Abstract
Many marine demosponges contain large amounts of phylogenetically complex yet highly sponge-specific microbial consortia within the mesohyl matrix, but little is known about how these microorganisms are acquired by their hosts. Settlement experiments were performed with the viviparous Caribbean demosponge Ircinia felix to investigate the role of larvae in the vertical transmission of the sponge-associated microbial community. Inspections by electron microscopy revealed large amounts of morphologically diverse microorganisms in the center of I. felix larvae, while the outer rim appeared to be devoid of microorganisms. In juveniles, microorganisms were found between densely packed sponge cells. Denaturing gradient gel electrophoresis (DGGE) was performed to compare the bacterial community profiles of adults, larvae, and juvenile sponges. Adults and larvae were highly similar in DGGE band numbers and banding patterns. Larvae released by the same adult individual contained highly similar DGGE banding patterns, whereas larvae released by different adult individuals showed slightly different DGGE banding patterns. Over 200 bands were excised, sequenced, and phylogenetically analyzed. The bacterial diversity of adult I. felix and its larvae was comparably high, while juveniles showed reduced diversity. In total, 13 vertically transmitted sequence clusters, hereafter termed “IF clusters,” that contained sequences from both the adult sponge and offspring (larvae and/or juveniles) were found. The IF clusters belonged to at least four different eubacterial phyla and one possibly novel eubacterial lineage. In summary, it could be shown that in I. felix, vertical transmission of microorganisms through the larvae is an important mechanism for the establishment of the sponge-microbe association.
Sponges (Porifera) are evolutionarily ancient metazoa that first appeared in Precambrian times nearly 600 million years ago (28). Today, an estimated 13,000 species, classified in three classes (Demospongiae, Calcarea, and Hexactinellida) populate virtually all aquatic habitats from shallow tropical reefs to the polar seas and the deep ocean and even freshwater lakes and rivers (20). A great diversity of sponges occurs on coral reefs, where they exhibit a wide range of shapes and colors. Sponges have a primitive morphology, as they lack true organs or tissues. Instead, sponges possess totipotent, mobile cells freely scattered in an extracellular matrix called the mesohyl, which is covered by a single cell layer, the pinacoderm. Inhalant and exhalant canals in the mesohyl build an aquiferous system through which water is pumped actively by flagellated choanocytes (3). As filter feeders, sponges efficiently take up nutrients like organic particles and microorganisms from the seawater, leaving the expelled water essentially sterile (37, 39, 57). Reproduction occurs either oviparously or viviparously. In oviparous sponges, gametes are released into the water during large, synchronized spawning events, with fertilization occurring outside the sponge body. Eggs of viviparous sponges are fertilized internally, and the brooding of embryos occurs within the mesohyl, often in specialized brooding chambers. After their release, larvae of viviparous sponges actively swim for a relatively short period of time (30) or creep over the substrate in search of suitable settlement sites. Upon settlement, they undergo a rapid metamorphosis to an early juvenile stage. Additionally, many sponges also reproduce asexually via budding or the formation of gemmulae.
The association of many demosponges with large microbial consortia is well recognized (16, 18, 19, 21). This complex microbial community differs significantly from seawater and sediments in both concentration and diversity (12, 17, 56). Approximately 40 to 60% of sponge biomass can consist of microorganisms, which are located mostly extracellularly in the mesohyl. This equals 108 to 1010 bacteria g−1 tissue and exceeds seawater concentrations by 2 to 4 orders of magnitude (11, 55). Morphological diversity of sponge-associated microorganisms was shown by electron microscopy. Several different morphotypes with unusual membrane features were described for various sponge species (12, 13, 53, 54, 58). More recently, classical cultivation as well as molecular studies based on 16S rRNA gene sequence information revealed a high phylogenetic diversity of sponge-associated microbial consortia. Members of eight phyla within the domain Bacteria (Proteobacteria [Alphaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria], Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Gemmatimonadetes, and Nitrospira) and one archaeal phylum (Crenarchaeota) have been detected (17, 34, 38, 49, 51, 56). Furthermore, a new candidate phylum, termed “Poribacteria,” has been detected in several high-microbial-abundance sponges (9, 10). This phylogenetically complex microbial consortium is highly sponge specific in that the corresponding phylogenetic lineages have been found in taxonomically and geographically different marine demosponges but have not been detected in seawater, sediments, other marine invertebrates, or a freshwater sponge (14).
Presently, little is known about how the unique and apparently stable sponge-microbe associations are established and maintained over time. Vertical transmission as a mechanism for bacterial passage between sponge generations was proposed by Levi and Porte in 1962 (26). Electron microscopy studies revealed the presence of microorganisms in oocytes of oviparous sponges and embryos/larvae of viviparous sponges (see reference 7 and references cited therein; 23-25, 40, 45). Vertical transmission of Cyanobacteria in developing eggs and sperm of the sponge Chondrilla australiensis (52) and transmission of a spiral bacterium through all stages of embryonic development in the sponge Halisarca dujardini (7) were reported. Similarly, Maldonado and coworkers (32) used electron microscopy to document the transmission of yeast in Chondrilla species. Enticknap et al. (6) were the first to phylogenetically identify alphaproteobacteria associated with larvae of the sponge Mycale laxissima and to confirm their presence by fluorescence in situ hybridization. Isolates JE061 to JE065 were most closely related to strain MBIC3368 that had previously been isolated from at least eight other demosponges (43). Sharp et al. (46) reported the vertical transmission of complex bacterial and archaeal consortia in the tropical sponge Corticium sp. Using fluorescently labeled 16S rRNA probes, it was possible to localize specific microbial lineages in the adult and in early and late stages of embryonic development.
For this study, the ball-shaped sponge Ircinia felix Duchassaing and Michelotti 1864 (order Dictyoceratida) (5) was chosen because it is a common and accessible species on shallow Caribbean coral reefs, grassbeds, and mangroves. It is easily identified by its gray to light brown color and hexagonally oriented, low white knobs that are interconnected by white lines. Ircinia species produce an array of low-molecular-weight volatile compounds that give them a strong, pungent smell (35). Ircinia sponges are viviparous and produce relatively large parenchymella-type larvae. In the Florida Keys, they are released in the early morning hours (0700 to 1000 h) in synchronized spawning events 1 to 2 days following the full moon, typically in May and June (N. Lindquist, personal observation). Larvae swim briefly after spawning and settle within minutes to several hours and then metamorphose to a flattened early juvenile stage within a day. The aim of this study was to identify, localize, and phylogenetically characterize the bacterial community in adults, larvae, and early-stage juveniles of I. felix. The different developmental stages were analyzed by a combination of molecular (denaturing gradient gel electrophoresis [DGGE]) and visual (electron microscopy) techniques.
