Abstract
We investigated the genetic diversity of symbiotic bacteria associated with two newly discovered species of Osedax from Monterey Canyon, CA, at 1,017-m (Osedax Monterey Bay sp. 3 “rosy” [Osedax sp. MB3]) and 381-m (Osedax Monterey Bay sp. 4 “yellow collar”) depths. Quantitative PCR and clone libraries of 16S rRNA gene sequences identified differences in the compositions and abundances of bacterial phylotypes associated with the newly discovered host species and permitted comparisons between adult Osedax frankpressi and juveniles that had recently colonized whalebones implanted at 2,891 m. The newly discovered Osedax species hosted Oceanospirillales symbionts that are related to Gammaproteobacteria associated with the previously described O. frankpressi and Osedax rubiplumus (S. K. Goffredi, V. J. Orphan, G. W. Rouse, L. Jahnke, T. Embaye, K. Turk, R. Lee, and R. C. Vrijenhoek, Environ. Microbiol. 7:1369-1378, 2005). In addition, Osedax sp. MB3 hosts a diverse and abundant population of additional bacteria dominated by Epsilonproteobacteria. Ultrastructural analysis of symbiont-bearing root tissues verified the enhanced microbial diversity of Osedax sp. MB3. Root tissues from the newly described host species and O. frankpressi all exhibited collagenolytic enzyme activity, which covaried positively with the abundance of symbiont DNA and negatively with mean adult size of the host species. Members of this unusual genus of bone-eating worms may form variable associations with symbiotic bacteria that allow for the observed differences in colonization and success in whale fall environments throughout the world's oceans.
Bone-eating worms of the genus Osedax (Polychaeta: Siboglinidae) are among the most unusual of all marine animals. Although the genus was recently discovered, four named and five unnamed species are now known from whalebones found worldwide at depths ranging from 25 to 3,000 m (7, 9, 12, 26; Y. Fujiwara, personal communication; C. E. Braby, S. B. Johnson, W. J. Jones, G. W. Rouse, and R. C. Vrijenhoek, unpublished data). These mouthless and gutless worms possess an unusual adaptation, highly branched, posterior “roots” that extract nutrients from sunken bones. Goffredi et al. (14) previously proposed that this nutritional strategy relies on intracellular heterotrophic microbes of the gammaproteobacterial order Oceanospirillales, which are known to metabolize complex carbon compounds. Their study revealed evidence from stable isotopes and lipid analysis for carbon transfer from whalebones through the endosymbiont to the worm host, but the specific biodegradative and catabolic role of the symbionts remains unknown. Nonetheless, together, the host and symbiont can rapidly remineralize recalcitrant carbon compounds from mammalian bones and recycle them to the surrounding seafloor community.
Although morphologically similar, Osedax species differ ecologically. For example, Osedax rubiplumus, a rapid colonizer of new whalebones, was completely replaced by Osedax frankpressi in less than 10 months (12; Braby et al., unpublished). Additionally, novel species of Osedax have been found at experimental whale falls implanted at various depths in Monterey Bay, each experiencing unique methane, sulfide, and oxygen levels and temperature regimens (Braby et al., unpublished; V. J. Orphan, personal communication). Thus, it is reasonable to suggest that physiochemical differences may play a role in the ecological distribution of Osedax species as they do for other species in Monterey Bay (10).
In this study, we examine symbiont diversity in Osedax isolates from Monterey Bay. Our goals were to characterize the symbiont identity and abundance and to explore the mechanisms of symbiont and nutrient acquisition in these worms. This study was stimulated by the discovery of two new Osedax species on whalebones from different depths: Osedax Monterey Bay species 3 (Osedax sp. MB3) from 1,017-m and Osedax Monterey Bay species 4 (Osedax sp MB4) from 381-m depths. The new species differ morphologically and genetically from previously described Osedax species, and formal descriptions are under way (G. Rouse, unpublished data). For now, we refer to the new worms as Osedax sp. MB3 and Osedax sp. MB4 (also called “rosy” and “yellow collar,” respectively [Braby et al., unpublished]). Herein, we characterize the diversity and abundance of symbionts associated with the new species and compare them with associations previously observed for both O. rubiplumus and O. frankpressi. To learn more about the early development of Osedax symbiosis, we investigated juveniles of Osedax frankpressi that had recently colonized experimentally deployed whalebones. Additionally, we conducted a comparative study of all three species to determine their capacities for degrading collagen, a primary protein constituent of bony tissues.
MATERIALS AND METHODS
Specimens.
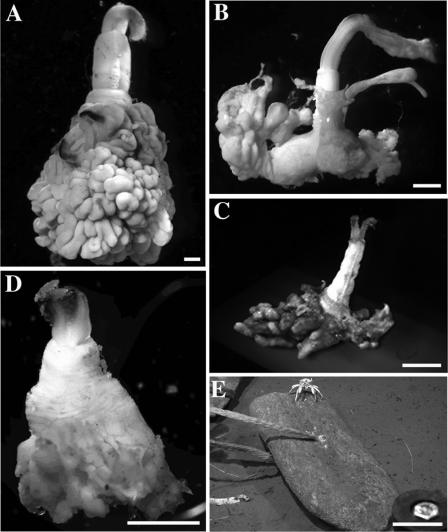
All specimens were collected in Monterey Canyon from May 2004 to January 2006 (Table 1 and Fig. 1). Osedax sp. MB3 was collected at 36.78°N, 122.08°W, at a 1,017-m depth, during R/V Tiburon dives T776, T916, and T931 (Fig. 1B). Osedax sp. MB4 was collected at 36.79°N, 121.89°W, at a 381-m depth, during R/V Tiburon dive T933 (Fig. 1C). Osedax frankpressi “recruits” were collected at 36.63°N, 122.43°W, at a 2,891-m depth, during R/V Tiburon dive T769 (Fig. 1D). Recruits colonized whalebones that were implanted ∼10 m from the natural whale fall at 2,891 m (Fig. 1E) (14). Osedax frankpressi adults (Fig. 1A) (collected at 36.63°N, 122.43°W at a 2,891-m depth, R/V Tiburon dives T742 and T932) were included for molecular and enzymatic comparisons. Representative samples were surface treated with 100% ethanol and frozen at −80°C or preserved directly in paraformaldehyde for fluorescence in situ hybridization (FISH)microscopy (see specifics below).
TABLE 1.
