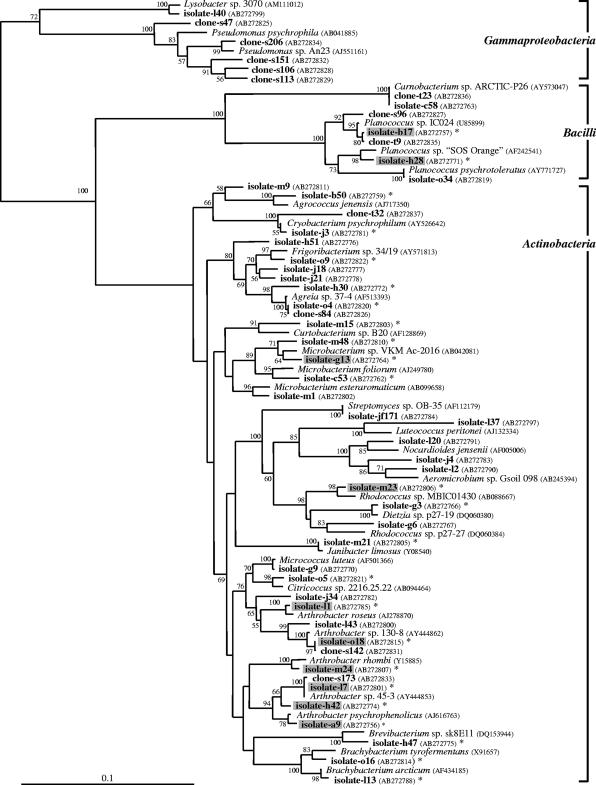
A radiocarbon date of 24,884 ± 139 years BP (before 1950 AD; data number NUTA2-3477) was obtained from methane collected from the sampled ice wedge. The stable carbon isotopic ratio was −84.651‰, which differs from that of atmospheric methane, demonstrating that any contamination by atmospheric air was negligible. Bacterial colonies grew at concentrations of 105 to 106 CFU/ml of melted ice. In total, 270 aerobic or facultatively anaerobic bacteria were isolated. Most of the isolates were non-spore-forming bacteria. When the sequences with greater than 98% similarity were treated as the same species, isolates and 273 clonal types were grouped into 41 operational taxonomic units (OTUs) and 12 OTUs, respectively. The number of OTUs was determined by the DOTUR program (http://www.plantpath.wisc.edu/fac/joh/dotur.html) (28). A phylogenetic tree of representatives of OTUs and their closest relatives was constructed with distance data by a neighbor-joining method (26). Bootstrap analyses for 1,000 replicates were performed. OTUs of both the isolates and clones were affiliated with three different classes, Actinobacteria, Bacilli, and Gammaproteobacteria (Fig. 2). Similar topologies of the OTUs were observed from the trees generated by the maximum-likelihood and maximum-parsimony methods (data not shown). The 36 OTUs of isolates and 4 clonal OTUs were affiliated with the order Actinomycetales, and many were closely related to the genera Arthrobacter, Brachybacterium, Cryobacterium, Microbacterium, and Rhodococcus. In the Bacilli branch, 4 OTUs of isolates and 3 OTUs of clones were closely related to the genera Planococcus and Carnobacterium. The dominant taxon of clones was Gammaproteobacteria (93.1% of the total number of clones). In this class, isolates and clones were closely related to Lysobacter and Pseudomonas, respectively. Actually, the strains which were identical to representative clonal type no. 206 in 16S rRNA gene sequences had been isolated mainly from MME-2 agar plates; however, unfortunately, all of these isolates could not be subcultured. All of the isolates that were examined for their sensitivity to temperature grew at 4°C and 20°C but not 37°C. Ten isolates which were closely related to the genera Arthrobacter, Planococcus, Microbacterium, and Rhodococcus could grow at −5°C after 3 months of cultivation (Fig. 2).
FIG. 2.
Phylogenetic relationship of the representative isolates, clonal types (bold), and their closest relatives based on partial 16S rRNA gene sequences. Bootstrap values that were above 50% are shown at the nodes. The scale bar represents 1 substitution/10 nucleotides. E. coli (accession no. X80725) was used as the outgroup. Asterisks and shaded clusters indicate the representative isolates that were examined for their sensitivity to temperature and those that grew at −5°C, respectively. The accession numbers shown were previously reported in references 2, 4, 5, 11, 12, 14, 15, 18, 20, 22, 24, 25, 27, 29, 30, 34, and 37.