MATERIALS AND METHODS
Larva settlement experiments.
Experiments were performed on a shallow tropical patch reef (25°01.610′N, 80°23.671′W) offshore of Key Largo, FL, in June and August 2004 using the NOAA's National Undersea Research Center facilities and vessels. Sponge larvae were collected in situ from four individual sponges (sponges 2, 3, 4, and 5) using methodology described previously by Lindquist et al. (30) and transferred into 3-liter holding trays containing natural seawater. Additionally, tissue samples were taken from the corresponding adult specimens from which larvae had been collected. Individual larvae were captured from the holding trays with glass Pasteur pipettes and were either transferred into 150 μl sterile filtered seawater for DNA extraction and stored at −80°C or fixed in 2.5% glutaraldehyde-double-distilled water for transmission electron microscopy and stored at 4°C. Larvae not used for microscopy or DNA extraction were settled by placing them into sealed plastic containers (∼75-ml volume) with NITEX (100-μm) “windows” to allow the flow of ambient seawater into the containers. I. felix larvae settled onto the nylon mesh (i.e., pantyhose) that held the larval collection containers on a rack at a depth of 9 m. During this setup process, larval and juvenile contact with air was avoided. Juveniles were recovered 1 to 3 days postsettlement, along with control pieces of mesh, which did not contain settled sponges. These samples were preserved as described above for DNA and microscopic analysis. Fluorescence in situ hybridization experiments could not be reliably performed because of the small larval size and limited juvenile biomass.
Transmission electron microscopy.
The adult sponge samples that had been preserved in 2.5% glutaraldehyde-double-distilled water were cut into small pieces of about 1 mm3. All samples (adults, larvae, and juveniles) were then washed five times in cacodylate buffer (50 mM, pH 7.2), fixed in 2% osmium tetroxide for 90 min, washed again five times in double-distilled water, and incubated overnight in 0.5% uranyl acetate. After dehydration in an ethanol series (30, 50, 70, 90, 96, and three times at 100% for 30 min, respectively), samples were incubated three times for 30 min in 1× propylene oxide, maintained overnight in 1:1 (vol/vol) propylene oxide-Epon 812 (Serva), incubated twice for 2 h in Epon 812, and finally embedded in Epon 812 for 48 h at 60°C. Samples were then sectioned with an ultramicrotome (OM U3; C. Reichert, Austria) and examined by transmission electron microscopy (EM 10; Zeiss, Germany).
DGGE.
Frozen pieces of adult sponge tissue were ground in liquid nitrogen with a mortar and pestle, and DNA was extracted using the Fast DNA Spin kit for soil (Q-Biogene, Heidelberg, Germany) in accordance with the manufacturer's instructions. Larva and juvenile DNA were extracted from three, five, or seven individual larvae or juveniles, which had been pooled immediately after collection, by heating in 150 μl of double-distilled water in a water bath for 10 min at 100°C. The solution was used as a template for PCR. The juvenile DNA was extracted together with a small piece of nylon on which the sponge had grown because it was impossible to remove the tiny sponge from its substratum without losing too much biomass. The universal primers 341f, with a GC clamp, and 907r (33) were used for PCR amplification of bacterial 16S rRNA genes. Cycling conditions on a Mastercycler gradient (Eppendorf, Hamburg, Germany) were as follows: an initial denaturing step at 95°C for 5 min and 30 cycles of denaturing at 95°C for 1 min, primer annealing at 54°C for 1 min, and elongation at 72°C for 45 s, followed by a final extension step at 72°C for 10 min. DGGE was performed with a Bio-Rad (München, Germany) DCode Universal mutation detection system on a 10% (wt/vol) polyacrylamide gel in 1× Tris-acetate-EDTA and using a 0 to 90% denaturing gradient; 100% denaturant corresponded to 7 M urea and 40% (vol/vol) formamide. Electrophoresis was performed for 6 h at 150 V and 60°C. Gels were stained for 30 min with SYBR gold (Molecular Probes) and scanned on a Typhoon 8600 scanner (Amersham Biosciences). DGGE banding pattern similarities were determined by cluster analysis using Quantity One (Bio-Rad, München, Germany). Selected bands were excised with an ethyl alcohol-sterilized scalpel and incubated in 25 μl double-distilled water overnight at 4°C. Four microliters of eluted DNA was subsequently used for reamplification with primers 341f and 907r under the PCR conditions described above. PCR products were ligated into the pGEM-T Easy vector (Promega) and transformed by electroporation into competent Escherichia coli XL1-Blue cells. Plasmid DNA of up to four different clones per excised band was isolated by standard miniprep procedures, and the correct insert size was verified by using agarose gel electrophoresis following restriction digestion (41). Sequencing was performed using an ABI 377XL automated sequencer (Applied Biosystems). Sequences were edited with the ContigExpress Tool in Vector NTI suite 6.0 (InforMax, Inc.).
Phylogenetic sequence analysis.
Sequences were checked for chimeras with the program Pintail (1) and for other amplification and sequencing artifacts. Following the removal of chimeras from the data set, percent similarities (p distances) between sequences from the same source (adult, larva, or juvenile) were determined with the editor Align (Multicolor Sequence Alignment Editor; D. Hepperle [http://wwwuser.gwdg.de/∼dheppner/]), and those with identities above 99% were grouped together in operational taxonomic units (OTUs). Only one randomly chosen sequence per OTU was used for further analysis. As a first approximation, the phylogenetic affiliation was determined for each OTU by comparison against sequences available in GenBank using BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Sequences obtained in this study together with reference sequences (all nearest BLAST matches and, moreover, representatives of the respective phylum) were aligned automatically with ClustalX (50), and the alignment was subsequently corrected manually using Align (Multicolor Sequence Alignment Editor; D. Hepperle [http://wwwuser.gwdg.de/∼dheppner/]). Phylogenetic trees were constructed with the ARB software package (31). Initially, neighbor-joining (Jukes-Cantor correction) and maximum parsimony trees were calculated with nearly full-length sequences (>1,250 bp) and 100 pseudoreplicates. Subsequently, partial sequences were added to the trees without changing the topology by the use of the parsimony-interactive method in ARB. Finally, 50% majority rule consensus trees were constructed.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study were deposited in the EMBL/GenBank/DDBJ database under accession numbers DQ661746 to DQ661857.
RESULTS
Transmission electron microscopy.
