Abstract
The deep anoxic shelf of the northwestern Black Sea has numerous gas seeps, which are populated by methanotrophic microbial mats in and above the seafloor. Above the seafloor, the mats can form tall reef-like structures composed of porous carbonate and microbial biomass. Here, we investigated the spatial patterns of CH4 and CO2 assimilation in relation to the distribution of ANME groups and their associated bacteria in mat samples obtained from the surface of a large reef structure. A combination of different methods, including radiotracer incubation, beta microimaging, secondary ion mass spectrometry, and catalyzed reporter deposition fluorescence in situ hybridization, was applied to sections of mat obtained from the large reef structure to locate hot spots of methanotrophy and to identify the responsible microbial consortia. In addition, CO2 reduction to methane was investigated in the presence or absence of methane, sulfate, and hydrogen. The mat had an average δ13C carbon isotopic signature of −67.1‰, indicating that methane was the main carbon source. Regions dominated by ANME-1 had isotope signatures that were significantly heavier (−66.4‰ ± 3.9 ‰ [mean ± standard deviation; n = 7]) than those of the more central regions dominated by ANME-2 (−72.9‰ ± 2.2 ‰; n = 7). Incorporation of 14C from radiolabeled CH4 or CO2 revealed one hot spot for methanotrophy and CO2 fixation close to the surface of the mat and a low assimilation efficiency (1 to 2% of methane oxidized). Replicate incubations of the mat with 14CH4 or 14CO2 revealed that there was interconversion of CH4 and CO2. The level of CO2 reduction was about 10% of the level of anaerobic oxidation of methane. However, since considerable methane formation was observed only in the presence of methane and sulfate, the process appeared to be a rereaction of anaerobic oxidation of methane rather than net methanogenesis.
Submarine gas seeps and subsurface methane-sulfate transition zones are special types of ocean habitats where microbial life is based on anaerobic oxidation of methane (AOM) as an energy and carbon source (2, 3, 9, 12, 26, 28, 30, 40, 43, 45, 46). A peculiar type of methane-driven microbial habitat is the large gas seepage area in the anoxic part of the northwestern slope of the Crimean Peninsula in the Black Sea (23). Here, giant accumulations of microorganisms have been found to develop from subsurface aggregations into reef-forming microbial mats that have dimensions up to decimeters to meters (44). Previous investigations have shown that the reef-forming microbial mats in the Black Sea are dominated by anaerobic methanotrophs (ANME-1 and -2) and a variety of Crenarcheota and have relatively high bacterial diversity (1, 19, 23, 34, 35, 40, 42, 44).
The identification of methanotrophic archaea has been based primarily on detection of a 13C-depleted isoprenoid lipid biomarker and corresponding 16S rRNA gene sequences representing distinct phylogenetic clusters related to the Methanosarcinales. Today, four different clusters are known, three of which depend on sulfate as the final electron acceptor (ANME-1 to -3) (2, 15, 27) and one of which depends on nitrite (33). Microscopy after whole-cell hybridization assays with 16S rRNA-targeted fluorescent probes revealed that these ANME groups are generally associated with a specific bacterial partner, which also shows distinct 13C depletion of biomarker lipids. The sulfate-reducing bacterial (SRB) partners associated with ANME-1 and -2 usually belong to the Desulfosarcina-Desulfococcus cluster, and the partner associated with ANME-3 belongs to the Desulfobulbus cluster of the Deltaproteobacteria. The associations between ANME cells and SRB have diverse morphologies, but usually one type dominates in a habitat (51). So far, the mat type association has been found only in the Black Sea, the largest anoxic marine basin on Earth. The discovery of such large biomasses of anaerobic methanotrophs has permitted metagenomics- and proteomics-based studies of the enigmatic functioning of AOM (17, 22, 38). Since no ANME or SRB partner strain is available in culture yet, so far in vitro feeding studies of methanotrophic mats have been the main complementary approach for improving our understanding of AOM (24, 25, 37).
In the Black Sea, methanotrophic mats were found below the chemocline at water depths of 100 to 450 m on the northwestern Crimean and Romanian shelves (19, 20, 23; B. B. Jørgensen et al., Poseidon cruise POS 317-3 2004, METROL project; http://www.metrol.org/index.php?bereich=3), as well as in subsurface sediments of the Sorokin Trough mud volcanoes at water depths of around 2,000 m (40). The dense accumulation of microorganisms and the high rates of AOM apparently cause precipitation of carbonates within the mats. Subsurface methanotrophic mats can reach thicknesses of decimeters and form nodules (44) or cemented crusts (40). It has been suggested that the subsurface mats grow upward into the anoxic water column following the more or less vertical pathway of rising gas bubbles (44). At actively gas-emitting seeps, such as those on the northwestern Crimean shelf, the mats then form reef-like structures composed of cemented basal plates with chimney-like morphology extending vertically into the water column. These vertical structures are stabilized by interior porous carbonates partially or fully covered by nodule-shaped mats that reach thicknesses of centimeters to decimeters. They are up to 4 m high and 1 m in diameter, and methane bubbles rise vertically through their porous calcified interiors. Two key factors facilitating the development of such enormous methanotrophic biomasses appear to be the continuous release of methane gas and the permanent anoxic conditions in the water column (20, 44). However, it is not known how methanotrophic cells in central parts of the thick mats overcome the probable methane and sulfate limitation that might be caused by fast consumption of the substrates in peripheral mat layers. Microscopic analyses of thin sections of mats have suggested that channels in the mats may function as pathways for gas and solute transport (23). Molecular comparisons of mat samples between and within reef structures have revealed various proportions of anaerobic methanotrophs, as well as various proportions of their sulfate-reducing partner bacteria and other microbes (1, 15, 23, 34, 35, 44). However, it is not known which environmental factors select for the relative levels of the different microbial populations in the mats. One goal of this investigation was to look for patterns in the distribution of the dominant methanotrophs and sulfate reducers in relation to methane uptake by mats at micrometer to millimeter scales. Furthermore, it is unclear under what thermodynamic conditions AOM is connected with growth and if other carbon substrates besides methane (e.g., CO2) are utilized by the microbial assemblages in the mats. Therefore we combined radiotracer rate measurements with two-dimensional approaches, including beta microimaging, catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH), and secondary ion mass spectrometry (SIMS). Incubation of mats with 14C-labeled methane and subsequent two-dimensional beta microimaging provided information about 14C uptake under in vitro conditions. By investigating the distribution of methanotrophic organisms by CARD-FISH using adjacent thin sections of mat, the 14C assimilation patterns could be linked to community structure. The additional use of SIMS provided the two-dimensional distribution of stable carbon isotopes in the mat biomass. This approach allowed us to obtain an integrative view of methane assimilation over the lifetimes of cell colonies in a mat and answered questions concerning the relevance of other carbon sources for growth.
Anaerobic hydrocarbon degraders usually exhibit much slower growth than their aerobic counterparts. Generation times of 2 to 5 months have been determined for incubation of anaerobic methane oxidizers belonging to the ANME-2 cluster retrieved from methane seeps (4, 5, 24). Samples of Black Sea mats have been used for in vitro physiological studies, but no growth has been observed in incubations lasting weeks to several months (16, 25, 37). The results obtained so far suggest that microorganisms performing AOM have very low growth yields, with <2% assimilation of the methane oxidized (24). In comparison, SRB growing on conventional substrates, such as acetate or lactate, assimilate approximately 10% of the carbon substrate (49). Here we obtained concurrent measurements of AOM and carbon uptake from methane and CO2 to determine the carbon assimilation rates of actively methane-oxidizing mat sections. Beyond methanotrophs, an even greater diversity of microorganisms in the Black Sea mats may profit from methane seepage as commensals utilizing inorganic and organic matter derived from AOM, as indicated by the δ13C signature (−72.2‰ δ13C) of bulk mat samples (23). However, analyses of specific lipid biomarkers extracted from Black Sea mats suggested that mat microorganisms have different metabolic activities, ranging from pure methanotrophy to detritus-based feeding (41). Hence, another goal of this investigation was to document the range of micrometer-scale carbon isotope signatures along defined transects by SIMS in order to detect microbial assemblages potentially utilizing carbon sources not derived from methane (e.g., phytodetritus, dissolved organic carbon, or chemoautotrophic fixation of seawater bicarbonate). It has also been proposed that methanogenesis could be an additional source of energy in the Black Sea mats (19, 32, 37, 42). However, the cooccurrence of methanogenesis and AOM in the same niche appears unlikely due to thermodynamic constraints. Labeling studies with [14C]methane in cultures of methanogenic archaea suggested that methanogenesis is reversible to a small extent during net formation of methane (54, 55). Here we examined whether AOM is the dominant process in the methanotrophic mats and if it is reversible by measuring 14CH4 formation from 14CO2 in vitro under different thermodynamic conditions.
MATERIALS AND METHODS
Subsampling of methanotrophic reefs.
Samples of methanotrophic reefs were obtained by the submersible JAGO (MPI Seewiesen, Germany) during the cruise of the Russian R/V Professor Logachev to the northwestern Black Sea in June and July 2001 (GHOSTDABS station no. 55; water depth, 230 m; 44°46.48N, 31°59.54E) (23). A single reef structure (height, 150 cm; diameter, 30 cm) (Fig. 1) was sampled by grabbing a large carbonated piece (ca. 2 to 3 kg) covered with a thick microbial mat (thickness, 5 to 8 cm) from the base of the reef with the manipulator of the submersible. The sample was stored in a watertight box filled with bottom water to maintain anoxic conditions during recovery. On board subsamples of the mat were obtained under anoxic conditions with cut-off plastic tubes (inside diameter, 1.5 cm; length, 10 cm). Cylindrical mat pieces, including pieces of the mat surface (black layer) and the central section (pinkish layer), were obtained from the reef sample (Fig. 1E). Samples of the calcified interior could not be obtained by this method. Plastic tubes containing mat pieces were transferred to a 500-ml bottle and submerged in 250 ml of anoxic bottom water. The bottle was sealed with a rubber stopper and a screw cap and flushed with methane for several minutes to ensure that there was a pure methane headspace. The bottle was stored at 12°C in the dark. In the home laboratory additional subsamples were obtained under an anoxic atmosphere (N2-CO2, 4:1) using an anoxic glove box (Mecaplex).