Our results demonstrate that the Fox tunnel ice wedge has remained continuously frozen for the past 25,000 years. From the ice we collected, living bacteria were reproduced at concentrations as high as 106 CFU/ml of melted ice. Although bacteria are reported to be rarely recovered from ice wedges (8, 10), this study clearly demonstrates the existence of viable bacteria within ice wedge ice. We could easily recognize soil particles in the ice wedge sample melt, suggesting that these suspended solids might be a habitat that protected cells from ice crystals. The bacteria isolated from Siberian permafrost on LB medium were affiliated with Actinobacteria, Bacilli, and Alpha-, Beta-, and Gammaproteobacteria (31). On the contrary, Proteobacteria were not isolated from our ice wedge sample on the same medium (data not shown), indicating that the higher taxonomic levels of the ice wedge isolates were less diverse. This is consistent with the results of molecular analysis. No clonal type affiliated with Alpha- and Betaproteobacteria appeared in the clone library. In general, the bacterial community can be distorted by several biases such as DNA extraction (23) or PCR (33). However, phylogenetic diversity among the 16S rRNA gene clones was considered to be remarkably low. To assess if the number of screened clones was sufficient for an estimation of diversity in the clone library, rarefaction analysis was performed by the DOTUR program. The expected number of OTUs was plotted against the number of clones at various distance levels. The calculated rarefaction curves of clonal OTUs nearly reached an asymptote at a distance level of greater than 1%, indicating that the number of clones screened was enough (see Fig. S1 in the supplemental material). On the basis of the finding that some of these ice wedge isolates were able to grow at −5°C, i.e., at the in situ temperature, we assumed that these bacteria accomplished better strategies for surviving in the ice wedge. Similarly, Psychrobacter sp. isolated from a Siberian permafrost cryopeg was reported to grow at −10°C, the temperature of cryopegs (1). Although it is still unknown whether the organisms are active or dormant in situ, these results suggest that bacteria that were adapted to ice wedge conditions have survived for thousands of years. Our investigation of these adapted bacteria not only provides novel information about adaptation or survival mechanisms under extreme conditions but also may lead to a wide variety of biotechnological applications that had not previously been explored.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the representative isolates and clones reported in this study were deposited in GenBank under accession numbers AB272756 to AB272838.
Supplementary Material
Footnotes
Published ahead of print on 9 February 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bakermans, C., A. I. Tsapin, V. Souza-Egipsy, D. A. Gilichinsky, and K. H. Nealson. 2003. Reproduction and metabolism at −10°C of bacteria isolated from Siberian permafrost. Environ. Microbiol. 5:321-326. [DOI] [PubMed] [Google Scholar]
- 2.Behrendt, U., A. Ulrich, and P. Schumann. 2001. Description of Microbacterium foliorum sp. nov. and Microbacterium phyllosphaerae sp. nov., isolated from the phyllosphere of grasses and the surface litter after mulching the sward, and reclassification of Aureobacterium resistens (Funke et al. 1998) as Microbacterium resistens comb. nov. Int. J. Syst. Evol. Microbiol. 51:1267-1276. [DOI] [PubMed] [Google Scholar]
- 3.Bowman, J. P., S. A. Mccammon, M. V. Brown, D. S. Nichols, and T. A. Mcmeekin. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63:3068-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, M. D., P. A. Lawson, N. Nikolaitchouk, and E. Falsen. 2000. Luteococcus peritonei sp. nov., isolated from the human peritoneum. Int. J. Syst. Evol. Microbiol. 50:179-181. [DOI] [PubMed] [Google Scholar]
- 5.Doumbou, C. L., V. Akimov, M. Cote, P. M. Charest, and C. Beaulieu. 2001. Taxonomic study on nonpathogenic streptomycetes isolated from common scab lesions on potato tubers. Syst. Appl. Microbiol. 24:451-456. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 2005. PHYLIP (phylogeny inference package) version 3.65. Department of Genome Sciences, University of Washington, Seattle.
- 7.French, H. M. 1976. The periglacial environment, p. 309. Longman, London, United Kingdom.
- 8.Gilichinsky, D. A. 2002. Permafrost model of extraterrestrial habitats, p. 125-142. In G. Horneck and C. Baumstark-Khan (ed.), Astrobiology: the quest for the conditions of life. Springer Verlag, New York, NY.