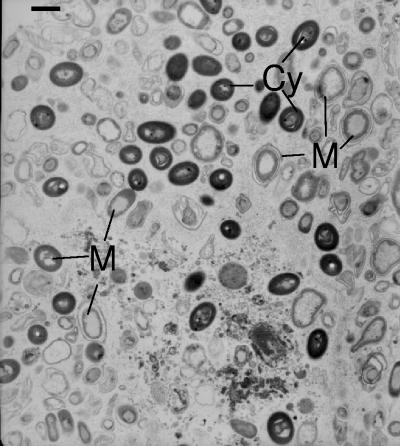
Electron microscopy of adult I. felix specimens 4 and 5 revealed large and complex microbial communities (Fig. 1). Microorganisms are embedded extracellularly and are scattered equally in the collagen matrix of the mesohyl. Bacterial sizes were between 1 and 2.5 μm in length and 0.5 to 1 μm in diameter and showed different shapes like cocci, rods, and other, irregular forms. Cyanobacteria are easily identified by their characteristic internal thylakoid membrane stacks (Fig. 1). The different morphotypes resemble subtypes from other sponge species described previously by Vacelet (53), Wilkinson (58), and Friedrich et al. (12). Previously described prokaryotic cells with nucleoid-like structures (9, 13) could also be detected in I. felix.
FIG. 1.
Transmission electron microscopy of microorganisms in the I. felix adult mesohyl. Cy, Cyanobacteria; M, microorganisms. Scale bar, 1 μm.
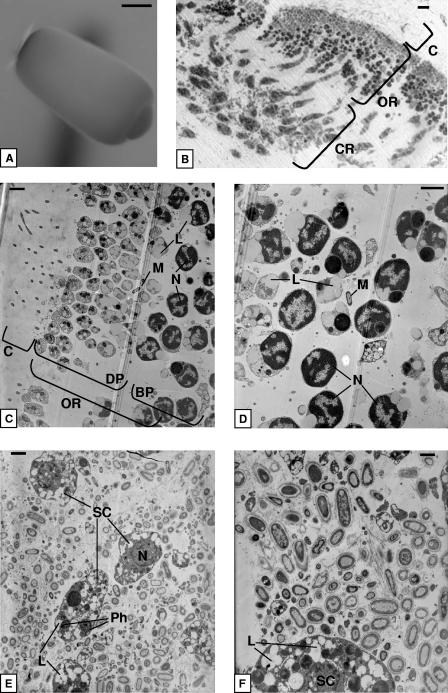
I. felix individuals 4 and 5 released parenchymella-type larvae that were light gray, about 500 μm in length (Fig. 2A), and completely covered with a carpet of small, equally long cilia as seen under the dissecting microscope. Much longer cilia were present at the dark pigmented posterior pole. Figure 2B gives an overview of a larval cross section. The ciliary region, the outer region, and the central region form three distinct layers. The ciliary region is followed by seemingly two layers of small sponge cells (Fig. 2C and D). In fact, because of diagonal sectioning, the two layers represent the distal and basal parts of the same elongated, pear-shaped sponge cells, which build an external ciliated epithelium. The distal part of these cells contains cell organelles like the Golgi apparatus and mitochondria (data not shown). The enlarged basal part contains the nucleus with condensed chromatin (Fig. 2D). These cells contain large amounts of lipids (A. Ereskovsky, St. Petersburg State University, personal communication). In the central region of the larvae, electron-transparent amoeboid sponge cells are loosely embedded in the extracellular matrix (Fig. 2E and F). They contain nuclei, phagosomes, and many lipids. Visual inspection of at least five different individuals revealed that the outer region of the larvae is almost free of bacteria, whereas the central region of the larva contains numerous microorganisms. These microorganisms look similar in size, shape, and membrane structure to those found in adult samples. Some electron micrographs also show phagosomes containing digested bacterial remnants (Fig. 2E) and bacterial cell division (data not shown).
FIG. 2.
(A) Dissection microscopy of a parenchymella larva of I. felix. (B) Light microscopy of a larva cross section showing the ciliary, outer, and center regions. (C and D) Transmission electron microscopy of the ciliary and outer regions showing the distal and the basal parts of sponge cells. (E and F) Transmission electron microscopy of the central part of the larva with electron-transparent sponge cells surrounded by microorganisms. M, microorganisms; BP, basal part; C, ciliary region; CR, central region; DP, distal part; L, lipids; N, nucleus; OR, outer region; Ph, phagosome; SC, sponge cell. Scale bar, 100 μm (A), 10 μm (B), 2 μm (C, D, and E), and 1 μm (F).
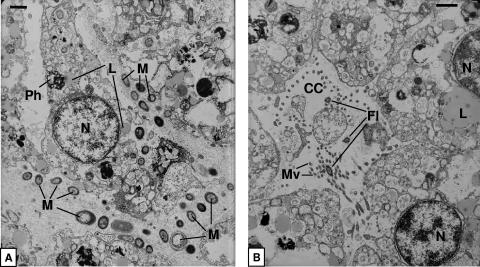
The juveniles of sponges 4 and 5 showed densely packed electron-transparent amoeboid sponge cells (Fig. 3). In contrast to the sponge cells of the larvae, juvenile cells contain fewer lipids but more phagosomes and vesicles (data not shown). Microorganisms in juveniles are located between sponge cells (Fig. 3A), which show phagosomes, indicating microbial phagocytosis. The presence of choanocyte chambers indicates that a functional aquiferous system has already been established (Fig. 3B).
FIG. 3.
(A) Transmission electron microscopy of microorganisms in the I. felix juvenile mesohyl. (B) Transmission electron microscopy of a choanocyte chamber in a juvenile. M, microorganisms; CC, choanocyte chamber; Fl, flagella; L, lipids; Mv, microvilli; N, nucleus; Ph, phagosome. Scale bar, 1 μm.
DGGE.