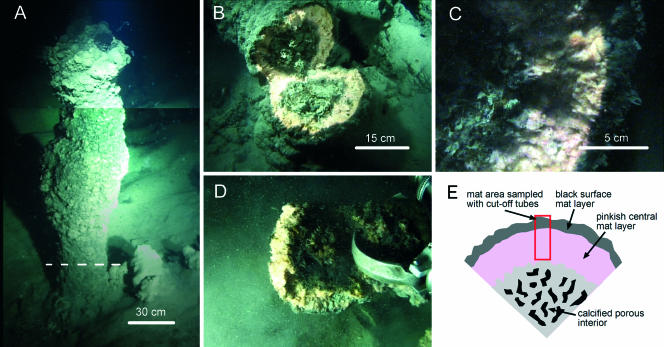
FIG. 1.
Reef structure sampled in this study. (A) Complete reef structure (total height, approximately 150 cm). The dashed line indicates the approximate height at which the structure was cut down. The image consists of two single images which were assembled to show the complete structure. (B) Reef structure after it was cut down. The inner gray core consists of porous carbonates, through which gas bubbles were rising. The carbonated core is covered by a voluminous microbial mat with a thick pink inner layer and a thin black outer layer. (C) Close-up of the carbonated core and the microbial mat. (D) Reef sample, from which subsamples were obtained for this study, grabbed by the submersible arm. (E) Diagram of the mat and carbonate zones in the reef and the area of where subsamples were obtained. The images were obtained from a video sequence filmed by the JAGO Team and were made available through the courtesy of W. Michaelis, GHOSTDABS Project.
Incubation setup for combined radiotracer rate measurement, beta microimaging, SIMS, and CARD-FISH (experiments 1a and b).
We conducted two long-term experiments with mat pieces including both the surface and central sections of the mat (Fig. 1E). In the first experiment (experiment 1a) we incubated mat pieces with the radiotracers 14CH4, H14CO3−, and 35SO42− to measure AOM, methane formation via CO2 reduction, and sulfate reduction, respectively. The mat pieces were subsequently analyzed by beta microimaging to measure the two-dimensional incorporation of methane- and CO2-derived 14C into mat biomass. In the second experiment (experiment 1b) we repeated the incubation of mat pieces with the 14CH4 and H14CO3− radiotracers. In this experiment the mat pieces were investigated to determine (i) methane oxidation and formation using radiotracer techniques, (ii) the two-dimensional distribution of 14CH4 and 14CO2 assimilated into mat biomass using beta microimaging, (iii) the two-dimensional distribution of stable carbon isotopes in the mat using SIMS, and (iv) the two-dimensional distribution of methanotrophic cells in the mat using CARD-FISH.
For these investigatons the mat pieces were removed from their storage tubes and cut into smaller pieces (about 1.5 by 1.0 by 0.3 cm) having a wet weight between 0.2 and 0.5 g. The mat sections included the formerly seawater-exposed outer surface of the mat (black layer) and the central pinkish mat (Fig. 1E). In experiment 1a replicate mat pieces were prepared for measurement of methane oxidation (n = 4), methane formation (n = 4), and sulfate reduction (n = 4). For each assay, one of the mat pieces was fixed for 24 h in 4% formaldehyde prior to incubation and served as a negative control. Mat pieces were transferred into separate Hungate tubes (16 ml) filled with 14 ml artificial seawater medium (50) (17 mM sulfate, 30 mM HCO3−). The tubes containing control pieces were filled with 13 ml medium and 1 ml formaldehyde (37%) to prevent microbial activity. The tubes were sealed, and the headspace (ca. 2 ml) was flushed with methane for 1 min. After flushing, the gas in the headspace was changed to 100% methane at 2 × 105 Pa, resulting in a dissolved methane concentration of about 2.8 mM (52) at the beginning of the experiment. Then the following radiotracers were injected into replicates and into control tubes for measurement of methane oxidation, methane formation, and sulfate reduction: 55 kBq 14CH4 (gaseous; 50 μl; free of 14CO and 14CO2), 185 kBq H14CO3− (dissolved in water; 100 μl), and 1,000 kBq carrier-free 35SO42− (dissolved in water, 50 μl), respectively.
In experiment 1b replicate mat pieces were prepared for methane oxidation (n = 3) and methane formation (n = 3) and transferred into separate Hungate tubes (19.5 ml) filled with 17 ml artificial seawater medium. The headspace (ca. 2.5 ml) was 100% methane at 2 × 105 Pa. Then 600 kBq 14CH4 (gaseous; 80 μl; free of 14CO and 14CO2) and 150 kBq H14CO3− (dissolved in water; 100 μl) were injected into replicate tubes for measurement of methane oxidation and methane formation, respectively.
In these two experiments the tubes were stored horizontally in the dark for 62 days (experiment 1a) and for 109 days (experiment 1b) at 9°C, which was the in situ temperature during sampling. The tubes were gently shaken twice per week. After incubation, the headspace was removed from each sample using a gas-tight syringe, with simultaneous replacement with distilled water using a second syringe. The incubation media for methane oxidation and methane formation samples were transferred into 50-ml glass vials filled with 25 ml sodium hydroxide (2.5%, wt/wt). The media for sulfate reduction samples were transferred into 50-ml plastic centrifuge vials filled with 20 ml zinc acetate (20%, wt/wt). All vials were closed immediately with rubber stoppers (methane oxidation and methane formation) or plastic caps (sulfate reduction), and the headspaces of the samples were reinjected subsequently. After addition of the media, the vials were shaken thoroughly, and activities were measured as described below. The remaining mat pieces for methane oxidation and methane formation incubation experiments were prepared for beta microimaging (experiments 1a and b), SIMS, and CARD-FISH (experiment 1b) as described below.
Incubation setup for investigation of methane formation in the mat (experiments 2a and b).
In experiments 2a and b we studied methane formation from CO2 in mat pieces in the presence or absence of methane and sulfate (experiment 2a), as well as hydrogen (experiment 2b).
In experiment 2a three replicate mat pieces per assay (between 0.3 and 0.5 g) were incubated in Hungate tubes (19.5 ml) with 17 ml artificial seawater medium (30 mM HCO3−) and 150 kBq of the H14CO3− radiotracer (dissolved in water; 100 μl) as follows: (i) methane headspace and sulfate-rich seawater medium (17 mM sulfate), (ii) methane headspace and sulfate-free seawater medium, (iii) nitrogen headspace and sulfate-rich seawater medium (17 mM sulfate), and (iv) nitrogen headspace and sulfate-free seawater medium. For each assay, one control tube without mat material was incubated simultaneously. The incubation time was 32 days. The tubes were gently shaken every day. Other treatments were the same as those used for methane formation samples, as described above (experiment 1).
In experiment 2b no further large mat material was available. Instead, a mixture of small mat pieces (volume of each piece, <10 mm3) from the reef sample was used (total weight for each replicate, 0.3 to 0.6 g). The incubation setup was the same as that in experiment 2a except that 1 ml of an H2-CO2 mixture (4:1) was added to each headspace. For each assay, one control tube without mat pieces was incubated simultaneously. The incubation time was 9 days. The tubes were gently shaken every day. Other treatments were the same as those used for methane formation samples, as described above (experiment 1).
Mat dry weight and density.
Mat dry weight was determined by weighing three replicate mat pieces before and after freeze drying for 48 h. Mat density was determined by weighing three replicate mat pieces whose volume was known (1 cm−3). Water drops were gently removed from the mat pieces by draining with a cellulose filter before weighing.
Microbial turnover rates. (i) AOM.
AOM was analyzed by using 14CH4 combustion and 14CO2 acidification as described by Treude et al. (45). The amount of CH4 at time zero was calculated from the headspace volume and pressure. AOM rates were calculated as follows:
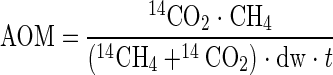 |
(1) |
where AOM is the rate of methane oxidation (μmol g [dry weight]−1 day−1), 14CO2 is the radioactivity of the microbially produced radioactive carbon dioxide (dpm), CH4 is the total amount of methane in the Hungate tube at time zero (μmol), 14CH4 is the radioactivity of the residual radioactive methane (dpm), dw is the dry weight of the mat (g), and t is the incubation time (days).
(ii) Methane formation.
The analytical procedures used for methane formation were the same as those used for AOM. The amount of HCO3− at time zero was calculated from the concentration in the artificial seawater medium. Methane formation rates were calculated as follows:
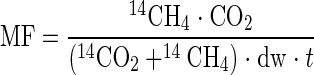 |
(2) |
where MF is the rate of methane formation from CO2 (μmol g [dry weight]−1 day−1), 14CH4 is the activity of the microbially produced radioactive methane (dpm), CO2 is the total amount of carbon dioxide in the Hungate tube at time zero (μmol), 14CO2 is the activity of the residual radioactive carbon dioxide (dpm), dw is the dry weight of the mat piece (g), and t is the incubation time (days).
(iii) Sulfate reduction.
Sulfate reduction was analyzed using the cold-chromium distillation method described by Kallmeyer et al. (13). The amount of sulfate at time zero was calculated from the concentration in the defined artificial seawater medium. Sulfate reduction rates were calculated as follows:
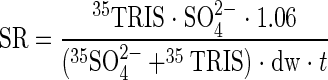 |
(3) |
where SR is the rate of sulfate reduction (μmol g [dry weight]−1 day−1), SO42− is the total amount of sulfate in the Hungate tube at time zero (μmol), 35TRIS is the radioactivity of the metal mono- and disulfides and elemental sulfur produced (dpm), 1.06 is the correction factor for the expected isotopic fractionation (11), 35SO42− is the radioactivity of the residual radioactive sulfate (dpm), dw is the dry weight of the mat (g), and t is the incubation time (days).
Beta microimaging.