- 9.Gilichinsky, D. A., E. Rivkina, C. Bakermans, V. Shcherbakova, L. Petrovskaya, S. Ozerskaya, N. Ivanushkina, G. Kochkina, K. Laurinavichuis, S. Pecheritsina, R. Fattakhova, and J. M. Tiedje. 2005. Biodiversity of cryopegs in permafrost. FEMS Microbiol. Ecol. 53:117-128. [DOI] [PubMed] [Google Scholar]
- 10.Gilichinsky, D. A., S. Wagener, and T. A. Vishnivetskaya. 1995. Permafrost microbiology. Permafrost Periglacial Process 6:281-291. [Google Scholar]
- 11.Groudieva, T., M. Kambourova, H. Yusef, M. Royter, R. Grote, H. Trinks, and G. Antranikian. 2004. Diversity and cold-active hydrolytic enzymes of culturable bacteria associated with Arctic sea ice, Spitzbergen. Extremophiles 8:475-488. [DOI] [PubMed] [Google Scholar]
- 12.Inagaki, F., M. Suzuki, K. Takai, H. Oida, T. Sakamoto, K. Aoki, K. H. Nealson, and K. Horikoshi. 2003. Microbiol. communities associated with geological horizons in coastal subseafloor sediments from the sea of Okhotsk. Appl. Environ. Microbiol. 69:7224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansen, N., S. L. Huang, and N. B. Aughebbaugh. 1988. Alaska's CRREL permafrost tunnel. Tunneling Underground Space Technol. 3:19-24. [Google Scholar]
- 14.Jussila, M. M., G. Jurgens, K. Lindström, and L. Suominen. 2006. Genetic diversity of culturable bacteria in oil-contaminated rhizosphere of Galega orientalis. Environ. Pollut. 139:244-257. [DOI] [PubMed] [Google Scholar]
- 15.Kleinsteuber, S., V. Riis, I. Fetzer, H. Harms, and S. Mullar. 2006. Population dynamics within a microbial consortium during growth on diesel fuel in saline environments. Appl. Environ. Microbiol. 72:3531-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackay, J. R. 1972. The world underground ice. Ann. Assoc. Am. Geogr. 62:1-22. [Google Scholar]
- 18.Malik, A., M. Sakamoto, S. Hanazaki, M. Osawa, T. Suzuki, M. Tochigi, and K. Kakii. 2003. Coaggregation among nonflocculating bacteria isolated from activated sludge. Appl. Environ. Microbiol. 69:6056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margesisn, R., P. Schumann, C. Sproer, and A. M. Gounot. 2004. Arthrobacter psychrophenolicus sp. nov., isolated from an alpine ice cave. Int. J. Syst. Evol. Microbiol. 54:2067-2072. [DOI] [PubMed] [Google Scholar]
- 20.Martin, K., P. Schumann, F. A. Rainey, B. Schuetze, and I. Groth. 1997. Janibacter limosus gen. nov. sp. nov., a new actinomycete with meso-diaminopimelic acid in the cell wall. Int. J. Syst. Bacteriol. 47:529-534. [DOI] [PubMed] [Google Scholar]
- 21.Miteva, V. I., P. P. Sheridan, and J. E. Brenchley. 2004. Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mocali, S., E. Bertelli, F. D. Cello, A. Mengoni, A. Sfalanga, F. Viliani, A. Caciotti, S. Tegli, G. Surico, and R. Fani. 2003. Fluctuation of bacteria isolated from elm tissues during different seasons and from different plant organs. Res. Microbiol. 154:105-114. [DOI] [PubMed] [Google Scholar]
- 23.More, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osorio, C. R., J. L. Barja, R. A. Hutson, and M. D. Collins. 1999. Arthrobacter rhombi sp. nov., isolated from Greenland halibut (Reinhardtius hippoglossoides). Int. J. Syst. Evol. Microbiol. 49:1217-1220. [DOI] [PubMed] [Google Scholar]
- 25.Reddy, G. S. N., J. S. S. Prakash, G. I. Matsumoto, E. Stackebrandt, and S. Shivaji. 2002. Arthrobacter roseus sp. nov., a psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int. J. Syst. Evol. Microbiol. 52:1017-1021. [DOI] [PubMed] [Google Scholar]
- 26.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstruction phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Saul, D. J., J. M. Aislabie, C. E. Brown, L. Harris, and J. M. Foght. 2005. Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol. Ecol. 53:141-155. [DOI] [PubMed] [Google Scholar]
- 28.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubert, K., W. Ludwig, N. Springer, R. M. Kroppenstedt, J. P. Accolas, and F. Fiedler. 1996. Two coryneform bacteria isolated from the surface of French Gruyere and Beaufort cheeses are new species of the genus Brachybacterium: Brachybacterium alimentarium sp. nov. and Brachybacterium tyrofermentans sp. nov. Int. J. Syst. Bacteriol. 46:81-87. [DOI] [PubMed] [Google Scholar]
- 30.Sheridan, P. P., and J. E. Brenchley. 2000. Characterization of a salt-tolerant family 42 β-galactosidase from a psychrophilic Antarctic Planococcus isolate. Appl. Environ. Microbiol. 66:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi, T., R. H. Reeves, D. A. Gilichinsky, and E. I. Friedmann. 1997. Characterization of viable bacteria from Siberian permafrost by 16S rRNA gene sequencing. Microb. Ecol. 33:169-179. [DOI] [PubMed] [Google Scholar]
- 32.Steven, B., R. Leveille, W. H. Pollard, and L. G. Whyte. 2006. Microbial ecology and biodiversity in permafrost. Extremophiles 10:259-267. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiago, I., A. P. Chung, and A. Verissimo. 2004. Bacterial diversity in a nonsaline alkaline environment: heterotrophic aerobic populations. Appl. Environ. Microbiol. 70:7378-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vishnivetskaya, T. A., M. A. Petrova, J. Urbance, M. Ponder, C. L. Moyer, D. A. Gilichinsky, and J. M. Tiedje. 2006. Bacterial community in ancient Siberian permafrost as characterized by culture and culture-independent methods. Astrobiology 6:400-414. [DOI] [PubMed] [Google Scholar]
- 36.Xiang, S., T. Yao, L. An, B. Xu, and J. Wang. 2005. 16S rRNA sequences and differences in bacteria isolated from the Muztag Ata glacier at increasing depths. Appl. Environ. Microbiol. 71:4619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon, J. H., S. T. Lee, and Y. H. Park. 1998. Inter- and intraspecific phylogenetic analysis of the genus Nocardioides and related taxa based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 48:187-194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.