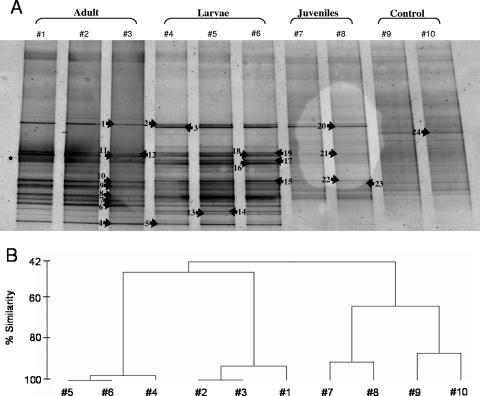
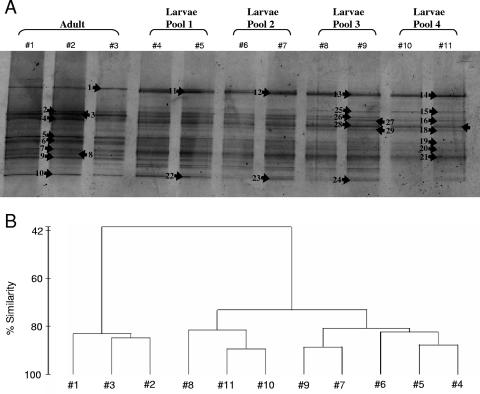
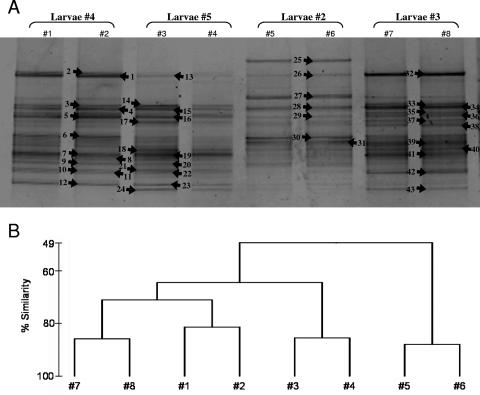
DGGE was used to fingerprint the bacterial community of I. felix adult, larval, and juvenile stages. Each lane on a gel represents an independent PCR. Banding patterns of the same sample were highly similar, indicating little or no PCR bias between PCRs of the same DNA extraction. Total band numbers were similar for adult 4 (average number of 21) and its larvae (average number of 22.7) but increased in juveniles (average number of 27.5) and the nylon control (average number of 27) (Fig. 4A). Cluster analysis of DGGE banding patterns placed the adult and larva samples together, whereas the juvenile samples clustered with the control sample (Fig. 4B). DGGE band analysis of different larval pools released by the same adult individual revealed highly similar banding patterns that corresponded to that of the adult (Fig. 5A and B). In contrast, comparison of DGGE bands of larvae released by different adult individuals revealed a higher variability (Fig. 6A and B). The similarity of DGGE banding patterns from different larva pools from the same individual ranged down to 70%, whereas that of larva pools from different individuals was even lower (Fig. 5B and 6B). On the gel in Fig. 6, some bands were present (e.g., DGGE band 25) or absent (e.g., DGGE bands 24 and 43 are missing in larva pool 4) only in a given sample, yet the majority of bands were still shared between the different samples (e.g., DGGE bands 1, 13, 26, and 32).
FIG. 4.
(A) Denaturing gradient gel electrophoresis (gel A) of the adult, larva, and juvenile samples (individual 4). Three independent PCRs each were run for adult and larva samples, and two each were run for the juvenile and the control samples. A piece of nylon without sponge tissue taken after the settlement experiment was used as a control. Arrows indicate excised and sequenced bands. (B) Dendrogram showing percent similarity of banding patterns. Numbers correspond to lanes in the gel.
FIG. 5.
(A) Denaturing gradient gel electrophoresis (gel E) of I. felix adult 4 compared to different larva pools released by the same adult specimen. Three independent PCRs were run for the adult, and two each were run for pooled larvae. Arrows indicate excised and sequenced bands. (B) Dendrogram showing percent similarity of banding patterns. Numbers correspond to lanes in the gel.
FIG. 6.
(A) Denaturing gradient gel electrophoresis (gel F) of four larva pools released by four different adult specimens (specimens 2, 3, 4, and 5). Two independent PCRs each were run. Arrows indicate excised and sequenced bands. (B) Dendrogram showing the percent similarity of banding patterns. Numbers correspond to lanes in the gel.
Phylogenetic sequence analysis.
In total, 218 sequences were derived from excised DGGE bands. After the removal of 54 chimeras (Pintail) and five sequences with undefined sequencing artifacts, the remaining 159 sequences were maintained separately according to their source (adult, larva, or juvenile). Each set of sequences was grouped into OTUs based on a >99% similarity cutoff. One sequence of each of the altogether 112 unique OTUs was chosen, and accordingly, 41 adult, 53 larval, and 18 juvenile sequences were used for further phylogenetic analysis. The sequences fell into seven different phyla of the domain Bacteria (Table 1). One additional sequence cluster that could not be affiliated with any of the publicly available sequence entries was identified (GenBank, February 2007). The adult I. felix samples contained members of the phyla Proteobacteria (Alphaproteobacteria [n = 8], Gammaproteobacteria [n = 8], and Deltaproteobacteria [n = 2]), Chloroflexi (n = 7), Acidobacteria (n = 6), Gemmatimonadetes (n = 5), and Cyanobacteria (n = 3) and members of the cluster of uncertain affiliation (n = 2). The DGGE-derived sequences obtained from I. felix larvae represented the Acidobacteria (n = 13), Gammaproteobacteria (n = 13), Alphaproteobacteria (n = 9), Chloroflexi (n = 6), members of the uncertain cluster (n = 5), Deltaproteobacteria (n = 2), Actinobacteria (n = 2), Bacteroidetes (n = 2), and the Gemmatimonadetes (n = 1). Cyanobacteria were absent in larval samples. The phylogenetic diversity of the I. felix juveniles revealed members of three phyla, which were the Proteobacteria (Alphaproteobacteria [n = 10] and Gammaproteobacteria [n = 3]), Actinobacteria (n = 2), Cyanobacteria (n = 2), and members of the cluster of uncertain affiliation (n = 1). It is important to note that the numbers do not reflect the in vivo abundances in the sponge. A total of 63%, 55%, and 28% of all sequences obtained from adult, larva, and juvenile samples, respectively, had a sponge-derived sequence as their closest relative (BLAST analysis, as a first approximation).
TABLE 1.
Phylogenetic diversity of bacteria associated with adult, larval, and juvenile I. felix
| Bacterial phylum | Resulta
|
||
|---|---|---|---|
| Adult | Larvae | Juveniles | |
| Acidobacteria | + | + | − |
| Actinobacteria | − | + | + |
| Bacteroidetes | − | + | − |
| Chloroflexi | + | + | − |
| Cyanobacteria | + | − | + |
| Gemmatimonadetes | + | + | − |
| Alphaproteobacteria | + | + | + |
| Gammaproteobacteria | + | + | + |
| Deltaproteobacteria | + | + | − |
| Uncertain affiliation | + | + | + |
+, present; −, absent.
Clusters of vertically transmitted bacterial groups.