Osedax species used in this studya
| Species | Depth (m) | Latitude, longitude | Date (dive) | Individual no. | Intraspecific symbiont variation |
|---|---|---|---|---|---|
| O. frankpressi | 2,895 | 36.613, −122.434 | 9/2004 (T742), adults | 7 | 0.020 |
| 10 | |||||
| 11/2004 (T769), juveniles | 1 | ||||
| 2 | |||||
| 3 | |||||
| Osedax sp. MB3 | 1,017 | 36.771, −122.083 | 1/2005 (T776) | 19 | 0.012 |
| 28 | |||||
| 29 | |||||
| 30 | |||||
| 1/2006 (T931) | 1 | ||||
| 2 | |||||
| Osedax sp. MB4 | 381 | 36.790, −121.887 | 1/2006 (T933) | 1 | 0.040 |
| 2 |
FIG. 1.
Osedax spp. (A) Adult O. frankpressi. (B) Osedax sp. MB3 (“rosy”). (C) Osedax sp. MB4 (“yellow collar”). (D) Juvenile O. frankpressi recruited to implanted whalebones. (E) Implanted whalebones (partial jawbone of a blue whale). All bars are 1 mm, except in E (bar, 70 cm).
DNA analysis.
The DNEASY kit (QIAGEN, Valencia, CA) was used to extract total DNA from frozen tissue. DNA concentrations were determined fluorometrically using a Hoefer DyNA Quant 200 (Amersham Pharmacia Biotech) and Hoechst dye 33258. DNA samples (2 μl) were added to 2 ml fresh assay solution (containing 1 μg ml−1 H33258 in TNE buffer [0.2 M NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 7.4]), and concentrations were compared against a standard concentration curve generated using salmon testis DNA (10 to 1,000 μg ml−1 in TNE buffer). For PCR amplification, a 1,465-bp fragment of the 16S rRNA gene was generated using bacterium-specific primers (27F and 1492R) (18). PCR products were cloned (Invitrogen Topo TA cloning technology) and sequenced (CEQ 8800 [Beckman Coulter] or ABI 3100 [Applied Biosystems]). Bacterial clone libraries were constructed from the roots of six individual worms of Osedax sp. MB3 (n = 3 of each, collected from dives 12 months apart), two individual worms of Osedax sp. MB4, and three individual recruits of O. frankpressi (Tables 1 and 2 and Fig. 2). Discrepancies in the number of clones examined for each host species were a consequence of the differences in bacterial diversity among species (Table 2). For example, the larger number of clones (n = 259) investigated for Osedax sp. MB3 was necessary to achieve saturation for the total microbial ribotypes observed for that particular species. Intervening gaps and ambiguities were excluded from the analysis, resulting in the inclusion of 1,236 bp. Additional sequences were obtained from GenBank and compiled and aligned with our sequences using ARB Fast Aligner and Sequencher v4.5 (Gene Codes). 16S rRNA gene clone libraries were screened for chimeric sequences using the program Mallard (3).
TABLE 2.
Bacterial ribosomal 16S rRNA clone library results for Osedax spp.a
| Microbial group and subgroup | No. of clones
|
Representative ribotype ID (GenBank accession no.) | ||
|---|---|---|---|---|
| Osedax frankpressib (2,895 m) | Osedax sp. MB3 (1,017 m) | Osedax sp. MB4 (381 m) | ||
| Oceanospirillales | 42 | 95 | 85 | |
| Phylotype 1 | 95 | T776.29.G3 (DQ911537) | ||
| T931.1.B1 (DQ911539) | ||||
| Phylotype 4 | 42 | T769r.1.B10 (DQ911535) | ||
| Phylotypes 5 and 6 | 85 | T933.1.G2 (DQ911544) | ||
| T933.2.H5 (DQ911547) | ||||
| Epsilonproteobacteria | 4 | 111 | 1 | |
| Arcobacter-related | 3 | 53 | T776.29.15 (EF100882) | |
| T776.29.C8 (EF100883) | ||||
| T931.1.A8 (EF100886) | ||||
| Sulfurimonas-related | 43 | T776.29.D3 (EF100884) | ||
| T776.29.H8 (EF100885) | ||||
| T931.1.H4 (EF100889) | ||||
| Sulfurospirillum-related | 15 | 1 | T776.29.11 (EF100881) | |
| T931.1.D7 (EF100888) | ||||
| Other groups | 4 | 53 | 5 | |
| CFB | 3 | 28 | 2 | T933.1.E3 (EF117251) |
| Low-G+C gram positive | 0 | 8 | 1 | T776.30.A8 (EF117250) |
| Gammaproteobacteria | 0 | 5 | 1 | T776.28.F8 (EF117249) |
| Unknown/other | 1 | 12 | 1 | NA |
| Total clones sequenced | 50 | 259 | 91 | |
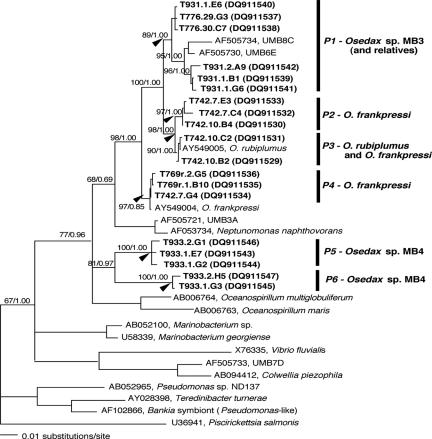
Only dominant representative ribotypes are listed. Total numbers for each are shown in boldface type. There were three individual O. frankpressi juveniles, six individuals for Osedax sp. MB3, and two individuals for Osedax sp. MB4. Osedax sp. MB3 individuals were collected in January 2005 (dive T776) and January 2006 (dive T931). Phylotypes (P1 to P6) are internal clades within the Oceanospirillales-related symbionts of Osedax (Fig. 2). Ribotype names (dive.individual.clone) correspond to taxon labels in Fig. 2 and 3. NA, not applicable.
Osedax frankpressi juveniles from dive T769.
FIG. 2.
Phylogenetic relationships, based on 16S rRNA, between free-living members of the Oceanospirillales (Gammaproteobacteria) and the symbiont ribotypes associated with Osedax worms (boldfaced taxa designated by dive.individual.clone) (Table 2). The number following the dive represents an individual worm, and the following letter/number is a unique 16S sequence from clone libraries. GenBank accession numbers for sequences acquired during this study are DQ911529 to DQ911547. The maximum likelihood tree was generated from sequences treated with the GTR+I+G model. Arrows at nodes indicate support for the internal clade (phylotypes P1 to P6) designations. The numbers at the nodes represent bootstrap values (maximum likelihood)/posterior probability values (Bayesian).