The two-dimensional distribution of precipitated and assimilated 14C within the mat was analyzed using beta microimaging. Mat pieces incubated for determination of methane oxidation (14CH4 tracer added) and methane formation (14CO2 tracer added) in experiments 1a and b were fixed for 24 h with 3% paraformaldehyde (in seawater) immediately after incubation. After fixation, the pieces were washed two times with 1× phosphate-buffered saline and transferred into 1× phosphate-buffered saline-ethanol (1:1). The mat pieces were embedded in Tissue Tec OCT compound (Ted Pella Inc., California) and sectioned using a cryostat (HM505 E; Microm, Walldorf, Germany) as described previously (36). Sections that were 20 μm thick (experiment 1a) and 10 μm thick (experiment 1b) (replicate and control samples; 24 to 30 sections per mat piece) were prepared from surface and central parts of the mat pieces reaching ca. 1.5 cm into the mat. The sections were transferred to gelatin-coated glass slides (experiment 1a) and uncoated glass slides (experiment 1b), dried at room temperature, and subsequently used for high-resolution two-dimensional beta microimaging (18) with a beta imager (Biospace Mésures, Paris, France). Scannig of each section was performed for 12 h. Subsequently, the sections were washed with 0.1 M glycine buffer (pH 2.0) to remove precipitated [14C]carbonate and scanned again for 12 h. Glass slides without mat sections were scanned for 12 h to determine the background radioactivity.
In experiment 1a the radioactivity in beta imager micrographs was determined using the Biospace (Paris, France) software β-vision (version 4.2) provided by the manufacturer. For rough comparisons of replicate, control, and background samples the total radioactivity of scanned micrographs (32 by 23 mm) was determined. In order to determine the radioactivity in restricted areas of the micrographs (i.e., in mat sections or in distinct zones of mat sections), areas were manually selected and the radioactivity was calculated using the software. The calculations were corrected for the radioactivity in background areas that were the same size. Finally, the total amount of assimilated radioactivity per mat sample (or zone in the mat sample) was calculated as follows:
 |
(4) |
The radioactivity of mat pieces was determined before and after acidification to differentiate between 14C precipitated as carbonate and 14C assimilated into biomass. The volumes of mat sections were calculated by using area and thickness; the volumes of mat pieces were calculated by using weight and density.
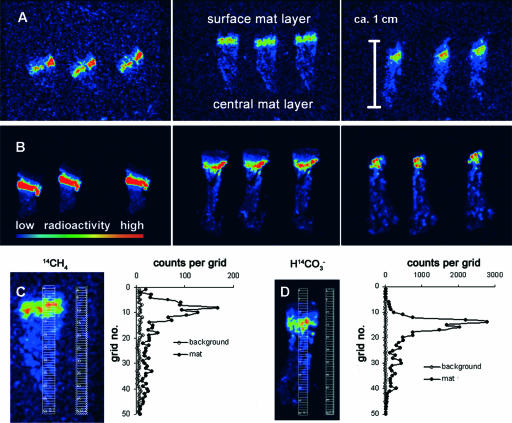
Additionally, the amount of assimilated 14C was determined for areas of 1-by-50 grids (∼3 mm2 per grid) (Fig. 2C and D) vertically crossing mat sections in micrographs to determine the distribution of 14C in the mat. The radioactivity of each grid area was calculated and plotted against the depth in the mat (i.e., ascending grid numbers). The data were corrected for the background radioactivity.
FIG. 2.
(A and B) Beta imager micrographs of three replicate mat pieces incubated with 14CH4 (A) or H14CO2− (B). Each micrograph shows the distribution of 14C in three consecutive mat sections. The mat sections are oriented vertically with the former mat-water interface at the top. A high level of 14C radioactivity (expressed in counts per hour) is indicated by red, and a low level of radioactivity is indicated by blue, as shown by the radioactivity bar. (C and D) Vertical profile of 14C distribution in a mat section incubated with 14CH4 (C) or H14CO2− (D). For determinations, a grid was placed on the section as well as on the background (no mat section) in the beta imager micrograph, and activity was expressed as counts per grid cell. The grid cell numbers are low at the former mat-water interface and increase toward the center of the mat.
The rate of carbon assimilation from either CH4 or CO2 was calculated by using the following equation:
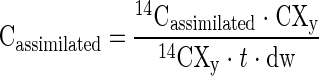 |
(5) |
where Cassimilated is the carbon assimilation rate (nmol C g [dry weight]−1 day−1), 14Cassimilated is the radioactivity of the 14C assimilated into the mat from either 14CH4 or 14CO2 at the end of the experiment (cpm), CXy is the total amount of either CH4 or CO2 in the Hungate tube at time zero (nmol), 14CXy is the activity of the 14CH4 or 14CO2 in the Hungate tube at time zero (cpm), t is the incubation time (days), and dw is the dry weight of the mat piece (g).
SIMS.
The two-dimensional distribution of stable carbon isotopes (i.e., the two-atom molecular ions 12C-12C and 13C-12C in the mat) was analyzed by SIMS using a CAMECA ims 1270 ion microprobe (NSF National Ion Microprobe Facility housed at the University of California, Los Angeles). The analysis was conducted with two selected mat sections from experiment 1b, one from a methane formation incubation (mat section 1) and one from a methane oxidation incubation (mat section 2). Both sections were analyzed by beta microimaging before this analysis (see above). In preparation for the SIMS analysis, the sections (thickness, 10 μm) were sputter coated with gold. Individual target regions that were approximately 15 to 20 μm in diameter were analyzed along latitudinal and longitudinal transects crossing zones where there was a high level of 14C incorporation in the mat (Fig. 3A and B). After analysis, the mat sections were photographed using a dissection microscope, and the exact location of each target within the mat was recorded.
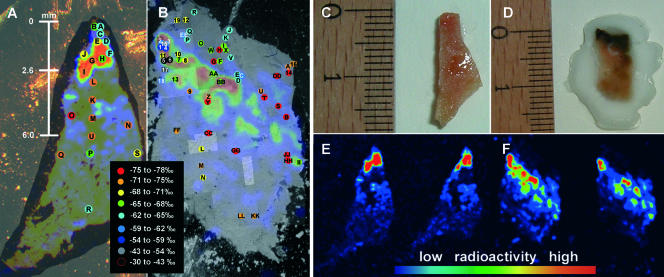
FIG. 3.
(A and B) Overlays of a photograph, a beta imager micrograph, and spots measured by SIMS of mat sections 1 (A) and 2 (B) incubated with H14CO2− and 14CH4, respectively. The mat sections are oriented vertically with the former mat-water interface at the top. The photograph includes the outer border of the mat section. A high level of 14C radioactivity (expressed in counts per hour) in the beta imager micrograph is indicated by red, and a low level of radioactivity is indicated blue, as shown by the radioactivity bar. Colored spots indicate the δ13C values of the bulk biomass determined by SIMS, as indicated in the chart. The precise δ13C value for each spot is shown in Tables 2 and 3. The scale bar in panel A (indicating depth) allows a comparison of activity and δ13C values with the distribution of dominant methanotrophic groups identified by CARD-FISH shown in Fig. 4. (C and D) Photographs of mat pieces in panels A and B, respectively, before sectioning. The scale is in centimeters. In panel D, the piece is embedded in Tissue Tek OCT compound for easier sectioning. (E and F) Beta imager micrographs of the mat sections in panels A and B, respectively. Each micrograph shows the distribution of 14C in two consecutive mat sections.
SIMS instrument configuration and δ13C standards.
For high-spatial-resolution 13C analysis of the microbial mat by SIMS, the ims 1270 instrument was configured in monocollection mode using dual Faraday cup detectors (measuring 12C-13C− and 12C2−) and a moderate primary Cs+ beam intensity setting of 2nA. This provided an average 12C2− count rate of 5 × 107 with precision greater than 1.5‰ for the majority of spots analyzed. Instrumental mass fractionation was determined by periodically analyzing Escherichia coli cells spotted onto a round glass piece during the ion microprobe session. For this study, an additional correction for a biological matrix effect of 9.9‰ was applied after we compared the average δ13C value for clusters of the E. coli standard (n = 9) measured with SIMS to the value obtained by bulk analysis of the same E. coli culture (δ13C, −22.3‰) using conventional gas source mass spectrometry as described by Orphan et al. (29).
CARD-FISH.
The two-dimensional distribution of anaerobic methanotrophs and SRB in the mat was analyzed by CARD-FISH. Thin sections (5 μm) of mat sections 1 and 2 from experiment 1b (see above) were produced as described above. CARD-FISH sections were obtained from areas next to sections used for beta microimaging and SIMS. CARD-FISH followed by tyramide signal amplification was performed according to the protocol of Pernthaler et al. (31) except for the cell wall permeabilization step. For permeabilization of tough archaeal cell walls, agarose-embedded sections were incubated in 0.1 M HCl for 1 min at room temperature, washed in MilliQ water, and dehydrated in absolute ethanol. For dual hybridizations, the CARD-FISH protocol was repeated two times with the same sections by using different probes and fluorescently labeled tyramide after the probe-delivered peroxidase from the first hybridization was inactivated by incubating the filter sections in 0.01 M HCl for 10 min at room temperature and washing the sections with 50 ml of MilliQ water. Horseradish peroxidase-labeled probes were purchased from biomers.net (Ulm, Germany). For dual hybridizations the following probes were used: (i) ANME-1-350 specific for ANME-1 archaea (2) combined with ANME-2c-622 specific for ANME-2 archaea belonging to subgroup c (15) (probes were specific using 60% [ANME-1-350] and 70% [ANME-2c-622] formamide, respectively) at a hybridization temperature of 35°C; (ii) DSS-658 specific for the Desulfococcus-Desulfosarcina group belonging to the Deltaproteobacteria (50% formamide) (21) and ANME-1-350 (40% formamide) at a hybridization temperature of 46°C; and (iii) DSS-658 (50% formamide) and EelMS932 specific for ANME-2 archaea (50% formamide) (2) at a hybridization temperature of 46°C. Hybridized slides were analyzed with a confocal laser scanning microscope (Zeiss model LSM510). For the ANME-1-ANME-2c dual hybridization, images of 35 consecutive fields were used to generate a depth profile from the mat surface to a depth of 6 mm.
