Abstract
Two newly developed protocols for infective virus detection were compared to the plaque assay. An immunomagnetic separation procedure coupled with real-time reverse transcription-PCR of viral nucleic acids was developed to identify intact enteroviral particles, and a reporter cell system responding to viral replication based on fluorescent resonance energy transfer for detection of infectious enteroviruses was tested. Both new procedures detected infective viruses in environmental samples at the same level as the plaque assay.
In the United States, for regulatory purposes involving analysis of water (for example, drinking water sources) the total culturable virus assay is required for detection and enumeration of viruses. However, cell culture assays are expensive, labor-intensive, and time-consuming. Thus, simpler, more rapid detection methods based on detection of nucleic acids, such as PCR or reverse transcription-PCR (RT-PCR), are attractive. However, the results of methods based solely on PCR or RT-PCR often do not correlate with infectivity, as the presence of a viral genome does not necessarily indicate the presence of infectious viral particles, and thus these results are potentially false-positive results (9, 10). In addition, the need to concentrate large volumes (hundreds to thousands of liters) of water prior to analysis often results in accumulation of substances that inhibit PCR, which can cause false-negative results (5). Various techniques, such as immunomagnetic separation (IMS), have been proposed to avoid PCR inhibition (5, 7).
A protocol using IMS combined with a molecular beacon was developed previously (11). Magnetic beads (Dynabeads M-280 sheep anti-mouse immunoglobulin G; Dynal Biotech, Norway) coated with mouse anti-enterovirus monoclonal antibody (DakoCytomation, Denmark) were used to purify intact enteroviral particles, and the RNA genomes were analyzed by molecular beacon-based real-time RT-PCR to determine virus concentrations. A universal primer set (forward primer 5′-TCCTCCGGCCCCTGAATGCG-3′ and reverse primer 5′-ATGTCACCATAAGCAGCCA-3′) along with a molecular beacon probe was designed to target the conserved regions in the 5′ untranslated region in human enteroviruses (see the supplemental material). The probe (5′-CGAGCGGCGGAACCGACTACTTTGGGCGCTCG-3′; stem sequences underlined) was labeled with a fluorescent dye, 6-carboxyfluorescein, at the 5′ end and with a quencher, 4-(4′-dimethylaminophenylazo)benzoic acid, at the 3′ end. In order to assess the specificity of the IMS real-time RT-PCR, seven human enteroviruses were tested in duplicate. All enteroviruses were detected with a sensitivity of 1 PFU (average cycle threshold value, 38), while tests for adenovirus 2 (AdV2), AdV15, and hepatitis A virus (HAV) were negative (Table 1).
TABLE 1.
Specificity of the plaque assay, the reporter assay, and IMS RT-PCR
| Virus | Strain | Detection with:
|
||
|---|---|---|---|---|
| Plaque assaya | Reporter assayb | IMS RT-PCR | ||
| Adenovirus 2 | Sewage isolate | NAc | − | − |
| Adenovirus 15 | Sewage isolate | NA | − | − |
| Hepatitis A virus | GA76 | NA | − | − |
| Coxsackievirus B1 | Conn-5 | + | − | + |
| Coxsackievirus B3 | Nancy | + | − | + |
| Coxsackievirus B6 | Schmitt | + | − | + |
| Echovirus 11 | Gregory | + | + | + |
| Echovirus 17 | CHHE-29 | + | − | + |
| Echovirus 19 | Bruke | + | − | + |
| Poliovirus 1 | LscAb | + | + | + |
Plaque assay using BGMK cells as the host cells, which can detect 1 PFU per sample.
+, reporter assay detects 1 PFU per sample.
NA, not applicable.