Phylogenetic analyses, including maximum parsimony (heuristic search option with tree bisection reconnection branch-swapping algorithm) and maximum likelihood (general time-reversible plus proportion invariant plus gamma [GTR+I+G model), were performed using PAUP (v4.0b10) (30). To choose the most appropriate model for substitution, we tested a set of 30 models of sequence evolution with restrictions of the GTR model (31, 35), including six substitution classes, unequal base frequencies, and rate variations among sites. Rate variation was modeled as gamma-distributed rates, proportion of invariant sites, and site-specific rates based on the inferred (by manual alignment in ARB) stem-loop structure of the Escherichia coli 16S region. We assumed a neighbor-joining topology and maximized the likelihood function for the candidate models using standard procedures in PAUP (v4.0b10) (30). The relative fit of models was assessed by the Akaike information criterion, where the maximum likelihood score and the number of free parameters of the model were taken into account (1, 25). The GTR+I+G substitution model was the best-fit model for these data (Akaike information criterion score = 15,015) and was used for all parametric analyses. We estimated a Bayesian phylogenetic tree using MrBayes (version 3.0b4) (17) with Monte Carlo Markov chain lengths of 5.1 × 107 generations. The chains were sampled every 1,000 generations in order to reduce autocorrelation.
QPCR.
Quantitative PCR (QPCR) reactions for all DNA extracts (described above) were conducted in triplicate and contained 10 μl SYBR green PCR master mix (Applied Biosystems), 6.5 μl RNase- and DNase-free deionized water, 1 μl DNA sample (DNA extracts ranged from 10 to 160 ng μl−1 but were normalized to 10 ng μl−1 and diluted 100 times), and final primer concentrations of 300 nM for bacterial 16S rRNA and 200 nM for eukaryote 18S rRNA. The 154-bp partial bacterial 16S rRNA gene target was amplified using primers 303F (5′-CACCGGCAGTCTCCTTAGAG-3′) (melting temperature, 60.0°C) and 457R (5′-GAGATAGCTTGGTGCCTTCG-3′) (melting temperature, 60.0°C), designed using Primer3 (27). These primers were empirically determined to amplify all clones from a 16S rRNA gene library of Osedax sp. MB3 microbes (n = 96) (Table 1) and were thus considered to be adequate to assess microbially diverse populations within these species. In addition, a 181-bp partial eukaryote 18S rRNA gene target was amplified using primers Euk338F and Euk519R (15). Serial dilutions of the pCR4 plasmid (Invitrogen) that contained either a representative partial symbiont 16S rRNA gene sequence (isolated from Osedax sp. MB3 using primers 27F and 1492R) (18) or a partial eukaryote 18S rRNA gene sequence (isolated from Osedax sp. MB4 using general primers 82F and 1520R) (19) were used as standards for the QPCR assays. Plasmids used as standards for the QPCR assays were purified using a QIAGEN Mini plasmid prep kit (QIAGEN, Valencia, CA). Additionally, quantified E. coli DNA was used as a negative control for the eukaryote 18S rRNA gene assay and a positive control for the bacterial 16S rRNA gene assay. Slopes of standard curves (regression lines of cycle threshold [CT] versus log N, the log of the initial DNA concentration in standard templates) were used to estimate amplification efficiency in our QPCR assays. Primer set efficiencies were ∼120% for both sets (slope of CT versus concentration = −2.9). QPCR assays were run on an ABI Prism 7700 sequence detector system under the following thermal conditions: incubation for 2 min at 50°C and Taq activation for 10 min at 95°C, followed by 40 cycles of 15 s of denaturation at 95°C and 60 s of annealing/extension at 55°C. A dissociation curve from each QPCR reaction was examined to further ensure proper target sequence amplification. DNA abundance was calculated from the number of cycles necessary for the fluorescence to exceed a set threshold value (CT) relative to standard controls with known DNA concentrations. Average numbers of 16S rRNA operons per symbiont genome and genome copies per symbiont cell are not known for Osedax. For invertebrate hosts from reducing habitats, estimates of total symbiont rRNA gene copy abundance range from ∼1 × 1010 to 9 × 1010 g−1 (wet weight), while cell abundance estimates usually range from 109 to 1011 cells g−1 (wet weight) symbiont-containing tissue (20). In this study, the ratios of bacteria 16S rRNA to eukaryote 18S rRNA are given in Table 3, after subtracting the difference from the negative control.
TABLE 3.
Weights, QPCR results, and collagenolytic activity for whole individuals of various Osedax speciesa
| Osedax speciesb | Mean ± SD (no. of individuals)
|
|||
|---|---|---|---|---|
| Wt (g) | QPCR ratio of bacterium/host | Collagenolytic activity (U g−1 [wet wt])
|
||
| Type I | Type IV | |||
| O. frankpressi adult | 0.34 ± 0.17 (10) | 0.38 ± 0.13 (7) | 5.9 ± 4.4 (3) | 0.9 ± 0.3 (2) |
| O. frankpressi recruit | NM | 0.44 ± 0.06 (2) | NM | NM |
| Osedax sp. MB3 | 0.03 ± 0.01 (10) | 0.57 ± 0.15 (6) | 6.8 ± 4.3 (3) | 1.9 ± 1.6 (2) |
| Osedax sp. MB4 | 0.01 ± 0.01 (6) | 1.63 ± 0.21 (4) | 18.4c (1) | 5.8c (1) |
QPCR results are shown as ratios of bacterial 16S versus eukaryote 18S rRNA abundance. NM, not measured.
Activities listed are for whole individuals.
Due to their small size, activity measurements for collagen types I and IV were made on separate individuals of Osedax sp. MB4.
FISH microscopy.