RESULTS
AOM with sulfate.
The AOM and sulfate reduction rates of whole mat sections utilized for experiments 1a and b were the same order of magnitude as the rates reported in previous studies of mats (Table 1). At dissolved methane concentrations of approximately 2.8 mM and sulfate concentrations of 17 mM the AOM rates ranged from 7.8 to 20.9 μmol g (dry weight)−1 day−1, and the sulfate reduction rates ranged from 4.3 to 10.9 μmol g (dry weight)−1 day−1. No AOM or sulfate reduction activity was found in the fixed control samples. The AOM and sulfate reduction rates were the same order of magnitude. The relationship between AOM and sulfate reduction in the mat was extensively studied in previous experiments, which revealed a 1:1 ratio for the two processes in samples taken from the same study site (23, 25, 44). In comparisons with other studies no substantial differences in AOM and sulfate reduction rates were found when short- or long-term incubations were compared (Table 1).
TABLE 1.
Comparison of AOM and sulfate reduction rates in Black Sea microbial mats determined in this and other studiesa
| Reference | AOM rate (μmol g [dry wt]−1 day−1) | Sulfate reduction rate (μmol g [dry wt]−1 day−1) | Methane-dependent sulfate reduction rate (μmol g [dry wt]−1 day−1) | Incubation time (days) |
|---|---|---|---|---|
| This study (expt 1a) | 20.9 | 10.9 | 62 | |
| 9.4 | 7.3 | |||
| 8.2 | 4.3 | |||
| This study (expt 1b) | 13.9, 12.4, 7.8 | 109 | ||
| 23 | 18 ± 12 (3)b | 19 ± 1 (3)b | 5 | |
| 34 ± 5.8 (6)b | 60 | |||
| 25 | 41 ± 16 (38)b | 60 | ||
| 37 | 9.1 ± 1.6 (3)b | 242 |
AOM and (gross) sulfate reduction rates were determined by incubation with radiotracers. Methane-dependent sulfate reduction rates were determined from the difference in the sulfide production rates with and without methane and were equal to AOM rates.
Mean ± standard deviation (n).
Distribution of methanotrophic archaea.
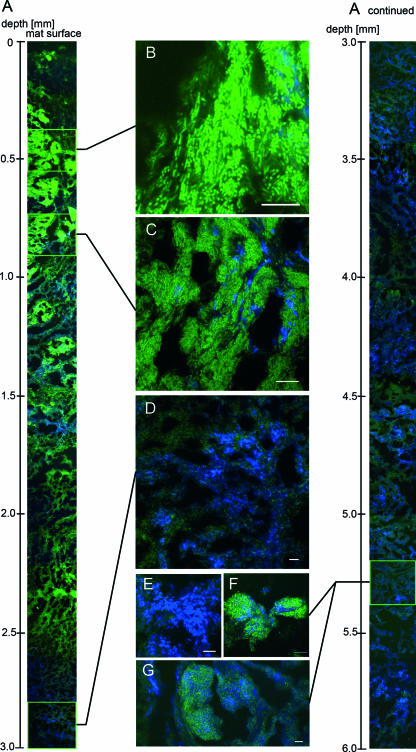
Dual CARD-FISH assays with probes specific for the anaerobic methanotrophic ANME-1 and ANME-2 archaea were performed to investigate the relationship between the patterns of microbial composition and methane assimilation in the mat. Within the ANME-2 group we focused on subgroup c, since no other subgroups were detected previously in similar mats (15; K. Knittel, unpublished data). Microscopic analysis of hybridized sections from the two mat pieces used, mat sections 1 and 2, showed that there was zonation of ANME-1 and ANME-2c signals from the mat surface down to a depth of 6 mm (Fig. 4). Whereas ANME-1 cells were very dominant and active in areas close to the former mat-water interface, ANME-2 cells were absent in these regions but were dominant in the mat in regions toward the interior. There were no obvious differences in microbial distribution between the two mat pieces investigated. We analyzed mat section 1 in greater detail and found that ANME-1 strongly dominated the microbial biomass at depths between 0.4 and 2.6 mm, covering the zone where the level of 14C assimilation was highest (compare Fig. 4 with Fig. 3A). ANME-1 cells had the typical morphology; they were rectangular and 2 to 3 μm long. The brightest fluorescence of ANME-1 cells was found at depths between 0.4 and 1.2 mm close to the former mat surface, indicating that the cellular rRNA content in this region was greater than that in the deeper layers of the mat. High numbers of ANME-1 cells were also present in the deep layers, but the fluorescence intensity of FISH probes decreased at depths below 2.6 mm. These ANME-1 cells could be detected only on the basis of their unique rectangular cell morphology with overexposure of the picture. Generally, ANME-1 and ANME-2 cells appeared to be homogenously mixed with SRB belonging to the Desulfococcus-Desulfosarcina cluster (Fig. 5B and D). However, we found some areas that were up to 400 μm in diameter where ANME-1 cells formed dense colonies without direct cell-to-cell contact with their sulfate-reducing partners (Fig. 5A).
FIG. 4.
Confocal laser scanning micrographs of a mat section (consistent with the mat sample shown in Fig. 3A, C, and E) hybridized with horseradish peroxidase-labeled probes specific for methanotrophic ANME-1 (green) and ANME-2c (blue) archaea. (A) Profile through the mat. The depth below the mat surface is indicated on the left. (B to G) Close-ups of ANME-1 and ANME-2c distributions and aggregations at different depths of the mat. Scale bars = 10 μm.
FIG. 5.
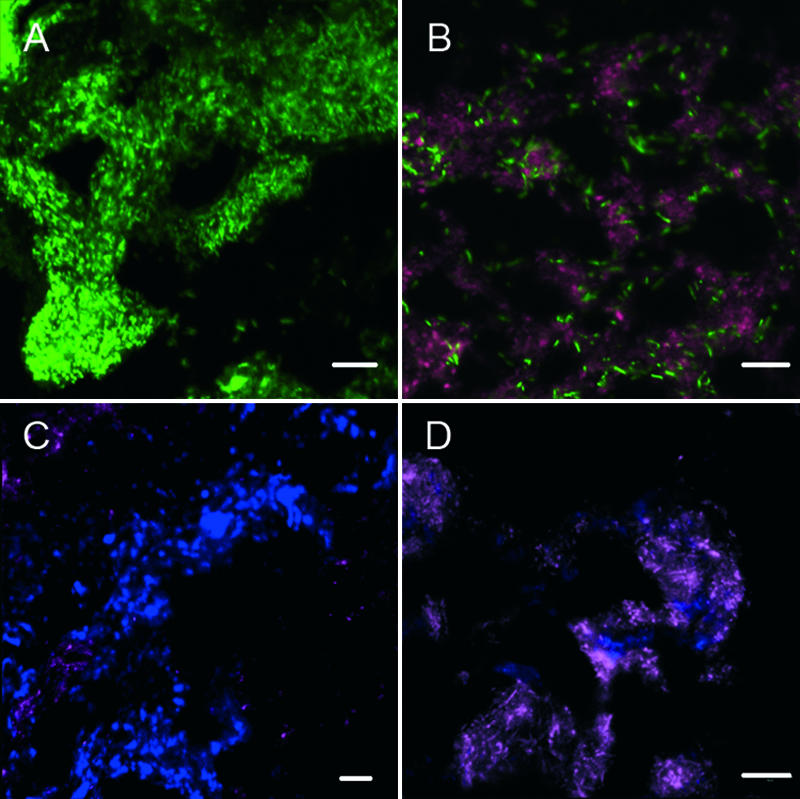
Confocal laser scanning micrographs of a mat section (consistent with the mat sample shown in Fig. 3A, C, and E) hybridized with horseradish peroxidase-labeled probes specific for methanotrophic ANME-1 archaea (green) and sulfate-reducing Desulfococcus and Desulfosarcina spp. (purple) (A and B) and for methanotrophic ANME-2 archaea (blue) and sulfate-reducing Desulfococcus and Desulfosarcina spp. (purple) (C and D). Scale bars = 10 μm.
Very few small ANME-2c zones were detected in the surface of the mat (Fig. 4B and C). However, in the central part at depths below 2.6 mm, large areas of highly fluorescent ANME-2c cells were identified (Fig. 4D and E). The cells were cocci which mostly occurred in clusters. In contrast to the previous study of Knittel et al. (15), typical ANME-2c—Desulfococcus-Desulfosarcina aggregates were rarely detected in these mat sections, except for one area in mat section 2 (data not shown). Instead, the ANME-2c cells were found mostly embedded in a thick matrix without any tightly associated bacterial partner (Fig. 5C) or in large aggregates that were ca. 20 to 50 μm in diameter in close contact with ANME-1 cells (Fig. 4F and G).
Assimilation of carbon from 14C-labeled methane and CO2.
The levels of 14C activity revealed by beta imager micrographs (32 by 23 mm) of mat sections incubated with 14CH4 or H14CO3− in experiment 1a (for 14CH4, 1,927 ± 259 counts per h [mean ± standard deviation; n = 26]; for H14CO3−, 11,334 ± 3,102 counts per h [mean ± standard deviation; n = 26]) were substantially higher than the levels revealed by micrographs of formalin-killed control sections (for 14CH4, 1,076 ± 112 counts per h [mean ± standard deviation; n = 16]; for H14CO3−, 1,176 ± 189 counts per h [mean ± standard deviation; n = 8]) or the levels on slides without a mat (background glass slides; 1,115 ± 57 counts per h [mean ± standard deviation; n = 4]). Killed mat and background micrographs revealed the same level of 14C activity, indicating that no 14C was assimilated or precipitated into control mat pieces.
In most mat pieces, acidification with glycine buffer (pH 2.0) did not result in a significant loss of 14C activity through degassing of 14CO2 (P > 0.05, as determined by a paired-sample t test for nonacidified and acidified sections of mat pieces); hence, the 14C detected in mat sections was apparently assimilated into biomass. Only one mat piece incubated with H14CO3− exhibited a significant loss of 14C activity after acidification (15%; P = 0.02; n = 9), which most likely represented 14C precipitated as carbonate.