Another recently developed method (4), in which a genetically engineered reporter cell line is used for detection of infectious viruses based on fluorescence resonance energy transfer (FRET), was tested. This reporter assay was performed as described previously (4). Briefly, cells were cultured in 16-well slide chambers to confluence, and monolayers containing approximately 105 cells were infected with viruses as indicated below. Reporter cells without viruses were used as the negative control. The cells were challenged with AdV2, AdV15, and HAV (100 or 1,000 PFU) to test substrate specificity of the 2A protease. Samples were mounted on glass slides 7.5 h postinfection and examined using a confocal microscope (Leica TCS SP2/UV). To observe the in vivo FRET phenomenon, the cells were visualized by fluorescent microscopy at excitation and emission wavelengths of 440 and 480 nm, respectively, for enhanced cyan fluorescent protein and at excitation and emission wavelengths of 480 and 520 nm, respectively, for enhanced yellow fluorescent protein. For microscopic examination of the infected cells, the well surface was divided into 20 image-capturing areas; each observation frame contained approximately 2,500 cells. The cells with an increased enhanced cyan fluorescent protein signal, indicating active virus replication, were counted and identified as infectious units. The specificity of the reporter assay was evaluated by challenging the reporter cells with the enteroviruses listed in Table 1, and only echovirus 11 (EV11) and poliovirus 1 (PV1) were detected with a detection limit of 1 PFU due to the high level of conservation in their 2Apro cleavage sequences.
Surface water from various sites in southern California was used to evaluate the performance of the two protocols. Water concentrates from several localities were pooled and confirmed to be negative for human enteroviruses and HAV by a plaque assay (3, 12). PV1 was added to the concentrate at a concentration of 1, 10, 100, or 1,000 PFU per 1-ml sample; each 1-ml sample was analyzed using IMS RT-PCR, the reporter assay, and the plaque assay in parallel. Dilutions of the PV1 stock were prepared, and the threshold cycles, determined by real-time RT-PCR, were regressed against the corresponding number of PFU, as determined by the plaque assay. Each data point was generated with five independent replications.
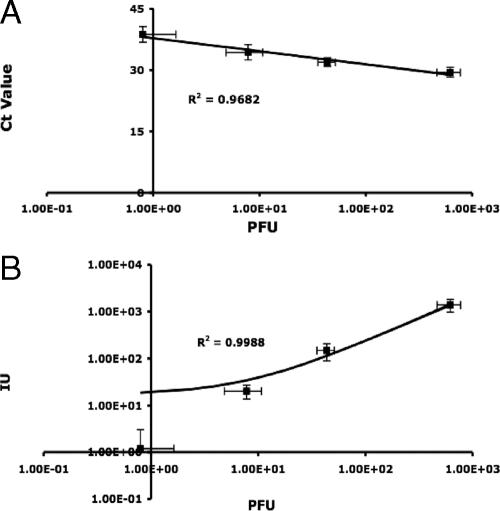
The same set of samples was used to evaluate the potential for environmental application of the reporter assay. Numbers of infected reporter cells, determined by microscopy as described above, were regressed against the corresponding number of PFU, as determined by the plaque assay. Each data point was generated from five independent experiments. Both IMS RT-PCR and the reporter assay exhibited a high level of correlation (R2 > 0.9) with the conventional plaque assay, indicating that the two methods performed as well as the plaque assay (Fig. 1), although we cannot state conclusively that the RT-PCR detected only infective particles. Analysis of variance confirmed that there was no significant difference between the three methods tested (P = 0.3982).
FIG. 1.
Pairwise comparison of IMS RT-PCR, reporter assay, and plaque assay. A standard curve of the viral concentration, determined by the standard plaque assay, versus the threshold cycle, obtained by real time RT-PCR, was generated (A). The correlation between the reporter assay and the plaque assay was established by challenging the reporter cells with 10-fold serial dilutions of the poliovirus stock, and the numbers of infected cells were plotted against the numbers of PFU (B). IU, infectious units; Ct, cycle threshold.
Nucleic acid amplification techniques involving RT-PCR have been used to detect enteroviruses rapidly (2). Various attempts to address the issue of infectivity, notably with integrated cell culture PCR and IMS, have been reported previously (1, 5, 8). To our knowledge, this is the first report of the development of an IMS RT-PCR using molecular beacon-based real-time PCR for the detection of human enteroviruses in environmental water samples. The IMS procedure using monoclonal antibodies provided a high degree of sensitivity and specificity (Fig. 1). The anti-enterovirus monoclonal antibody reacts with most of the enteroviruses; there is no cross-reaction with adenovirus, rhinovirus, or HAV (6). The one-step real-time RT-PCR using a universal primer set and the molecular beacon probe targeting all enteroviruses is a simplified procedure with excellent specificity (see the supplemental material). The IMS RT-PCR assay was designed to target a wide range of human enteroviruses; it identified all seven enteroviruses tested with the same sensitivity (Table 1). The reporter assay was designed for identification of infectious PV1, but it also responded to viruses with a similar protease cleavage site, such as EV11 (4). The reporter assay selectively detected infectious PV1 and EV11 but not other human enteroviruses tested in this study (Table 1). To our knowledge, this is the first report of using FRET for the detection of infectious viruses in environmental water samples. By taking advantage of the specificity between the viral protease and its cleavage sequence, it is possible to tailor a reporter cell line that responds exclusively to a given viral infection. It should also be possible to design a reporter cell line responding to all human enteroviruses by using a consensus linker sequence that is recognized by the 2A protease of all enteroviruses.