Osedax species ovisac/root tissue was prepared for in situ hybridizations by an initial 3-h preservation at 4°C in phosphate-buffered 4% paraformaldehyde and subsequent transfer to 1× phosphate-buffered saline-ethanol (1:1). Tissue samples were embedded in Steedman's wax (1 part cetyl alcohol and 9 parts polyethylene glycol 400 distearate, mixed at 60°C) (28) according to the protocol described previously by Pernthaler and Pernthaler (24) and sectioned (6 to 40 μm thick on Superfrost Plus slides; Fisher Scientific) using a Leica RM2125 manual microtome. Slides were dewaxed by three rinses in 100% ethanol (10 min each), followed by rehydration in 70% ethanol (10 min). Hybridization buffers and wash buffers were made as described previously by using 35% formamide in the hybridization buffer and 450 mM NaCl in the wash solution (8). Formamide stringency was determined previously (14). Hybridizations were conducted at 46°C for 2 to 8 h, followed by a 15-min wash at 48°C. Sections were stained with a dilute 4′6′-diamidino-2-phenylindole (DAPI) solution (5 μg ml−1) for 1 min and examined under epifluorescence microscopy using a DeltaVision restoration microscopy system (Applied Precision). The Cy3-labeled oligonucleotide probe specific to Osedax endosymbionts was designed from the 16S rRNA gene and designated sym435 (sequence, 5′-CTTTCCTCACWGCTGAA-3′; E. coli positions 435 to 452, modified slightly from sym435_I described previously by Goffredi et al. [14] to accommodate all known Osedax endosymbionts). A Cy3-labeled bacterial universal probe set (EUB338, EUB338 II, and EUB338 III, E. coli positions 338 to 355) (2) and a gammaproteobacterial universal probe, Gam42a (21), were used as positive controls. A Cy5-labeled general probe targeting the 16S rRNA gene of eukaryotes, Euk516, was also used (2). An additional probe (CF319a [5′-TGGTCCGTGTCTCAGTAC-3′]) (21) was used to test for the presence of Cytophaga-related bacteria associated with Osedax sp. MB3. FISH microscopy was conducted on three individual worms of Osedax sp. MB3, two individual worms of Osedax sp. MB4, and two individual recruits of O. frankpressi (Fig. 3 and 4).
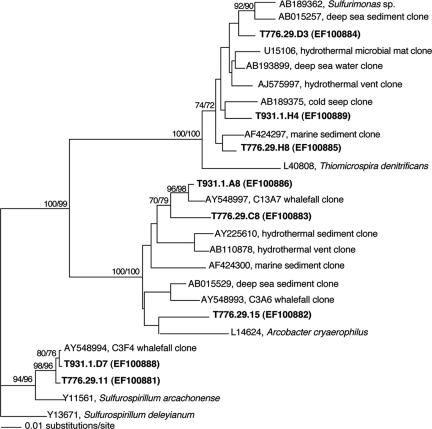
FIG. 3.
Phylogenetic relationships, based on 16S rRNA, between free-living members of the Epsilonproteobacteria and bacterial ribotypes associated with Osedax sp. MB3 (boldfaced taxa designated by dive.individual.clone) (Table 2). The number following the dive represents an individual worm, and the following letter/number is a unique 16S sequence from clone libraries. GenBank accession numbers for sequences acquired during this study are EF100881 to EF100889. The numbers at the nodes represent bootstrap values (neighbor joining/parsimony).
FIG. 4.
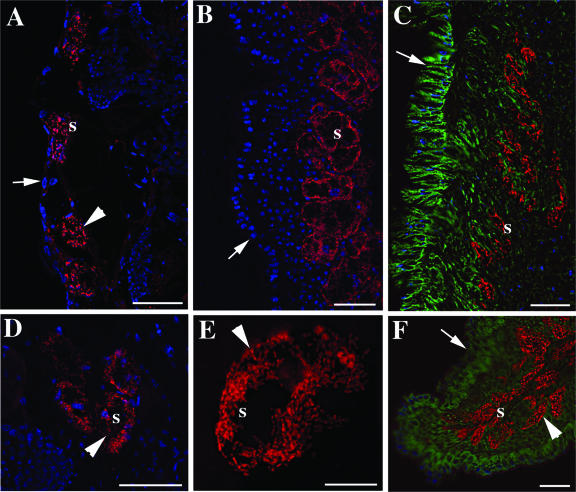
Osedax species symbionts. Images of FISH microscopy of the primary symbiont within the roots of Osedax sp. MB3 (A and D), Osedax sp. MB4 (B and E), and juvenile recruits of O. frankpressi (C and F) are shown. A to F are hybridizations with the symbiont-specific sym435 probe labeled with Cy3 (shown in red). C and F are dual stained with a eukaryote-specific Cy5-labeled probe (shown in green). All images are embedded sections (7 to 20 μm thick) that were also stained with DAPI prior to imaging (shown in blue, except in D). Images were captured with softWoRx 3.4.4 (Applied Precision). Intact bacteriocytes (arrowheads) and the external surface of root tissue (arrows) are shown. s, symbiont. All bars are 50 μm, except in E (bar, 10 μm).
Collagenolytic activity assays.
Collagenolytic activity was measured using the EnzChekGelatinase/Collagenase assay kit (Molecular Probes, Eugene, OR). Reaction mixtures contained 20 μl substrate, including both type I and type IV collagen conjugated with fluorescein (initial concentration, 1 mg ml−1), 80 μl 1× reaction buffer (containing 0.5 M Tris-HCl, 1.5 M NaCl, 50 mM CaCl2, 2 mM sodium azide, pH 7.6), and 100 μl diluted sample (homogenized in 20 to 80 volumes [vol/wt] of reaction buffer) or 100 μl reaction buffer as the negative control. Negative controls also included samples exposed to a general inhibitor of metalloproteinases (1,10-phenanthroline [30 mg ml−1]) or samples boiled for 10 min. Commercially available collagenase isolated from Clostridium histolyticum was used (0.05 to 0.2 U ml−1) to generate standard curves. Enzyme activity was measured in U g−1 (wet weight), with 1 U equal to the amount of enzyme necessary to liberate 0.2 μmol glycine h−1. Reaction mixtures were incubated in darkness between 2 and 18 h, and fluorescence intensity was measured using a SynergyHT microplate reader (Bio-Tek Instruments, Inc., Winooski, VT) set for excitation at 485 ± 20 nm and emission detection at 540 ± 25 nm.
Statistics.
The Wilcoxon Mann-Whitney test was used to test for differences in distribution between data sets for QPCR, collagenolytic activity, and weight measurements. Similarly, Kendall's τ nonparametric test was used to test for correlations between data sets (JMP v6.0; SAS Institute Inc.). Simpson's diversity index (D) was used to estimate overall microbial diversity associated with each host species.
Nucleotide sequence accession numbers.
GenBank accession numbers for sequences acquired during this study are DQ911529 to DQ911547, EF100881 to EF100889, and EF11724 to EF117251.
RESULTS
Size differences among Osedax species.