All mat sections in experiments 1a and b incubated with 14CH4 and H14CO3− had a distinct layer in which there was strongly elevated 14C assimilation (Fig. 2A and B and 3E and F). In all cases this layer was located close to the original mat surface (i.e., the former mat-water interface) at a depth of approximately 1 to 2 mm, as indicated by profiling the 14C activity through the mat (Fig. 2C and D). The layer comprised only 15% ± 3% (mean ± standard deviation; n = 6) of the volume of each mat piece but had 52% ± 6% (mean ± standard deviation; n = 6) of the 14C activity. The assimilation rates in this distinct zone were 239 ± 219 nmol C g (dry weight)−1 day−1 (mean ± standard deviation; n = 3) for CH4 and 1,423 ± 929 nmol C g (dry weight)−1 day−1 (mean ± standard deviation; n = 3) for CO2. Based on a comparison of the methane turnover and assimilation rates of methane-derived carbon in this zone, 1.6% ± 0.6% (mean ± standard deviation; n = 3) of the carbon of the CH4 consumed was assimilated into mat biomass.
Patterns of δ13C isotope signatures.
For mat section 1 the δ13C values determined with SIMS ranged from −62.4 to −77.3‰, with a mean ± standard deviation of −68.9‰ ± 4.4 ‰ (n = 20) and a median ± median deviation of −69.8‰ ± 3.7 ‰ (Fig. 3A and Table 2). For mat section 2 the values ranged from −31.3 to −77.7‰, with a mean ± standard deviation of −66.5‰ ± 9.3‰ (n = 56) and a median ± median deviation of −67.4‰ ± 6.5‰ (Fig. 3B and Table 3). The average for both mat sections was −67.1‰ ± 8.3 ‰ (mean ± standard deviation; n = 76). We compared the distribution of ANME-1 and ANME-2c cells in detail with the distribution of δ13C values in mat section 1 (Fig. 3A). For the ANME-1-dominated zone (depth, 0.4 to 2.6 mm), the δ13C values for the mat biomass were −66.4‰ ± 3.9 ‰ (mean ± standard deviation) (spots C to H and J), whereas for the ANME-2c-dominated zone (depth, 2.6 to 6.0 mm) the values were −72.9‰ ± 2.2 ‰ (mean ± standard deviation) (spots I, K to O, and U). The difference between the δ13C values for the ANME-1- and ANME-2c-dominated zones was significant (P < 0.005, as determined by a paired-sample t test). These findings correspond to the results of previous SIMS (29) and membrane lipid (1) analyses showing an offset of −10 to −20‰ in the isotopic values between ANME-1 biomass and ANME-2 biomass, with more depleted δ13C values in the ANME-2 biomass. We also compared the distributions of δ13C in three different areas defined on the basis of the beta imager micrographs as the less active surface layer (at the mat-water interface with a low level of 14C incorporation), the highly active subsurface layer (the distinct subsurface layer with a high level of 14C incorporation), and the less active central layer (the deeper mat layer below the highly active subsurface layer) in mat sections 1 and 2. Tables 2 and 3 show the results for selected spots. For both mat sections, similar δ13C values were obtained for the less active surface layer and the highly active subsurface layer; the values were −65.9‰ ± 4.4 ‰ (mean ± standard deviation; n = 4) for the less active surface layer and −66.6‰ ± 4.0 ‰ (mean ± standard deviation; n = 5) for the highly active subsurface layer in mat section 1 and −67.5‰ ± 4.9 ‰ (mean ± standard deviation; n = 19) for the less active surface layer and −64.0‰ ± 11.5 ‰ (mean ± standard deviation; n = 29) for the highly active subsurface layer in mat section 2. Lower δ13C values were found in the deeper less active central layer; the values were −71.0‰ ± 3.7 ‰ (mean ± standard deviation; n = 11) in mat section 1 and −73.1‰ ± 2.7 ‰ (mean ± standard deviation; n = 8) in mat section 2.
TABLE 2.
δ13C values and standard errors for spots determined by SIMS in mat section 1 shown in Fig. 3
| Spot in mat section | Area in beta imager micrographa | δ13C (‰) | SE |
|---|---|---|---|
| A | LASL | −62.4 | 0.9 |
| B | LASL | −67.5 | 0.9 |
| C | LASL | −62.4 | 0.7 |
| D | HASSL | −62.6 | 0.7 |
| E | HASSL | −69.9 | 0.5 |
| F | HASSL | −63.4 | 0.6 |
| G | HASSL | −71.5 | 1.0 |
| I | HASSL | −65.4 | 0.6 |
| J | LASL | −71.4 | 3.8 |
| K | LACL | −69.7 | 1.7 |
| L | LACL | −72.1 | 1.0 |
| M | LACL | −71.3 | 0.6 |
| N | LACL | −74.2 | 0.6 |
| O | LACL | −71.4 | 0.6 |
| P | LACL | −77.3 | 0.7 |
| Q | LACL | −65.8 | 1.0 |
| R | LACL | −73.4 | 1.2 |
| S | LACL | −64.3 | 0.9 |
| T | LACL | −69.0 | 0.7 |
| U | LACL | −72.8 | 0.7 |
| E. coli (n = 9) | −22.3b | 0.9 |
Locations of the spots in the beta imager micrograph. The mat was divided into the less active surface layer (LASL), the highly active subsurface layer (HASSL), and the less active central layer (LACL). For further explanation see the text.
Average δ13C value for clusters of the E. coli standard.
TABLE 3.
δ13C values and standard errors for spots determined by SIMS in the mat section shown in Fig. 3
| Spot in mat section | Area in beta imager micrographa | δ13C | SE |
|---|---|---|---|
| A | LASL | −74.7 | 0.5 |
| B | HASSL | −75.5 | 0.6 |
| C | HASSL | −60.7 | 1.4 |
| D | HASSL | −63.8 | 0.6 |
| E | HASSL | −62.9 | 1.3 |
| F | LASL | −66.2 | 1.5 |
| G | LASL | −77.0 | 1.7 |
| I | LASL | −66.7 | 0.8 |
| J | LASL | −64.5 | 0.9 |
| K | LASL | −64.3 | 0.5 |
| L | LACL | −69.1 | 0.9 |
| M | LACL | −72.6 | 0.8 |
| N | LACL | −70.4 | 1.1 |
| O | LASL | −65.3 | 0.8 |
| P | LASL | −63.1 | 0.8 |
| Q | LASL | −63.4 | 1.1 |
| R | LASL | −63.0 | 0.8 |
| S | HASSL | −76.3 | 0.8 |
| T | HASSL | −75.8 | 0.8 |
| U | HASSL | −72.2 | 1.0 |
| V | LASL | −63.7 | 0.7 |
| W | LASL | −65.3 | 0.9 |
| X | LASL | −66.2 | 2.4 |
| Y | HASSL | −75.0 | 0.9 |
| Z | HASSL | −72.2 | 1.1 |
| AA | HASSL | −65.6 | 0.8 |
| BB | HASSL | −66.2 | 0.8 |
| CC | LACL | −76.0 | 0.8 |
| DD | LASL | −76.0 | 0.5 |
| EE | LASL | −60.7 | 1.1 |
| FF | LACL | −71.4 | 0.9 |
| GG | LACL | −76.6 | 2.2 |
| HH | HASSL | −75.7 | 0.9 |
| II | HASSL | −66.8 | 0.8 |
| JJ | HASSL | −77.7 | 0.6 |
| KK | LACL | −73.7 | 0.5 |
| LL | LACL | −74.6 | 0.7 |
| 1 | HASSL | −57.6 | 1.3 |
| 2 | HASSL | −60.2 | 1.3 |
| 3 | HASSL | −60.4 | 1.7 |
| 4 | HASSL | −56.0 | 2.6 |
| 5 | HASSL | −39.9 | 4.6 |
| 6 | HASSL | −31.3 | 5.4 |
| 7 | HASSL | −67.8 | 2.6 |
| 8 | HASSL | −68.0 | 2.1 |
| 9 | HASSL | −73.3 | 1.2 |
| 10 | HASSL | −68.8 | 1.8 |
| 11 | HASSL | −68.7 | 1.0 |
| 12 | LASL | −69.0 | 0.6 |
| 13 | HASSL | −67.1 | 1.9 |
| 14 | LASL | −75.4 | 0.9 |
| 15 | HASSL | −47.3 | 2.3 |
| 16 | LASL | −70.9 | 1.6 |
| 17 | HASSL | −43.5 | 1.4 |
| 18 | HASSL | −60.7 | 1.4 |
| 19 | LASL | −67.7 | 0.7 |
Locations of the spots in the beta imager micrograph. The mat was divided into the less active surface layer (LASL), the highly active subsurface layer (HASSL), and the less active central layer (LACL). For further explanation see the text.
Formation of methane from bicarbonate.