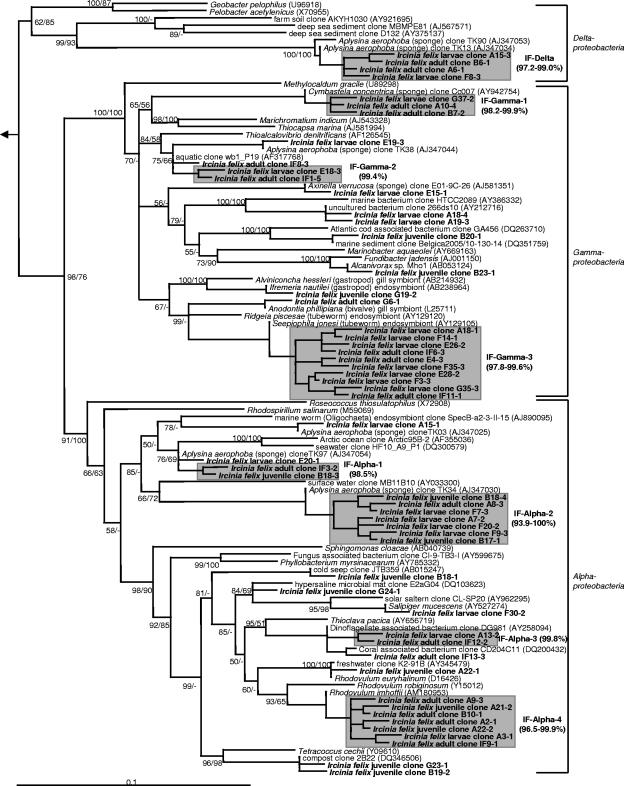
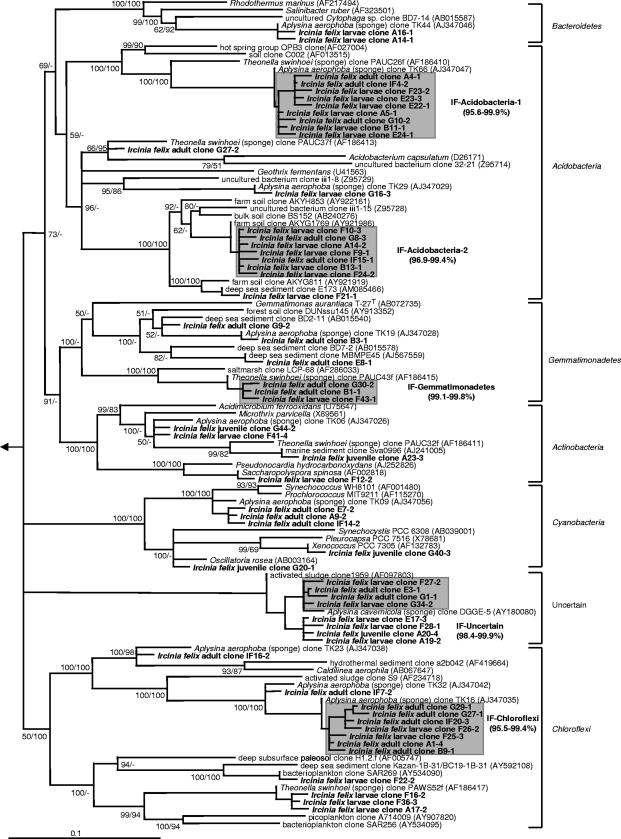
Vertically transmitted phylotypes are defined as monophyletic clusters of two or more sequences that were recovered from both the adult sponge and offspring (larvae and/or juveniles) of I. felix (hereafter termed IF clusters). Altogether, 13 monophyletic sequence clusters were identified (Fig. 7 and 8), which belonged to four different bacterial phyla and one additional lineage of uncertain affiliation. More than 60% of all I. felix-derived DGGE sequences fell into these clusters. The in-cluster similarity was above 97% for eight clusters and above 95.5% for 12 clusters. Only cluster IF-Alpha-2 had a relatively low in-cluster similarity of 93.9%. Two clusters (IF-Alpha-2 and IF-Alpha-4) contained sequences of all three developmental stages (adults, larvae, and juveniles).
FIG. 7.
Phylogenetic distance tree calculated with 16S rRNA gene proteobacterial DGGE sequences recovered from I. felix. Neighbor-joining and maximum parsimony (100 pseudoreplicates) bootstrap values are provided. Sequences obtained in this study are in boldface type. The clones are coded as follows: DGGE gel (capital letter)-excised band (first number)-clone (second number). Gray boxes depict monophyletic clusters of sequences that originated from both adults and offspring (larvae and/or juveniles) of I. felix. The scale bar indicates 10% divergence. Arrow, to outgroup (Geothrix fermentans U41563, Holophaga foetidae X77215, and Acidobacterium capsulatum D26171). GenBank accession numbers are shown in parentheses.
FIG. 8.
Phylogenetic distance tree calculated with 16S rRNA bacterial DGGE sequences recovered from I. felix. Neighbor-joining and maximum parsimony (100 pseudoreplicates) bootstrap values are provided. Sequences obtained in this study are in boldface type. The clone labels are coded as follows: DGGE gel (capital letter)-excised band (first number)-clone (second number). Gray boxes depict monophyletic clusters of sequences that originated from both adults and offspring (larvae and/or juveniles) of I. felix. The scale bar indicates 10% divergence. Arrow, to outgroup (Pyrobaculum calidiformis AB078332, Sulfolobus metallicus D85519, and Desulfurococcus mobilis M36747). GenBank accession numbers are shown in parentheses.
DISCUSSION
Vertical transmission, the passage of microbial symbionts to the next host generation through the reproductive cell lines, is a hallmark of evolutionarily ancient symbioses. Vertical transmission has been documented in terrestrial insects such as ants (42) and aphids (2) as well as in marine invertebrates such as bryozoans (29) and bivalves (4, 47). Since the passage of symbionts via the reproductive stages is highly selective, this process frequently leads to cospeciation between the host and symbiotic lineages, resulting in congruent phylogenetic trees (36). The presence of bacteria in the reproductive stages has also been demonstrated in representatives of all three sponge classes by electron microscopy, suggesting that vertical transmission is a widespread mechanism in this phylum (see reference 7 and references cited therein). In comparison to the well-studied symbioses mentioned above, the microbial associations of sponges are different in several respects. Rather than the known one (few)-symbiont-one-host types of associations, the microbial consortia of sponges are exceedingly complex, containing sponge-specific representatives of at least eight different bacterial phyla and one archaeal phylum (18). Second, the microbial biomass within the bacteriosponges (high-microbial-abundance sponges) is massive, contributing up to 40 to 60% of the animal's biomass (53, 59). To our knowledge, no other animal phylum tolerates such amounts of internal, freely dispersed microorganisms. The enormous microbial biomass in vertebrate intestines is also located internally but is contained within specialized organs such as the rumen. Third, because of the characteristic anatomy of sponges, there are no physical barriers such as organs or tissues that separate the sites of reproduction from the mesohyl microbiota. In other symbioses, symbionts are frequently contained in specialized cells (bacteriocytes) or modified host organs (i.e., the trophosome of the tubeworm Riftia pachyptila and the glands of Deshayes of the shipworm Bankia setacea). It was therefore of interest to document the passage of microorganisms via the reproductive stages of a Caribbean demosponge and to phylogenetically characterize the bacterial consortia within them. A combination of visual (electron microscopy) and molecular (DGGE) techniques was used towards this goal.