Osedax sp. MB3 and MB4 lack functional mouths and guts and instead possess a large posterior ovisac sheathed in green symbiont-containing tissue, as seen in other Osedax species (Fig. 1) (14). The new species differ significantly in weight (P = 0.0039, Wilcoxon test), with means of 0.03 ± 0.01 g (∼6 mm in height) for Osedax sp. MB3 and 0.01 ± 0.01 g (∼4 mm in height) for Osedax sp. MB4 (Table 3). They are much smaller than adult O. frankpressi (0.34 ± 0.17 g; ∼15 mm in height) and O. rubiplumus (1.2 ± 0.5 g; ∼25 mm in height) isolates from Monterey Canyon but comparable to Osedax mucofloris isolates recovered from off the coast of Sweden (∼5.5 mm in height) (9). The root tissue of recruits of O. frankpressi appeared to be morphologically different from those of adults. Juvenile root tissues were yellow, instead of the brilliant green, as seen in adults, and the structural complexity of this tissue appeared to be less developed (e.g., fewer convolutions and less vascularization) (Fig. 1D versus an adult in A).
Oceanospirillales-symbiont diversity.
Osedax sp. MB3 and MB4 hosted bacterial 16S rRNA ribotypes (Table 2) that were 91 to 98% similar to Oceanospirillales ribotypes previously associated, intracellularly, with O. frankpressi (Osedax_sym1) (GenBank accession number AY549004) and O. rubiplumus (Osedax_sym2) (GenBank accession number AY549005). Together, these ribotypes, along with several free-living ribotypes (for example, UMB6E and UMB8C, isolated from Boston Harbor), form a group within the Gammaproteobacteria (Fig. 2). Within this group, we refer to well-supported internal clades as phylogenetic types or phylotypes (P1 to P6) (Fig. 2). The Oceanospirillales phylotypes (P5 and P6) recovered from Osedax sp. MB4 (dive T933) differed by 6 to 9% from other Oceanospirillales compared in this study (Fig. 2). Together, Oceanospirillales phylotypes comprised 94% of the 90 16S ribotypes recovered from Osedax sp. MB4 (Table 2). The Oceanospirillales phylotype (P1) recovered from Osedax sp. MB3 (dives T776 and T931) differed by 2 to 4% from the previously described phylotypes (Fig. 2), but they comprised only 37% of the total bacterial diversity observed in this host species (other microbes are considered below). Oceanospirillales recovered from juvenile O. frankpressi (T769r) differed by <0.4% from the Osedax_sym1 phylotype (P4; from adult O. frankpressi) and comprised 84% of the clones recovered from this species (Table 2). Phylotypes P2, P3, and P4 represent diversity observed in O. frankpressi symbionts (dive T742, September 2004) collected after the initial individuals described previously by Goffredi et al. (dive T486, October 2002) (14).
The diversity of Oceanospirillales found in Osedax worms varied among host species and across time. The Oceanospirillales associated with Osedax sp. MB4 were the most divergent (average intraspecific divergence of 4%) (Table 1 and Fig. 2). For example, both individuals T933_1 and T933_2 hosted two phylotypes (P5 and P6) (Fig. 2). Osedax sp. MB3 hosted ribotypes (within phylotype P1) that differed by as much as 3.2% in individual T931_1 (average intraspecific divergence of 1.2%) (Table 1 and Fig. 2). Additionally, Osedax sp. MB3, obtained in January 2006 (dive T931), possessed Oceanospirillales ribotypes that differed by ∼2.0% 16S rRNA divergence from the previous collection (January 2005, dive T776), with the exception of one ribotype (T931_1_E6). Individual worms from dive T776 did not reveal evidence of multiple infections (i.e., divergence) by Oceanospirillales. Similarly, Osedax frankpressi juveniles sampled for this study (T69r) also exhibited no evidence of multiple infections, but ribotypes found in the juveniles differed from those of adult O. frankpressi individuals sampled from the same whale carcass 2 months earlier (T742). Two adult O. frankpressi individuals (T742_7 and T742_10) were infected by Oceanospirillales strains that differed by as much as 3.3% for 16S rRNA (average intraspecific divergence of 2%) (Table 1 and Fig. 2).
Overall microbial diversity.
In addition to the “primary” Oceanospirillales symbiont, root tissues of Osedax sp. MB3 included a number of other microbial ribotypes, including members of the Epsilonproteobacteria (43% of clone library representatives), Cytophaga-Bacteroides (8%), and fusobacteria (3%) (Table 2). This pattern was observed in different individuals collected 1 year apart, in January 2005 (during dive T776) and January 2006 (T931) (comparison not shown), and for an individual adult of Osedax sp. MB3 that grew on cow bones deployed near the 1,081-m whale fall (30% Oceanospirillales, 44% Epsilonproteobacteria, and 16% Cytophaga-related bacteria, based on clone libraries) (W. J. Jones, personal communication). The majority of the epsilonproteobacterial ribotypes were related to sulfur-oxidizing Arcobacter (53 of 111 clones) and Sulfurimonas (45 of 111 clones) species and relatives of Sulfurospirillum (15 of 111 clones) (Table 2 and Fig. 3) (see reference 4 for groupings). Many of these ribotypes are most closely related to specific ribotypes previously recovered from the whale fall bones themselves (14), including the Arcobacter relatives C3A6 (GenBank accession number AY548993) and C13A7 (accession number AY548997) and the Sulfurospirillum-related strain C3F4 (accession number AY548994) (Fig. 3). Similarly, many Cytophaga strains recovered from the worms were most closely related to C3D9 (accession number AY548988) and C3C8 (accession number AY548984) and the low-G+C relative strain C3C9 (accession number AY548990). FISH microscopy, however, was negative for the presence of Cytophaga-related bacteria associated with Osedax sp. MB3 (Table 2).
Measures of Simpson's diversity index (D) reflected the increased overall microbial diversity within Osedax sp. MB3, the host species with the highest observed diversity of rRNA sequences (Simpson's D = 5.7) compared to Osedax sp. MB4 and juvenile O. frankpressi, with lower bacterial diversities and Simpson's D values of 1.1 and 1.4, respectively.
Symbiont abundance in host tissues.
The ratio of bacterial to eukaryote rRNA in whole Osedax individuals was estimated by QPCR analysis. Ratios varied from ∼0.4 to 1.6 among Osedax species (Table 3), with the lowest values observed in both adult and juvenile O. frankpressi individuals and the highest ratios found in Osedax sp. MB4. A significant inverse correlation existed (P = 0.0075, Kendall's τ) between the abundance of bacterial DNA and adult host size. The highest ratio (1.6) occurred in the smallest species, Osedax sp. MB4, an intermediate value (0.57) occurred in Osedax sp. MB3, and the lowest value (0.38) occurred in the largest species, O. frankpressi (Table 3). It should be noted that O. frankpressi recruits had a ratio similar to that of the adults (ratio of 0.44; P = 0.38, Wilcoxon test) (Table 3), despite their small size. Thus, the inverse correlation between adult size and bacterial loads appears to be a species-level trait and not directly a function of individual size.