Methanotrophic mats incubated with H14CO3− in long-term experiment 1 formed methane from CO2 in the presence of methane and sulfate at rates of 4.1 ± 3.5 μmol g (dry weight)−1 day−1 (mean ± standard deviation; n = 3) in experiment 1a and 1.0 ± 0.5 μmol g (dry weight)−1 day−1 (mean ± standard deviation; n = 3) in experiment 1b (Fig. 6A). No methane formation was observed in the fixed control samples. In a comparison with the AOM rates determined using the same experimental setups, the ratio of AOM to methane formation was approximately 5:1. In these long-term experiments (62 and 109 days) between 13 and 44% of the 14CH4 tracer was turned over in AOM incubations; i.e., the total dissolved methane concentrations were reduced to values ranging from approximately 1.6 to 2.4 mM. In methane formation incubations between 0.6 and 2.1% of the 14CO2 tracer was turned over; i.e., the concentrations of total dissolved inorganic carbon were reduced only insignificantly. In short-term experiment 2a (without hydrogen) substantial methane formation from CO2 was detected in incubations with approximately 1.4 mM methane; the rates were 2.3 ± 2.5 μmol g (dry weight)−1 day−1 (n = 3) in the presence of sulfate and 2.2 ± 1.2 μmol g (dry weight)−1 day−1 (n = 3) in the absence of sulfate (Fig. 6B). In the absence of methane, the rate of methane formation from CO2 was very low (≤0.01 μmol g [dry weight]−1 day−1) or not detectable. No methane formation was detected in control samples. The addition of hydrogen in experiment 2b did not result in an increase in methane formation from CO2. The rates of methane formation in incubations with methane and sulfate were similar to those in experiment 2a (1.7 ± 0.6 μmol g [dry weight]−1 day−1; n = 3) (Fig. 6C). No methane formation was detected in incubations without methane or in control samples. The only significant difference from experiment 2a was observed for incubations with methane but without sulfate. In this case, the rates were still considerable (0.16 ± 0.15 μmol g [dry weight]−1 day−1; n = 3), but they were about 1 order of magnitude lower than those obtained in experiment 2a. An explanation for this might be the different “morphotypes” of mat samples incubated. In experiment 2a we incubated one large mat piece per replicate, whereas in experiment 2b a number of very small broken mat pieces were included in each replicate. The disruption of the mat into smaller pieces might have led to the loss of sulfate stored in internal microchannels of the mat. Cell-free channels have been reported to comprise between 5 and 40% of the bulk mat volume (23; Knittel, unpublished data). Since the mats were stored in sulfate-rich medium prior to the experiments, the larger mat pieces used in experiment 2a might have utilized sulfate trapped within the mat matrix and internal microchannels during incubation without sulfate. The observed coupling of methane production to methane and sulfate has further implications for the interpretation of methane formation in the mat (see Discussion).
FIG. 6.
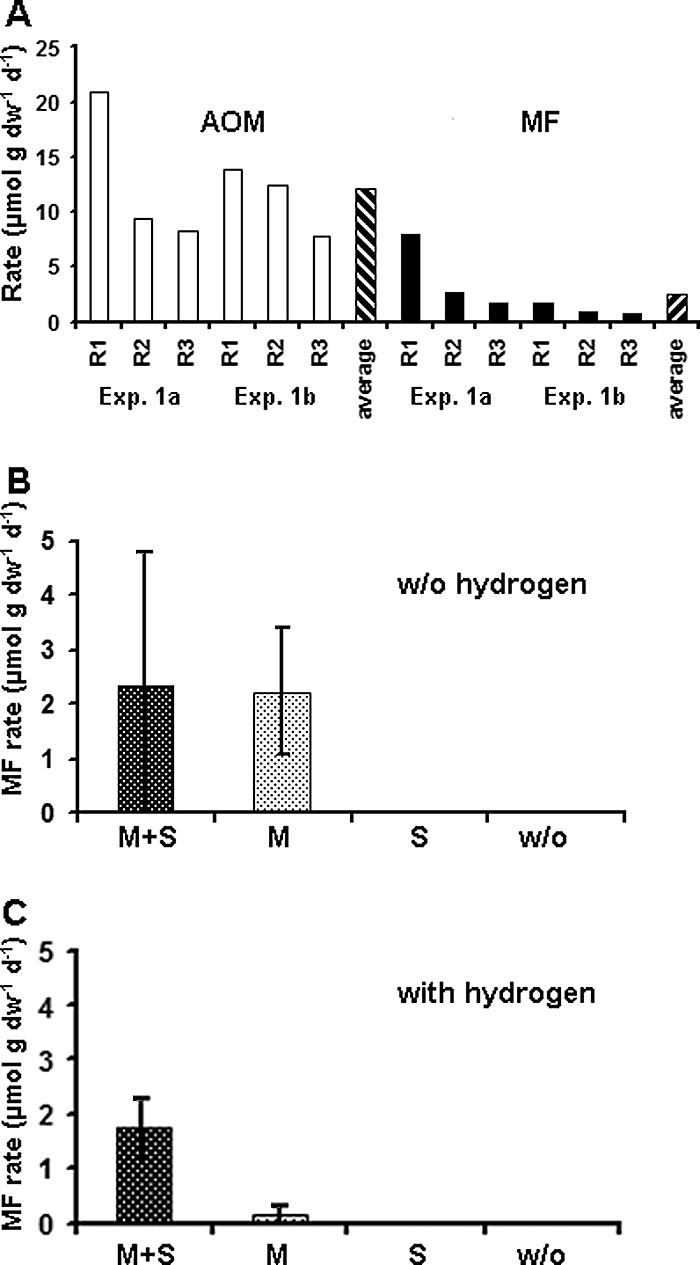
(A) Rates of AOM and methane formation (MF) determined in long-term experiment 1. R1 to R3 were replicate samples from either experiment 1a or experiment 1b. The AOM and MF rates were averaged to summarize the results of experiments 1a and b. (B and C) Methane formation rates determined in short-term experiment 2 in the presence or absence of methane, sulfate, and hydrogen. (B) Incubation without hydrogen added. (C) Incubation with hydrogen added. M+S, methane and sulfate; M, methane but no sulfate; S, sulfate but no methane; w/o, no methane and no sulfate. The error bars indicate standard deviations (n = 3). dw, dry weight.
DISCUSSION
Spatial patterns of methanotrophy and carbon assimilation in the mat.
The mat samples used in our long-term incubation experiments originated from a free-standing reef structure that was 30 cm in diameter (Fig. 1) with a mat that was approximately 5 to 8 cm thick. The samples were taken from the outer 1.5 cm of the mat, and the color changed from black to pink from the exterior to the interior of the mat. SIMS surveys of δ13C values throughout the ANME-dominated mat confirmed that assimilation of carbon derived from methane was the main biomass-building process. The mat had a δ13C carbon isotopic signature of −67.1‰ ± 8.3 ‰ (mean standard ± deviation; n = 76), which is in the same range as the isotopic signature of the gas emanating from the seeps of the northwestern Crimean shelf (−62.4 to −68.3‰ δ13C) (23) and is in accordance with previous measurements for bulk mat biomass from the same area (−72.2‰ δ13C) (23). Interestingly, the isotopic signatures of specific biomarker lipids of ANME microorganisms and their sulfate-reducing partners, which make up a large proportion of the mat, are substantially more depleted than the bulk biomass (23). Blumenberg et al. (1) found specific lipids with isotope signatures between −91 and −111‰ δ13C for archaea and between −79 and −85‰ δ13C for bacteria in mat samples from the same area that we investigated in this study. Here we studied the small-scale distribution of isotope signatures using spots that were approximately 15 to 20 μm in diameter using SIMS. The heaviest isotope value (−31‰ δ13C) and the lightest value (−77 ‰ δ13C) detected with SIMS were for individual spots for the same mat piece. However, most spots showed little variation from the median, around −67‰ δ13C. Hence, the δ13C values obtained (Tables 2 and 3 and Fig. 3) most likely represented a mixture of signals from different cells and cell colonies, including anaerobic methanotrophs with isotopically very light carbon signals, as well as organisms utilizing different carbon sources with enriched δ13C values. Furthermore, the two-dimensional mapping of SIMS values did not indicate the presence of distinct zones where there was similar isotopic depletion on a scale of 0.1 to 10 mm. The heavier and lighter spots were well mixed, and there was not a significant difference between the areas where there was less carbon assimilation and the areas where there was more carbon assimilation. This indicated that cells having different metabolisms were associated at locations that were within a few tenths of a micrometer from each other. The only significant pattern detected with regard to isotope signatures was found between areas dominated by ANME-1 cells (−66.4‰ ± 3.9 ‰ [mean standard deviation; n = 7]) and areas dominated by ANME-2 cells (−72.9‰ ± 2.2 ‰ [mean standard deviation; n = 7]).
The SIMS measurements can be utilized to speculate about the mixture of methanotrophic consortia with other types of microbial metabolism in the bulk mat. Typical values for organic matter in sediment traps and surface sediment samples from the Black Sea range from −25 to −28‰ δ13C according to Wakeham et al. (48), and microorganisms feeding on such carbon sources have an isotope signature slightly lighter than these values (−25 to −32‰ δ13C) (48). In contrast, methanotrophic cells have significantly lighter isotope signatures due to assimilation of methane-derived carbon (9). Hence, a bulk value close to −70‰ δ13C suggests that more than one-half of the mat biomass is composed of ANME cells, whereas the remaining community feeds on isotopically heavier carbon sources.
14C beta microimaging of methane and CO2 assimilation suggested that there was a carbon assimilation peak in a distinct layer that was approximately 1 to 2 mm thick adjacent to the mat surface dominated by ANME-1 microorganisms in close association with their partner bacteria (Fig. 3). The carbon assimilation rates detected in this layer ranged from 100 to 500 nmol C g (dry weight)−1 day−1 for methane and from 700 to 2,500 nmol C g (dry weight)−1 day−1 for CO2. The relative contribution of CO2 uptake is astonishingly high, indicating that the mat may have harbored a significant proportion of other chemoautotrophic microorganisms. Some of the CO2 could also have been utilized by the abundant methanotrophic cells since the synthesis of biomass from methane, which is the most reduced organic carbon compound, also requires the incorporation of a less reduced compound, such as CO2 (24). Of the methane consumed, only 1 to 2% was incorporated into mat biomass. This growth yield is similar to that obtained in in vitro experiments with enrichments of ANME-2 consortia (24). However, it is difficult to compare the values, since the contribution of ANME biomass to the total carbon dry weight of the mat is unknown.
Based on the spatial distribution of carbon assimilation we suggest that the growth of a mat covering a central carbonate pillar is radial, starting from small nodules and proceeding to larger structures internally supported by calcification of old mat (44). Older portions of the mat may be gradually overgrown because of more favorable energy availability at the outside. Methane probably advects from the inner methane-filled carbonate cavities of the reef to the periphery, but the sulfate supply may decrease considerably from the outer mat surface toward the interior due to microbial consumption. However, preliminary results for sulfate measurements in fluid taken from the inside of a reef structure revealed a sulfate concentration of 6 mM, which is still sufficient to enable AOM activity (T. Treude, unpublished data). Another problem may be that the older mat could be cut off from the energy supply by increasing calcification. Unfortunately, it was not possible with our sampling technique (plastic tubes) to penetrate into the more carbonated parts of the interior mat, and we do not have comparative measurements for methane oxidation and sulfate reduction in different parts of the reef structures. Hence, further studies are necessary to investigate methanotrophic activity and substrate supply in this transition zone.