Electron microscopy studies provided the first insights into the presence of microorganisms within the larvae of I. felix (Fig. 2). High amounts of morphologically diverse microorganisms were located extracellularly in the central part of the larvae, while the outer rim appeared to be almost free of bacteria. This is in agreement with the microscopic description of the Mediterranean species Ircinia oros, which also contained numerous bacteria in the inner part of the larvae but not in the peripheral region (8). In general, the larvae of both Ircinia species have a similar morphology including dark-pigmented cells and a ring of elongated cilia at the posterior pole, which might be important in the response to external stimuli during the planktonic life stage (27). Both Ircinia species have a ciliated epithelium consisting of densely packed sponge cells. This pear-shaped cell form and the location of the nucleus in the basal part are unusual, as the nucleus is located more distally in most parenchymella larvae (8). The central part of the I. felix larva contains only one loosely associated cell type, which has been identified as being archaeocytes in Ircinia oros (8) and other dictyoceratid species (24). Finally, both Ircinia larvae contain unusually high amounts of lipids, which is consistent with the lecithotrophic (nonfeeding) nature of this larvae. This reserve substance may allow for a long pelagic life phase and would increase the chance of survival during metamorphosis.
The bacterial community profile of the adult I. felix sponge strikingly resembles that of other high-microbial-abundance sponges that have been subject to molecular analysis of microbially diverse populations (i.e., see references 17, 44, 48, 51, and 56). I. felix contained sponge-specific sequences from seven of the eight previously reported bacterial phyla (Table 1). These are the Proteobacteria (Alphaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria), Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, and Cyanobacteria. Members of two sequence clusters previously reported as “uncertain affiliation-I and -II” (17), now known to belong to the phylum Gemmatimonadetes (60; this study), were also recovered from I. felix. Furthermore, a deeply routing sequence cluster that could not be affiliated with any of the known sequences available in the public libraries was identified in this study. The closest relatives are an unidentified clone from the Mediterranean sponge Aplysina cavernicola (GenBank accession number AY180080) (>97% similarity) (51) and an unidentified activated-sludge clone (GenBank accession number AF097803) (94.4% similarity) (Fig. 8). This sequence cluster might represent a novel clade of sponge-specific bacteria. Additionally, a gammaproteobacterial cluster (IF-Gamma-3) is noteworthy, as it forms a coherent clade with a number of 16S rRNA gene sequences from chemoautotrophic symbionts of deep-sea invertebrates including gastropods, bivalves, and tubeworms.
Interestingly, several previously reported sponge-specific clusters (17) could not be detected in this study (Fig. 7 and 8). These belong to the “Nitrospira-I” cluster of the phylum Nitrospira that contains exclusively nitrite-oxidizing bacteria. The gammaproteobacterial “Gamma-I” cluster that is most closely related to ammonia-oxidizing Nitrosococcus species was also not identified. If the coordinated metabolism of ammonia-oxidizing bacteria and nitrite-oxidizing bacteria is responsible for the process of nitrification in sponges, then the conspicuous absence of both clades would suggest that eubacterial nitrification is not an important process in I. felix. Nevertheless, nitrification mediated by Archaea, such as, for example, the Cenarchaeum symbiosum clade of sponge symbionts, remains a distinct possibility (15, 38). Members of the recently discovered candidate phylum “Poribacteria” and of the domain Archaea would not have been expected in this study because of mismatches in the PCR primer regions. Several lineages have so far exclusively been found in I. felix. These belong to the IF-Alpha-3 cluster related to a coral associated clone (GenBank accession number DQ200432), the IF-Alpha-4 cluster related to Rhodovulum species (GenBank accession number AM180953), and the IF-Acido-2 cluster related to acidobacterial clones from soil (GenBank accession numbers AY921986, AB240276, and AY922161). Whether these lineages are specific to the sponge I. felix remains to be seen as more sequences from other Ircinia sponges become available.
The bacterial diversity of I. felix larvae is comparable to that of the adult sponge with respect to the DGGE band numbers, patterns, and phylotypes recovered (Fig. 4, 7, and 8). The banding patterns of different larval pools from the same individual were more similar than those from different individuals (Fig. 5A and 6A), a fact that is also reflected in the cluster analysis (Fig. 5B and 6B). Deciphering the degree of fine-scale intraspecies variation poses a challenge for further studies. Sequencing and phylogenetic analysis of the DGGE bands showed the presence of all previously identified, sponge-specific bacterial phyla (18, 19, 21) in the larvae, with the exception of the Cyanobacteria. Since DGGE bands of the appropriate migration distance were also identified in the larval sample (Fig. 4), it is possible that the cyanobacterial phylotypes are present in the larvae but that the corresponding bands were not chosen for sequencing. Alternatively, since the larvae are brooded in the mesohyl and the Cyanobacteria are found predominantly on the outer surface, they might be incorporated into the larvae less efficiently. In fact, Usher et al. showed that only 25% of all larvae from Chondrilla australiensis contained Cyanobacteria (52).
The DGGE banding pattern of the I. felix juvenile sample contained more bands than the corresponding adult and larva samples. The dendrograms clustered the adult and larval samples together, while the juvenile formed one cluster with the control. The higher number of DGGE bands is probably an artifact from the extraction protocol, as the newly grown sponges were still attached to the nylon substrate at the time of DNA extraction. Hence, the DGGE banding pattern is a mixture of sponge-specific phylotypes and colonizers from seawater. Altogether, representatives of three bacterial phyla could be identified, including sponge-specific lineages of the phyla Actinobacteria and Alphaproteobacteria and the clade of uncertain affiliation. The reduced bacterial diversity might be due to methodological constraints, as more than twice as many sequences were gained from adult samples (n = 41) and from larvae (n = 53) than from juveniles (n = 18), which showed a generally more faint banding pattern. If more sequences would have been obtained from juveniles, the number of identified phyla that contain sponge-specific representatives might have been higher.
Alternatively, the reduced diversity in juveniles may be correlated to the feeding behavior of sponge larvae. Free-swimming larvae are unable to take up food particles from the water column (22). If the internal sponge symbionts would serve as food during the nonfeeding planktonic phase, then the microbial community within the larvae would be naturally reduced. This hypothesis is supported by electron microscopical observations that show evidence of phagocytosis in the larvae (Fig. 2E). Further experiments focusing on the juvenile stages are necessary to determine whether microbially diverse populations are truly reduced or whether the sponge-specific lineages are present in the larvae, albeit below the limit of detection. Conceivably, only a single bacterium of each lineage would be needed to inoculate the newly grown sponge.