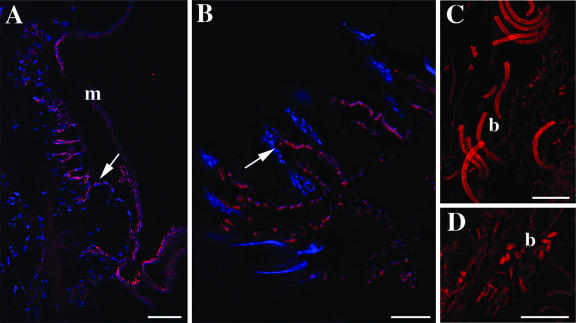
FISH microscopy, using the sym435 probe targeting the Oceanospirillales symbiont, corroborated the patterns seen in clone libraries and QPCR analysis. Root tissues from Osedax sp. MB4 demonstrated high numbers of symbiont-containing cells (i.e., bacteriocytes) (Fig. 4B and E). In contrast, the Oceanospirillales symbiont sparsely populated root tissues of Osedax sp. MB3, although it was concentrated within similar, yet scattered, bacteriocytes (Fig. 4A and D). For both of the new species and O. frankpressi juveniles, the Oceanospirillales symbionts were concentrated mostly in a distinct zone along the inner margins of the root tissue. This was particularly apparent when host tissue was stained in addition to the symbiont (Fig. 4C and F). Consistent with the clone libraries, Osedax sp. MB3 also had high numbers of bacteria that were unrelated to Oceanospirillales; negative results were obtained with the sym435 and Gam42 probes. These other bacteria were always external, some in intimate contact with the epithelial margin, but also concentrated in mucous surrounding the animal (Fig. 5A to D).
FIG. 5.
Additional bacteria associated with Osedax sp. MB3. All are hybridizations with a general bacterial EUB338 probe labeled with Cy3 (shown in red). Bars, 50 μm (A) and 10 μm (B to D). m, mucous layer; b, bacteria.
Collagenolytic enzyme activities.
An ability to degrade collagen was detected in all Osedax species examined in this study (Table 3). The reported collagenolytic enzyme activities are for whole organisms, which facilitated a comparison with QPCR results; however, only the symbiont-containing tissue (e.g., root tissue) had detectable collagenolytic enzyme activity. The ability to degrade collagen types I and IV corresponded with estimates of bacterial abundance (Table 3). Osedax sp. MB4, with the greatest abundance of the Oceanospirillales symbiont, had the highest collagenolytic activity (three to six times greater than those of the other two species).
Osedax sp. MB4 was significantly different from the other species when the degradations of collagen types I and IV were considered together (P < 0.09, Wilcoxon test). Osedax frankpressi and Osedax sp. MB3 possessed similar intermediate collagenolytic activities (P = 0.4 to 0.5, Wilcoxon test). The rate of collagenase activity in Clostridium, the positive standard, was three times faster than that of the Osedax species. This finding suggests that the enzymes responsible for this activity in Osedax may not be true collagenases but rather may be proteases with the capability of cleaving amino acids from the very tightly wound collagen molecule (32). Unfortunately, due to small sample sizes, general protease activity could not be measured directly in Osedax tissues.
DISCUSSION
For members within the genus Osedax, ecological differences exist, including general site preference/colonization success, temporal and spatial patterns, and more specific substrate preference (e.g., rib versus vertebral bones). For example, different kinds of bones (e.g., enamel-covered long bones versus vertebrae) seem to be favored by O. rubiplumus and O. frankpressi, respectively. Additional differences in depths of the carcass, temperature and pressure, and specific chemical conditions (sulfide-rich versus methane-rich) exist among Osedax environments (Braby et al., unpublished). Environmental boundaries experienced by these worms may be influenced, in large part, by the specific nature of their integration with local microbes. Two new species of Osedax appear to be very different in the ultrastructures, biomass, and potential metabolic capacities of their symbiotic populations. Variation among closely related species, including metabolism of both symbiont and host, symbiont biomass, and growth rates of both partners, has been shown to dramatically affect the ecology of symbiotic associations (10).
Although both new species of Osedax possess an Oceanospirillales-type symbiont within intracellular bacteriocytes, differences in bacterial abundance exist among the Osedax species investigated in this study. Osedax sp. MB4 is the smallest species in Monterey Bay, and symbiont abundance represented a proportionally greater fraction of the host. Symbiont abundance relative to the host varies dramatically in other associations and is influenced by a number of factors, including environmental variation, nutritional source, and developmental stage of the host (5, 6, 11, 34). Greater relative symbiont abundance might contribute to greater nutrition and more rapid growth rates, which might be critical for species that exploit unevenly distributed ephemeral resources. The transfer of symbiont nutrients to the host, whether via digestion of the symbionts directly or via translocation of nutrients from the symbionts, may also be related to microbial proliferation rates and abundance (33). Two previous studies provided evidence for the direct digestion of symbionts by Osedax. First, the incorporation of membrane-bound symbiont-produced fatty acids into host cellular components (e.g., wax esters), a process that would require symbiont digestion by the host, has been demonstrated previously (14). Second, there is new evidence of degradation of entire symbiont-containing bacteriocytes as an initiation to symbiont digestion in both O. rubiplumus and O. frankpressi (S. Katz and M. Bright, unpublished data).
Unlike Osedax sp. MB4, Osedax sp. MB3 possessed additional external bacteria that were distinct from the primary internal Oceanospirillales symbiont. FISH microscopy identified all internal bacteria as the Oceanospirillales symbiont, which was supported by a dominance of this ribotype in the clone libraries (Table 2). FISH microscopy, however, also revealed numerous bacteria associated with the outer mucous layer of Osedax sp. MB3 that did not hit with the gammaproteobacterial or Oceanospirillales symbiont-specific probes but that did hit with a general bacterial probe (Fig. 5). Clone libraries for whole animals revealed that ∼63% of the recovered ribotypes consisted of phylogenetically distinct groups, including Epsilonproteobacteria and Cytophaga/Fusobacterium (Table 2). These groups are possibly associated with the exterior of this particular worm species (although initial FISH microscopy using a Cytophaga-specific probe was negative); however, additional FISH microscopy is necessary to confirm this.