Distribution of ANME-1 and ANME-2c.
The ANME-1 and ANME-2c methanotrophic archaeal groups exhibited distinct zonation within the mat, with bright ANME-1 signals in the layer most active in carbon assimilation close to the mat surface and stronger ANME-2 signals toward deeper sections of the mat. However, members of both groups were present at all depths. This finding is different from the results of previous investigations, which showed that ANME-2 was completely absent in the reef pillar (1, 23) or that there was a clear differentiation between black ANME-2 and pinkish ANME-1 mat sections (35). This indicates that the Black Sea mats are rather inhomogeneous in terms of the relative abundance of populations and their spatial distributions. The community structure in the methanotrophic mats may be controlled by energetic constraints which are not yet understood; however, as in other biofilms, complex factors such as dispersal, competition, and succession may also influence the composition and distribution of microbes (44).
The close cell-to-cell contact of ANME-1 and ANME-2c cells found in some parts of the mat is very interesting and suggests that the two groups share the same niche. This finding contrasts with observations for some cold-seep sediments, where a clear vertical separation of the two groups was observed (15). Another interesting observation from this work is the lack of ANME-2/Desulfococcus-Desulfosarcina aggregates, which were abundant in other mat samples from the same location (1, 15, 35), as well as generally in cold-seep sediments (2, 14, 15, 27, 28). Monospecific ANME-2 aggregates have been found in gassy sediments in the Baltic Sea, where they occur beside monospecific DSS aggregates (45), as well as in methane seeps in the Eel River Basin (29). However, the factors controlling the aggregation or isolation of the two groups are unknown. The presence of high concentrations of ANME-1 cells without SRB belonging to the Desulfococcus-Desulfosarcina cluster in some parts of the mat again raises the question whether SRB form a syntrophic partnership with ANME-1 cells (29, 39). Indications that there were large mat sections with mostly empty sheaths of ANME-1 cells, as reported in a different study (>80% of the mat volume) (34), were not obtained in our investigation, since the CARD-FISH analyses showed the presence of intact ANME cells throughout the sections analyzed here. The lowest proportion of ANME cells was found in the mat surface layer (thickness, approximately 0.5 to 1 mm, but varying slightly between mat pieces). It is likely that the mat surface is utilized by colonizing microbes from the water column, which form a thin biofilm on top of the actual methanotrophic mat. In a parallel study ANME and Desulfococcus-Desulfosarcina cells accounted for only about 7 and 4%, respectively, of the total cell abundance in the top 1 mm of similar mats (K. Knittel, unpublished data).
Methane formation in the mat.
Concurrent with AOM, we detected substantial formation of methane from CO2 by the methanotrophic mat investigated here. However, we could not stimulate gross methanogenesis in radiotracer experiments by addition of hydrogen or removal of sulfate to inhibit SRB that usually compete with methanogens for hydrogen (53). Hence, it appears unlikely that the observed methanogenic activity was mediated by methanogens. Also, 16S rRNA gene library analysis did not reveal phylotypes related to methanogenic archaea other than members of the ANME-1 and ANME-2 groups (number of clones, >100) (15). Early studies of AOM using cultured methanogenic archaea revealed that methane consumption occurred simultaneously with methane production (54, 55). Here, the percentage of methane consumption during methanogenesis was up to 8% in incubations of methanogenic sludge. This observation and further studies using thermodynamics and molecular biology led to the hypothesis that AOM represents an enzymatic reversal in primarily methanogenic archaea (2, 8-10, 28, 47, 55). Recent molecular studies supported this hypothesis by revealing differences in nickel cofactor F430 of methyl-coenzyme M reductase in ANMEs from different environments, including samples from the Black Sea mats (6, 17). Methyl-coenzyme M reductase is the terminal enzyme of methanogenesis and hence is a likely candidate to initiate the first step in AOM. Furthermore, most of the unique genes involved in methanogenesis were found to be present in ANME-1 archaea (7). Hence, one hypothesis to explain the observed methane formation is the possible reversibility of AOM, if the enzymes of this process can react in both directions, like those involved in methanogenesis. It was difficult to determine the exact ratio of methane production during AOM from our long-term experiments as we did not ensure that there was a constant methane concentration throughout the incubation period and rereactions between the 14CH4 and 14CO2 tracers were possible. However, assuming that the mean methane formation rate was 2.0 ± 1.6 μmol g (dry weight)−1 day−1 (n = 6) measured during our short-term incubations with methane-sulfate and that the reported AOM rates on average range from 12 to 81 μmol g (dry weight)−1 day−1 (23, 25; this study), the level of methane production during AOM may range from 2 to 17% of the total methane turnover. These percentages are similar to findings of Orcutt et al. (26), who observed coinciding profiles for AOM and methanogenesis (from CO2-H2) in cold-seep sediments from the Gulf of Mexico and about 18% methane production from total methane turnover. In another study using mat material from the same reef that was investigated here, the ratio of AOM to methanogenesis was found to be 2:1 in long-term in vitro experiments (37). The results of previous studies of bacterial mat samples from the Black Sea reporting H2-CO2-based methane production rates that were on the order of three times higher than the rates of AOM are more difficult to explain (32). However, in these studies the mat samples were collected by trawling and exposed to air prior to anoxic incubation, which may have compromised the viability of the mats and skewed the relative rates of AOM, methanogenesis, and sulfate reduction.
Conclusions.
Here, we investigated the spatial patterns of stable carbon isotope signatures, as well as the spatial patterns of CH4 and CO2 assimilation, in relation to the distribution of ANME groups and their associated bacteria in mat samples obtained from the surface of a large methanotrophic reef structure. A combination of different methods, including radiotracer incubation, beta microimaging, SIMS, and CARD-FISH, was used with sections of mat obtained from a large reef structure in order to locate hot spots of methanotrophy and to identify the microbial consortia responsible. Furthermore, CO2 reduction to methane was investigated in the presence and absence of methane, sulfate, and hydrogen. The mat had an average δ13C carbon isotopic signature of −67.1‰, indicating that methane was the main carbon source. Regions dominated by ANME-1 had significantly heavier isotope signatures (−66.4‰) than the more central regions dominated by ANME-2 (−72.9‰). Incorporation of 14C from radiolabeled CH4 or CO2 revealed one hot spot for methanotrophy and CO2 fixation close to the surface of the mat. Replicate incubations of the mat with 14CH4 or 14CO2 revealed reduction of CO2 to CH4 that was about 10% of the AOM. However, since the methane formation observed required the presence of methane and sulfate and was not affected by addition of hydrogen, it appeared to be related to the AOM rather than to net methanogenesis.
Acknowledgments
We acknowledge F. Widdel for his advice concerning the interpretation of the methane formation experiments. We thank K. Nauhaus and N. Finke for fruitful discussions about methane formation in the mat. We thank the officers, crew, and shipboard scientific party of R/V Professor Logachev and the JAGO Team for excellent support during the Black Sea cruise in summer 2001. Special thanks are due to W. Michaelis. We acknowledge B. Ratunde, T. Wilkop, and I. Mueller for providing technical assistance. We thank the GHOSTDABS project and the JAGO Team for providing the reef pictures. We thank Kevin McKeegan for assistance with the SIMS configuration. Two anonymous reviewers are acknowledged for their comments.
The UCLA ion microprobe is supported by the W. M. Keck Foundation and by a grant from the National Science Foundation Instrumentation and Facilities Program (grant EAR 01-13563). Support for V. Orphan and C. House, as well as for the SIMS analyses, was provided by the NSF Microbial Interactions and Processes Program (grant 03488492). This is a publication of the MUMM (03G0554A) and GHOSTDABS (03G0559A) programs supported by the German Ministry of Education and Research (BMBF) and the German Research Foundation (DFG). Additional support was provided by the Max-Planck-Gesellschaft (Germany) and by the NASA Astrobiology Institute.
Footnotes
Published ahead of print on 2 February 2007.
Publication no. GEOTECH-258 of the R&D program GEOTECHNOLOGIEN.