In summary, it could be shown that in the sponge I. felix, the entire microbial consortium rather than individual phylogenetic lineages is passed on to the next generation via the reproductive stages. Accordingly, vertical transmission is specific in that the microorganisms of I. felix, but not those from seawater, are passed on but unselective in that there appears to be no differentiation between individual sponge-specific lineages. These data are congruent with those described previously by Sharp et al. (46), who reported on the vertical transmission of similarly complex phylogenetic lineages throughout the embryonic development of Corticium sp. This passage of diverse microorganisms to the next generation is probably due to the characteristic anatomy of sponges, where the reproductive elements are exposed to microorganisms in the mesohyl, whereas in higher animals, the reproductive elements are contained in specialized, bacterium-free reproductive organs. In conclusion, the ancient and widespread mechanism of vertical transmission is clearly important for the formation and maintenance of the phylogenetically complex yet highly sponge-specific microbial communities of many marine demosponges.
Acknowledgments
We gratefully acknowledge the staff of NOAA's National Undersea Research Center at Key Largo, FL, for professional work during field trips. We thank Melissa Southwell and Channing Jones (University of North Carolina at Chapel Hill) for help during sponge collection and settlement experiments and Andrey Vishnyakov and Alexander Ereskovsky (St. Petersburg State University) for helpful discussion of electron micrographs.
This research was supported by Deutsche Forschungsgemeinschaft grants HE3299/1-1 and HE3299/1-2 to U.H., UNCW/NURC (NA03OAR4300088) and NSF Chemical Oceanography Program (OCE 0351893) grants to C. S. Martens and N.L., and an NSF graduate fellowship to J.B.W.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, P., L. Baumann, C. Y. Lai, D. Rouhbakhsh, N. A. Moran, and M. A. Clark. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu. Rev. Microbiol. 49:55-94. [DOI] [PubMed] [Google Scholar]
- 3.Brusca, R. C., and G. J. Brusca. 1990. Phylum Porifera: the sponges, p. 181-210. In A. D. Sinauer (ed.), Invertebrates. Sinauer Press, Sunderland, MA.
- 4.Cary, S., and S. Giovannoni. 1993. Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. Proc. Natl. Acad. Sci. USA 90:5695-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duchassaing De Fonbressin, P., and G. Michelotti. 1864. Spongiaires de la mer Caraibe. Natuurk. Verh. Holl. Mij. Venetsch. Haarlem 21:1-124. [Google Scholar]
- 6.Enticknap, J. J., M. Kelly, O. Peraud, and R. T. Hill. 2006. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ereskovsky, A. V., E. Gonobobleva, and A. Vishnyakov. 2005. Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera, Demospongiae, Halisarca). Mar. Biol. 146:869-875. [Google Scholar]
- 8.Ereskovsky, A. V., and D. Tokina. 2004. Morphology and fine structure of the swimming larvae of Ircinia oros (Porifera, Demospongiae, Dictyoceratida). Invertebr. Reprod. Dev. 45:137-150. [Google Scholar]
- 9.Fieseler, L., M. Horn, M. Wagner, and U. Hentschel. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fieseler, L., A. Quaiser, C. Schleper, and U. Hentschel. 2006. Analysis of the first genome fragment from the marine sponge-associated, novel candidate phylum Poribacteria by environmental genomics. Environ. Microbiol. 8:612-624. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich, A. B., I. Fischer, P. Proksch, J. Hacker, and U. Hentschel. 2001. Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 12.Friedrich, A. B., H. Merkert, T. Fendert, J. Hacker, P. Proksch, and U. Hentschel. 1999. Microbial diversity in the marine sponge Aplysina cavernicola (formerly Verongia cavernicola) analyzed by fluorescence in situ hybridisation (FISH). Mar. Biol. 134:461-470. [Google Scholar]
- 13.Fuerst, J. A., R. I. Webb, M. J. Garson, L. Hardy, and H. M. Reiswig. 1998. Membrane-bounded nucleoids in microbial symbionts of marine sponges. FEMS Microbiol. Lett. 166:29-34. [Google Scholar]
- 14.Gernert, C., F. O. Glöckner, G. Krohne, and U. Hentschel. 2005. Microbial diversity of the freshwater sponge Spongilla lacustris. Microb. Ecol. 50:206-212. [DOI] [PubMed] [Google Scholar]
- 15.Hallam, S. J., T. J. Mincer, C. Schleper, C. M. Preston, K. Roberts, P. M. Richardson, and E. F. DeLong. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:520-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hentschel, U., L. Fieseler, M. Wehrl, C. Gernert, M. Steinert, M. Horn, and J. Hacker. 2003. Microbial diversity of marine sponges, p. 59-88. In W. E. G. Müller (ed.), Molecular marine biology of sponges. Springer-Verlag, Heidelberg, Germany. [DOI] [PubMed]
- 17.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentschel, U., K. M. Usher, and M. W. Taylor. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55:167-177. [DOI] [PubMed] [Google Scholar]
- 19.Hill, R. T. 2004. Microbes from marine sponges: a treasure trove of biodiversity for natural products discovery, p. 177-190. In A. T. Bull (ed.), Microbial diversity and bioprospecting. ASM Press, Washington, DC.
- 20.Hooper, J. N. A., and R. W. M. van Soest. 2002. Systema Porifera. A guide to the classification of sponges, volume 1. Plenum Publishers, New York, NY.