The presence of external bacteria on Osedax sp. MB3 was consistent for individual worms collected 12 months apart as well as for an individual adult of Osedax sp. MB3 that grew on cow bones deployed nearby, suggesting a persistent association (W. J. Jones, personal communication). This was demonstrated both microscopically and by the presence of non-Oceanospirillales bacteria in the clone libraries, with consistent percentages of the dominant groups, including Epsilonproteobacteria (43 to 44% of recovered ribotypes) and Cytophaga/Fusobacterium (8 to 16%). Previously, bacteria other than Oceanospirillales endosymbionts were found to comprise less than 10% of the microbes associated with Osedax hosts (14). The greater fraction of external bacteria associated with Osedax sp. MB3 is unique to this species. Many of the recovered bacterial phylotypes, however, are related to those recovered within the whale fall environment itself (i.e., bones and surrounding sediment) (14; V. J. Orphan, personal communication). It is not known whether these external bacteria confer an additional nutritional capability or another unknown influence on the worm. The association of invertebrate hosts with both internal and external bacteria is not unprecedented. For example, a recently discovered snail from hydrothermal vents in the Indian Ocean demonstrates partnerships with a diverse community of epibionts and a single endosymbiont (13). The epibionts of this snail belong to several distinct assemblages of bacteria, including Epsilonproteobacteria, while the endosymbiont is a member of the Gammaproteobacteria. Like Osedax, the exact role of these diverse bacteria, and their influence on the host, is not yet known.
Vestimentiferan tubeworms, siboglinid relatives of Osedax, are infected each generation with symbionts from the local environment in which the worm larvae settle (23; R. C. Vrijenhoek, M. Duhaime, and W. J. Jones, unpublished data). It is not known whether Osedax acquires symbionts in a similar manner; however, preliminary microscopic and molecular investigations have revealed negative evidence for the presence of symbionts associated with the eggs of mature Osedax females (S. K. Goffredi, unpublished data). Similarly, the diversity of Oceanospirillales symbiont ribotypes associated within and among individuals of three Osedax species is consistent with the hypothesis that these worms acquire their symbionts from the environment. Several individuals of Osedax sp. MB4 hosted symbiont ribotypes that differed by as much as 6% for the small-subunit (16S) rRNA. Furthermore, a recently discovered population of O. rubiplumus (at a depth of 1,700 m in Monterey Canyon) (unpublished data) possesses a gammaproteobacterial symbiont that is ∼20% divergent for 16S rRNA from the O. rubiplumus symbionts discovered in 2002 (Goffredi, unpublished).
Juvenile recruits of O. frankpressi appeared almost identical to adults in both the composition and identity of associated microbes. The root tissue of recruits, however, appeared to be morphologically different from those of adults, with less complexity in structure and color. The timing of symbiont acquisition is not known for Osedax, and we expect that future collections of these recruits may reveal a different stage of symbiont integration and development as the worms mature.
The Oceanospirillales symbionts associated with Osedax belong to a diverse bacterial group known for the heterotrophic degradation of complex organic compounds. Genes necessary for autotrophic CO2 fixation, including both Rubisco and ATP citrate lyase, were not detected in O. rubiplumus or O. frankpressi symbionts (14). A similar, presumably heterotrophic, strategy of organic carbon utilization by the symbionts is assumed for other Osedax species as well. The dominant carbon source available to the intracellular Oceanospirillales symbionts includes collagen and cholesterol, which are abundant in whalebones. Previously measured bulk δ13C values for O. frankpressi tissues (−12.5 to −13.5‰) were consistent with δ13C values for the collagen fraction in modern and fossil whalebones (−14 to −17‰) rather than the cholesterol fraction (−22 to −26‰) (29), suggesting a primary reliance on whalebone collagen.
Our observation of collagenolytic enzyme activity in three species of Osedax suggests the use of this protein as a source of organic carbon. Many pathogenic bacteria and free-living marine microbes, including Vibrio and Clostridium spp., are able to metabolize collagen (16, 22, 36). Most collagenase-like enzymes are zinc-containing metalloproteases that are localized extracellularly (32). Preliminary elemental analysis of the green symbiont-containing root tissue of O. rubiplumus revealed high concentrations of zinc (696 ppm) (Goffredi, unpublished), especially compared to other marine invertebrates (e.g., Mytilus californianus contains ∼20 ppm zinc) (J. Goetzl, Moss Landing Marine Labs, personal communication). We presently cannot exclude the possibility that the host produces the collagenase activity, as eukaryotes possess collagenases that are used for normal development. Nevertheless, two of our results suggest that the bacteria are responsible for this activity. First, collagenase activity was restricted to root tissues that house the bacteria, and second, collagenase activity generally corresponded with the abundance of Oceanospirillales symbionts in Osedax (Table 3). One can imagine a scenario in which an extracellular enzyme, like collagenase, might help the Osedax symbiosis exploit complex organic carbons in the external environment. Additional studies to investigate the specific sources of collagenolytic activity are under way, including an attempt to sequence the O. frankpressi symbiont genome (Moore Foundation, in progress).
Conclusion.
Despite apparently similar nutritional strategies in all known Osedax species, ecological differences exist. The two newly discovered species observed in this study (Osedax sp. MB3 and MB4) differ from each other and from previously described species, O. rubiplumus and O. frankpressi, in the identities and biomasses of their symbiotic populations. Differences in symbiont biomass can influence nutrient transfer and specific functional capabilities and could contribute to ecological differences among Osedax species. Conversely, Osedax lineages settling at different depths harbor distinct Oceanospirillales symbiont lineages (phylotypes). It is possible that ecological differences within and among the habitats occupied by Osedax species are responsible for the high diversity of Oceanospirillales symbionts observed in this study, especially if the symbionts are acquired each generation from the environment.
Osedax worms possess a large number of closely related symbionts from a single lineage of Gammaproteobacteria. This is similar to what has been previously shown for shipworms yet different for animals, such as mussels and gutless oligochaetes, that harbor multiple bacterial species. These invertebrates generally host phylogenetically distinct bacteria, commonly only a single phylotype of each symbiont species, i.e., a single thiotrophic or single methanotrophic 16S rRNA phylotype. The question then arises as to whether heterotrophic degradation of organic compounds, found in Osedax and shipworms, results in symbioses that are fundamentally different in diversity than those more well-studied, yet generally monospecific, chemoautotrophic associations.