REFERENCES
- 1.Blumenberg, M., R. Seifert, J. Reitner, T. Pape, and W. Michaelis. 2004. Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. USA 101:11111-11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Giesecke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 3.Elvert, M., E. Suess, J. Greinert, and M. J. Whiticar. 2000. Archaea mediating anaerobic methane oxidation in deep-sea sediments at cold seeps of the eastern Aleutian subduction zone. Org. Geochem. 31:1175-1187. [Google Scholar]
- 4.Girguis, P. R., A. E. Cozen, and E. F. DeLong. 2005. Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl. Environ. Microbiol. 71:3725-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girguis, P. R., V. J. Orphan, S. J. Hallam, and E. F. DeLong. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:5472-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. DeLong. 2004. Reverse methanognesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 8.Harder, J. 1997. Anaerobic methane oxidation by bacteria employing 14C-methane uncontaminated with 14C-carbon monoxide. Mar. Geol. 137:13-23. [Google Scholar]
- 9.Hinrichs, K.-U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. De Long. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 10.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediments: evidence for methanogen-sulphate reducer consortium. Global Biochem. Cycles 8:451-463. [Google Scholar]
- 11.Jørgensen, B. B., and T. Fenchel. 1974. The sulfur cycle of a marine sediment model system. Mar. Biol. 24:189-201. [Google Scholar]
- 12.Joye, S. B., A. Boetius, B. N. Orcutt, J. P. Montoya, H. N. Schulz, M. J. Erickson, and S. K. Logo. 2004. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem. Geol. 205:219-238. [Google Scholar]
- 13.Kallmeyer, J., T. G. Ferdelman, A. Weber, H. Fossing, and B. B. Jørgensen. 2004. A cold chromium distillation procedure for radiolabeled sulfide applied to sulfate reduction measurements. Limnol. Oceanogr. Methods 2:171-180. [Google Scholar]
- 14.Knittel, K., A. Boetius, A. Lemke, H. Eilers, K. Lochte, O. Pfannkuche, and P. Linke. 2003. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrates (Cascadia Margin, Oregon). Geomicrobiol. J. 20:269-294. [Google Scholar]
- 15.Knittel, K., T. Lösekann, A. Boetius, R. Kort, and R. Amann. 2005. Diversity and distribution of methanotrophic archaea (ANME) at cold seeps. Appl. Environ. Microbiol. 71:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krueger, M., T. Treude, H. Wolters, K. Nauhaus, and A. Boetius. 2005. Microbial methane turnover in different marine habitats. Palaeogeogr. Paleoclimatol. Palaeoecol. 227:6-17. [Google Scholar]
- 17.Krüger, M., A. Meyerdierks, F. O. Glöckner, R. Amann, F. Widdel, M. Kube, R. Reinhard, J. Kahnt, R. Böcher, R. K. Thauer, and S. Shima. 2003. A conspicuous nickel protein in microbial mats that oxidise methane anaerobically. Nature 426:878-881. [DOI] [PubMed] [Google Scholar]
- 18.Lanièce, P., Y. Charon, A. Cardona, L. Pinot, S. Maitrejean, R. Mastrippolito, B. Sandkamp, and L. Valentin. 1998. A new high resolution radioimager for quantitative analysis of radiolabelled molecules in tissue section. J. Neurosci. Methods 86:1-5. [DOI] [PubMed] [Google Scholar]
- 19.Lein, A. Y., M. V. Ivanov, N. V. Pimenov, and M. B. Gulin. 2002. Geochemical characteristics of the carbonate constructions formed during microbial oxidation of methane under anaerobic conditions. Mikrobiologiya 71:89-103. [PubMed] [Google Scholar]
- 20.Luth, C., U. Luth, A. V. Gebruck, and H. Thiel. 1999. Methane gas seeps along the oxic/anoxic gradient in the Black Sea: manifestations, biogenic sediment compounds and preliminary results on benthic ecology. Mar. Ecol. 20:221-249. [Google Scholar]
- 21.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. 1998. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microb. Ecol. 25:43-61. [Google Scholar]
- 22.Meyerdierks, A., M. Kube, T. Lombardot, K. Knittel, M. Bauer, F. O. Glöckner, R. Reinhard, and R. Amann. 2005. Insights into the genomes of archaea mediating the anaerobic oxidation of methane. Environ. Microbiol. 7:1937-1951. [DOI] [PubMed] [Google Scholar]
- 23.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, K. Knittel, A. Gieseke, K. Peterknecht, T. Pape, A. Boetius, A. Aman, B. B. Jørgensen, F. Widdel, J. Peckmann, N. V. Pimenov, and M. Gulin. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 24.Nauhaus, K., M. Albrecht, M. Elvert, A. Boetius, and F. Widdel. 2007. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ. Microbiol. 9:187-196. [DOI] [PubMed] [Google Scholar]
- 25.Nauhaus, K., T. Treude, A. Boetius, and M. Krüger. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I- and ANME-II-communities. Environ. Microbiol. 7:98-106. [DOI] [PubMed] [Google Scholar]
- 26.Orcutt, E., A. Boetius, M. Elvert, V. Samarkin, and S. B. Joye. 2005. Molecular biogeochemistry of sulfate reduction, methanogenesis and anaerobic oxidation of methane at Gulf of Mexico cold seeps. Geochim. Cosmochim. Acta 69:4267-4281. [Google Scholar]
- 27.Orphan, V. J., K.-U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Tayleor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. De Long. 2001. Methane-consuming Archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 29.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Acad. Nat. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancost, R. D., J. S. Sinninghe Damsté, S. De Lint, M. J. E. C. Van der Maarel, and J. C. Gottschal. 2000. Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic archaea and bacteria. Appl. Environ. Microbiol. 66:1126-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pimenov, N. V., I. I. Rusanov, M. N. Poglazova, L. L. Mityushina, D. Y. Sorokin, V. N. Khmelenina, and Y. A. Trotsenko. 1997. Bacterial mats on coral-like structures at methane seeps in the Black Sea. Microbiology 66:354-360. [Google Scholar]
- 33.Raghoebarsing, A. A., A. Pol, K. T. van de Pas-Schoonen, A. J. P. Smolders, K. F. Ettwig, W. I. C. Rijpstra, S. Schouten, J. S. S. Damsté, H. J. Op den Camp, M. S. Jetten, and M. Strous. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918-921. [DOI] [PubMed] [Google Scholar]
- 34.Reitner, J., J. Peckmann, M. Blumenberg, W. Michaelis, A. Reimer, and V. Thiel. 2005. Concretionary methane-seep carbonates and associated microbial communities in Black Sea sediments. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227:18-30. [Google Scholar]
- 35.Reitner, J., J. Peckmann, A. Reimer, G. Schumann, and V. Thiel. 2005. Methane-derived carbonate build-ups and associated microbial communities at cold seeps on the lower Crimean shelf (Black Sea). Facies 51:66-79. [Google Scholar]
- 36.Schramm, A., L. H. Larsen, N. P. Revsbech, N. B. Ramsing, R. Amann, and K.-H. Schleifer. 1996. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 62:4641-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seifert, R., K. Nauhaus, M. Blumenberg, M. Krueger, and W. Michaelis. 2006. Methane dynamics in a microbial community of the Black Sea traced by stable isotopes in vitro. Org. Geochem. 37:1411-1419. [Google Scholar]
- 38.Shima, S., and R. K. Thauer. 2005. Methyl-coenzyme M reductase and the anaerobic oxidation of methane in methanotrophic Archaea. Curr. Opin. Microbiol. 8:643-648. [DOI] [PubMed] [Google Scholar]
- 39.Sørensen, K. B., K. Finster, and N. B. Ramsing. 2001. Thermodynamic and kinetic requirements in anaerobic methane oxidizing consortia exclude hydrogen, acetate and methanol as possible electron shuttles. Microb. Ecol. 42:1-10. [DOI] [PubMed] [Google Scholar]
- 40.Stadnitskaia, A., G. Muyzer, B. Abbasa, M. J. L. Coolen, E. C. Hopmans, M. Baas, T. C. E. van Weeringa, M. K. Ivanov, E. Poludetkina, and J. E. Sinninghe Damste. 2005. Biomarker and 16S rDNA evidence for anaerobic oxidation of methane and related carbonate precipitation in deep-sea mud volcanoes of the Sorokin Trough, Black Sea. Mar. Geol. 217:67-96. [Google Scholar]
- 41.Thiel, V., J. Peckmann, H. H. Richnow, U. Luth, J. Reitner, and W. Michaelis. 2001. Molecular signals for anaerobic methane oxidation in Black Sea seep carbonates and microbial mat. Mar. Chem. 73:91-112. [Google Scholar]
- 42.Tourova, T. P., T. V. K. Kolganova, B. B. Kuznetsov, and N. V. Pimanov. 2002. Phylogenetic diversity of the archaeal component in microbial mats on coral-like structures associated with methane seeps in the Black Sea. Microbiologiya 71:196-201. [PubMed] [Google Scholar]
- 43.Treude, T., A. Boetius, K. Knittel, K. Wallmann, and B. B. Jørgensen. 2003. Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar. Ecol. Prog. Ser. 264:1-14. [Google Scholar]
- 44.Treude, T., K. Knittel, M. Blumenberg, R. Seifert, and A. Boetius. 2005. Subsurface microbial methanotrophic mats in the Black Sea. Appl. Environ. Microbiol. 71:6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treude, T., M. Krüger, A. Boetius, and B. B. Jørgensen. 2005. Environmental control on anaerobic oxidation of methane in the gassy sediments of Eckernförde Bay (German Baltic). Limnol. Oceanogr. 50:1771-1786. [Google Scholar]
- 46.Treude, T., J. Niggemann, J. Kallmeyer, P. Wintersteller, C. J. Schubert, A. Boetius, and B. B. Jørgensen. 2005. Anaerobic oxidation of methane in the sulfate-methane transition along the Chilean continental margin. Geochim. Cosmochim. Acta 69:2767-2779. [Google Scholar]
- 47.Valentine, D. L., and W. S. Reeburgh. 2000. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2:477-484. [DOI] [PubMed] [Google Scholar]
- 48.Wakeham, S. G., C. M. Lewis, E. C. Hopman, S. Schouten, and J. S. Sinninghe Damsté. 2003. Archaea mediate anaerobic oxidation of methane in deep euxinic waters of the Black Sea. Geochim. Cosmochim. Acta 67:1359-1374. [Google Scholar]
- 49.Widdel, F. 1988. Microbiology and ecology of sulfate- and sulfur-reducing bacteria, p. 469-585. In A. D. J. Zehnder (ed.), Biology of anaerobic microoganisms. John Wiley & Sons, New York, NY.
- 50.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer, New York, NY.
- 51.Widdel, F., A. Boetius, and R. Rabus. 2004. Anaerobic biodegradation of hydrocarbons including methane, p. 1028-1049. In The prokaryotes Online version. Springer Verlag, New York, NY.
- 52.Yamamoto, S., J. B. Alcauskas, and T. E. Crozier. 1976. Solubility of methane in distilled water and seawater. J. Chem. Eng. Data 21:78-80. [Google Scholar]
- 53.Zehnder, A. J. B. 1988. Biology of anaerobic microorganisms. Wiley, New York, NY.
- 54.Zehnder, A. J. B., and T. D. Brock. 1980. Anaerobic methane oxidation: occurrence and ecology. Appl. Environ. Microbiol. 39:194-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zehnder, A. J. B., and T. D. Brock. 1979. Methane formation and methane oxidation by methanogenic bacteria. J. Bacteriol. 137:420-432. [DOI] [PMC free article] [PubMed] [Google Scholar]