- 21.Imhoff, J. F., and R. Stöhr. 2003. Sponge-associated bacteria: general overview and special aspects of bacteria associated with Halichondria panicea, p. 35-56. In W. E. G. Müller (ed.), Molecular marine biology of sponges. Springer-Verlag, Heidelberg, Germany. [DOI] [PubMed]
- 22.Jaeckle, W. 1995. Transport and metabolism of alanine and palmitic acid by field-collected larvae of Tedania ignis (Porifera, Demospongiae): estimated consequences of limited label translocation. Biol. Bull. 189:159-167. [DOI] [PubMed] [Google Scholar]
- 23.Kaye, H. 1991. Sexual reproduction in four Caribbean commercial sponges. II. Oogenesis and transfer of bacterial symbionts. Invertebr. Reprod. Dev. 19:13-24. [Google Scholar]
- 24.Kaye, H. R., and H. M. Reiswig. 1991. Sexual reproduction in four Caribbean commercial sponges. III. Larval behaviour, settlement and metamorphosis. Invertebr. Reprod. Dev. 19:25-35. [Google Scholar]
- 25.Levi, C., and P. Levi. 1976. Embryogenese de Chondrosia reniformis (Nardo), demosponge ovipare, et transmission des bacteries symbiotiques. Ann. Sci. Nat. Zool. 18:367-380. [Google Scholar]
- 26.Levi, C., and A. Porte. 1962. Ètude au microscope électronique de l'éponge Oscarella lobularis Schmidt et de sa larve amphiblastula. Cah. Biol. Mar. 3:307-315. [Google Scholar]
- 27.Leys, S., and B. Degnan. 2001. Cytological basis of photoresponsive behavior in a sponge larva. Biol. Bull. 201:323-338. [DOI] [PubMed] [Google Scholar]
- 28.Li, C. W., J. Y. Chen, and T. E. Hua. 1998. Precambrian sponges with cellular structures. Science 279:879-882. [DOI] [PubMed] [Google Scholar]
- 29.Lim, G., and M. Haygood. 2004. “Candidatus Endobugula glebosa,” a specific bacterial symbiont of the marine bryozoan Bugula simplex. Appl. Environ. Microbiol. 70:4921-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindquist, N., R. Bolser, and K. Laing. 1997. Timing of larval release by two Caribbean demosponges. Mar. Ecol. Prog. Ser. 155:309-313. [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maldonado, M., N. Cortadellas, M. I. Trillas, and K. Rutzler. 2005. Endosymbiotic yeast maternally transmitted in a marine sponge. Biol. Bull. 209:94-106. [DOI] [PubMed] [Google Scholar]
- 33.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology, manual 3.4.4. Kluwer, Dordrecht, The Netherlands.
- 34.Olson, J. B., and P. J. McCarthy. 2005. Associated bacterial communities of two deep-water sponges. Aquat. Microb. Ecol. 39:47-55. [Google Scholar]
- 35.Pawlik, J. R., G. McFall, and S. Zea. 2002. Does the odor from sponges of the genus Ircinia protect them from fish predators? J. Chem. Ecol. 28:1103-1115. [DOI] [PubMed] [Google Scholar]
- 36.Peek, A. S., R. A. Feldman, R. A. Lutz, and R. C. Vrijenhoek. 1998. Cospeciation of chemoautotrophic bacteria and deep sea clams. Proc. Natl. Acad. Sci. USA 95:9962-9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pile, A. J. 1997. Finding Reiswig's missing carbon: quantification of sponge feeding using dual-beam flow cytometry, p. 1403-1410. In H. A. Lessios and I. G. Macintyre (ed.), Proceedings of the 8th International Coral Reef Symposium, vol. 2. Smithsonian Tropical Research Institute, Balboa, Panama. [Google Scholar]
- 38.Preston, C. M., K. Y. Wu, T. F. Molinski, and E. F. DeLong. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. USA 93:6241-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiswig, H. 1974. Water transport, respiration and energetics of three tropical marine sponges. J. Exp. Mar. Biol. Ecol. 14:231-249. [Google Scholar]
- 40.Rützler, K., R. W. M. van Soest, and B. Alvarez. 2003. Swenzea zeai, a Caribbean reef sponge with a giant larva, and Scopalina ruetzleri: a comparative fine-structural approach to classification (Demospongiae, Halichondrida, Dictyonellidae). Invertebr. Biol. 122:203-222. [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Sauer, C., D. Dudaczek, B. Holldobler, and R. Gross. 2002. Tissue localization of the endosymbiotic bacterium “Candidatus Blochmannia floridanus” in adults and larvae of the carpenter ant Camponotus floridanus. Appl. Environ. Microbiol. 68:4187-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheuermayer, M., S. M. Pimentel-Elardo, L. Fieseler, L. Grozdanov, and U. Hentschel. 2006. Microorganisms of sponges: phylogenetic diversity and biotechnological potential, p. 289-312. In P. Proksch and W. E. G. Müller (ed.), Frontiers in marine biotechnology, Horizon Scientific Press, London, United Kingdom.
- 44.Schirmer, A., R. Gadkari, C. Reeves, F. Ibrahim, E. DeLong, and C. Hutchinson. 2005. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 71:4840-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sciscioli, M., E. Lepore, M. Gherardi, and L. S. Liaci. 1994. Transfer of symbiotic bacteria in the mature oocyte of Geodia sydonium (Porifera, Demospongiae): an ultrastructural study. Cah. Biol. Mar. 35:471-478. [Google Scholar]
- 46.Sharp, K. H., B. Eam, D. J. Faulkner, and M. G. Haygood. 2007. Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73:622-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sipe, A., A. Wilbur, and S. Cary. 2000. Bacterial symbiont transmission in the wood-boring shipworm Bankia setacea (Bivalvia: Teredinidae). Appl. Environ. Microbiol. 66:1685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, M. W., P. J. Schupp, I. Dahllof, S. Kjelleberg, and P. D. Steinberg. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121-130. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, M. W., P. J. Schupp, R. de Nys, S. Kjelleberg, and P. D. Steinberg. 2005. Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environ. Microbiol. 7:419-433. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thoms, C., M. Horn, M. Wagner, U. Hentschel, and P. Proksch. 2003. Monitoring microbial diversity and natural products profiles of the sponge Aplysina cavernicola following transplantation. Mar. Biol. 142:685-692. [Google Scholar]
- 52.Usher, K. M., J. Kuo, J. Fromont, and D. C. Sutton. 2001. Vertical transmission of cyanobacterial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Hydrobiologia 461:15-23. [Google Scholar]
- 53.Vacelet, J. 1975. Étude en microscopie électronique de l'association entre bactéries et spongiaires du genre Verongia (Dictyoceratida). J. Microsc. Biol. Cell. 23:271-288. [Google Scholar]
- 54.Vacelet, J., and C. Donadey. 1977. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30:301-314. [Google Scholar]
- 55.Webster, N., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-proteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 56.Webster, N. S., K. J. Wilson, L. L. Blackall, and R. T. Hill. 2001. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wehrl, M., M. Steinert, and U. Hentschel. Bacterial uptake by the marine sponge Aplysina aerophoba. Microb. Ecol., in press. [DOI] [PubMed]
- 58.Wilkinson, C. R. 1978. Microbial associations in sponges. III. Ultrastructure of the in situ associations in coral reef sponges. Mar. Biol. 49:169-176. [Google Scholar]
- 59.Willenz, P., and W. D. Hartman. 1989. Micromorphology and ultrastructure of Caribbean sclerosponges. I. Ceratoporella nicholsoni and Stromatospongia norae (Ceratoporellidae-Porifera). Mar. Biol. 103:387-402. [Google Scholar]
- 60.Zhang, H., Y. Sekiguchi, S. Hanada, P. Hugenholtz, H. Kim, Y. Kamagata, and K. Nakamura. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating microorganism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53:1155-1163. [DOI] [PubMed] [Google Scholar]