Here we describe development of an assay using IMS real-time RT-PCR to detect human enteroviruses. This assay was directly compared with the reporter cell assay and plaque assay for the detection of infectious human enteroviruses. The performance of each of the methods was assessed using concentrated surface water samples, and it was found that the two protocols provided the same detection sensitivity as the conventional plaque assay. The results of this study can be used to assist in making decisions about the method that should be used when information concerning the presence and number of infectious enteroviruses in an environmental sample is needed.
Supplementary Material
Acknowledgments
We thank Xinping Cui for her assistance with the statistical analysis.
Yu-Chen Hwang is a recipient of a U.S. Environmental Protection Agency Greater Research Opportunity fellowship.
Footnotes
Published ahead of print on 2 February 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abd El Galil, K., M. El Sokkary, S. Kheira, A. Salazar, M. Yates, W. Chen, and A. Mulchandani. 2004. Combined immunomagnetic separation-molecular beacon-reverse transcription-PCR assay for detection of hepatitis A virus from environmental samples. Appl. Environ. Microbiol. 70:4371-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corless, C., M. Guiver, R. Borrow, V. Edwards-Jones, A. Fox, E. Kaczmarski, and K. Mutton. 2002. Development and evaluation of a “real-time” RT-PCR for the detection of enteroviruses and parechovirus RNA in CSF and throat swab samples. J. Med. Virol. 67:555-562. [DOI] [PubMed] [Google Scholar]
- 3.Flehmig, B. 1981. Hepatitis A virus in cell culture. II. Growth characteristics of hepatitis A virus in Frhk-4/R cells. Med. Microbiol. Immunol. 170:73-81. [DOI] [PubMed] [Google Scholar]
- 4.Hwang, Y., W. Chen, and M. Yates. 2006. Use of fluorescence resonance energy transfer for rapid detection of enteroviral infection in vivo. Appl. Environ. Microbiol. 72:3710-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jothikumar, N., T. Cromeans, V. Hill, X. Lu, M. Sobsey, and D. Erdman. 2005. Quantitative real-time PCR assay for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 71:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipson, S., K. David, F. Shaikh, and L. Qian. 2001. Detection of precytopathic effect of enteroviruses in clinical specimens by centrifugation-enhanced antigen detection. J. Clin. Microbiol. 39:2755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myrmel, M., E. Rimstad, and Y. Wasteson. 2000. Immunomagnetic separation of Norwalk-like viruses (genogroup I) in artificially contaminated environmental water samples. Int. J. Food Microbiol. 62:17-26. [DOI] [PubMed] [Google Scholar]
- 8.Olsvik, O., T. Popvic, E. Skjerve, K. Cudjoe, E. Hornes, J. Ugelstad, and M. Uhlen. 1994. Magnetic separation techniques in diagnostic microbiology. Clin. Microbiol. Rev. 7:43-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwab, K., R. De Leon, and M. Sobsey. 1995. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl. Environ. Microbiol. 61:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skraber, S., B. Gassilloud, L. Schwartzbrod, and C. Gantzer. 2004. Survival of infectious poliovirus-1 in river water compared to the persistence of somatic coliphages, thermotolerant coliforms and poliovirus-1 genome. Water Res. 38:2927-2933. [DOI] [PubMed] [Google Scholar]
- 11.Tyagi, S., and F. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 12.U.S. Environmental Protection Agency. 1984. Manual of methods for virology. U.S. Environmental Protection Agency publication EPA 600-4-84-013. Government Printing Office, Cincinnati, OH.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.