Acknowledgments
This work was supported by grants from the David and Lucile Packard Foundation, a Davidow grant to Caltech's Geological and Planetary Sciences division, and the U.S. National Science Foundation (MCB-0454860 to S.K.G.).
We thank the Tiburon pilots and Western Flyer crew, W. J. Jones for laboratory and shipboard support, R. Young for help with phylogenetic analyses, A. Pernthaler for advice regarding FISH analyses, J. Leadbetter for laboratory space at the California Institute of Technology, and V. Orphan for use of both sequencing and microscopy facilities (also at Caltech).
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Akaike, H. 1974. A new look at the statistical model identification. IEEE Trans. Autom. Contrib. 19:716-723. [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ekblad, A., H. Wallander, R. Carlsson, and K. Huss-Danell. 1995. Fungal biomass in roots and extramatrical mycelium in relation to macronutrients and plant biomass of ectomycorrhizal Pinus sylvestris and Alnus incana. New Phytol. 131:443-451. [DOI] [PubMed] [Google Scholar]
- 6.Fitt, W. K., F. K. McFarland, M. E. Warner, and G. C. Chilcoat. 2000. Seasonal patterns of tissue biomass and densities of symbiotic dinoflagellates in reef corals and relation to coral bleaching. Limnol. Oceanogr. 45:677-685. [Google Scholar]
- 7.Fujikura, K., Y. Fujiwara, and M. Kawato. A new species of Osedax (Annelida: Siboglinidae) associated with whale carcasses off Kyushu, Japan. Zool. Sci., in press. [DOI] [PubMed]
- 8.Glockner, F., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glover, A. G., B. Käilström, C. R. Smith, and T. G. Dahlgren. 2005. World-wide whale worms? A new species of Osedax from the shallow North Atlantic. Proc. Biol. Sci. 272:2587-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goffredi, S. K., and J. P. Barry. 2002. Species-specific variation in sulfide physiology between closely related vesicomyid clams. Mar. Ecol. Prog. Ser. 225:227-238. [Google Scholar]
- 11.Goffredi, S. K., J. P. Barry, and K. R. Buck. 2004. Vesicomyid symbioses from Monterey Bay (Central California) cold seeps. Symbiosis 36:1-27. [Google Scholar]
- 12.Goffredi, S. K., C. K. Paull, K. Fulton-Bennett, L. A. Hurtado, and R. C. Vrijenhoek. 2004. Unusual benthic fauna associated with a whale fall in Monterey Canyon, California. Deep Sea Res. I 51:1295-1306. [Google Scholar]
- 13.Goffredi, S. K., A. Warén, V. Orphan, C. L. Van Dover, and R. C. Vrijenhoek. 2004. Novel forms of structural integration between microbes and a vent gastropod from the Indian Ocean. Appl. Environ. Microbiol. 70:3082-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goffredi, S. K., V. J. Orphan, G. W. Rouse, L. Jahnke, T. Embaye, K. Turk, R. Lee, and R. C. Vrijenhoek. 2005. Evolutionary innovation: a bone-eating marine symbiosis. Environ. Microbiol. 7:1369-1378. [DOI] [PubMed] [Google Scholar]
- 15.Goffredi, S. K., W. J. Jones, C. A. Scholin, R. Marin, and R. C. Vrijenhoek. 2006. Molecular detection of marine invertebrate larvae. Mar. Biotechnol. 1:1-12. [DOI] [PubMed] [Google Scholar]
- 16.Harrington, D. J. 1996. Bacterial collagenases and collagen-degrading enzymes and their potential role in human disease. Infect. Immun. 64:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 19.Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Alio, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 20.Luyten, Y. A., J. R. Thompson, W. Morrill, M. F. Polz, and D. L. Distel. 2006. Extensive variation in intracellular symbiont community composition among members of a single population of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl. Environ. Microbiol. 72:412-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 22.Merkel, J. R., J. H. Dreisbach, and H. B. Ziegler. 1975. Collagenolytic activity of some marine bacteria. Appl. Microbiol. 29:145-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nussbaumer, A. D., C. R. Fisher, and M. Bright. 2006. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441:345-348. [DOI] [PubMed] [Google Scholar]
- 24.Pernthaler, A., and J. Pernthaler. 2005. Simultaneous fluorescence in situ hybridization of mRNA and rRNA for the detection of gene expression in environmental microbes. Methods Enzymol. 397:352-371. [DOI] [PubMed] [Google Scholar]
- 25.Posada, D., and K. A. Crandall. 2001. Selecting the best Wt model of nucleotide substitution. Syst. Biol. 50:580-601. [PubMed] [Google Scholar]
- 26.Rouse, G. W., S. K. Goffredi, and R. C. Vrijenhoek. 2004. Osedax: bone-eating marine worms with dwarf males. Science 305:668-671. [DOI] [PubMed] [Google Scholar]
- 27.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 28.Steedman, H. F. 1957. Polyester wax: a new ribboning embedding medium for histology. Nature 179:1345. [DOI] [PubMed] [Google Scholar]
- 29.Stott, A. W., R. P. Evershed, and N. Tuross. 1997. Compound specific approach to the δ13C analysis of cholesterol in fossil bones. Org. Geochem. 26:99-103. [Google Scholar]
- 30.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, MA.
- 31.Tavaré, S. 1986. Some probabilistic and statistical problems on the analysis of DNA sequences. Lect. Math. Life Sci. 17:57-86. [Google Scholar]
- 32.Watanabe, K. 2004. Collagenolytic proteases from bacteria. Appl. Microbiol. Biotechnol. 63:520-526. [DOI] [PubMed] [Google Scholar]
- 33.Whitehead, L. F., and A. E. Douglas. 1993. Populations of symbiotic bacteria in the parthenogenetic pea aphid (Acyrthosiphon pisum) symbiosis. Proc. Biol. Soc. Lond. B 254:29-32. [Google Scholar]
- 34.Wilkinson, T. L., D. Adams, L. B. Minto, and A. E. Douglas. 2001. The impact of host plant on the abundance and function of symbiotic bacteria in an aphid. J. Exp. Biol. 204:3027-3038. [DOI] [PubMed] [Google Scholar]
- 35.Yang, Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39:306-314. [DOI] [PubMed] [Google Scholar]
- 36.Yoshihara, K., O. Matsushita, J. Minami, and A. Okabe. 1994. Cloning and nucleotide sequence analysis of the colH gene from Clostridium histolyticum encoding a collagenase and a gelatinase. J. Bacteriol. 176:6489-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]