Abstract
In the crenarchaeote Sulfolobus islandicus REN1H1, a mobile element of 321 bp length has been shown to be active. It does not contain terminal inverted repeats and transposes by a replicative mechanism. This newly discovered element has been named SMN1 (for Sulfolobus miniature noninverted repeat transposable element).
Transposable elements are found in all three domains of life, i.e., Bacteria, Eukarya, and Archaea. Especially in the hyperthermophilic archaeon Sulfolobus solfataricus P2, a high number of insertion sequence (IS) elements have been identified (5, 6, 11, 13, 17), and so far four of them have been shown to be active in vivo (11). Besides these full-length IS elements, which carry a gene coding for a transposase and thus are able to catalyze their own excision and insertion, mobile elements that are too short to harbor a transposase gene also exist. These elements are called nonautonomous elements, because they can transpose only if a suitable transposase is supplied in trans by an autonomous IS element.
Nonautonomous mobile elements were first discovered to be present in Sulfolobus by use of a bioinformatic approach. They were identified by searching for small repeat elements in the genome sequence of S. solfataricus P2 (13). The detected short elements were classified as miniature inverted repeat transposable elements (MITEs), and the corresponding full-length IS elements were identified based on the similarity of the terminal inverted repeats. The first observation of an active MITE in Sulfolobus was made by Blount and Grogan (3), who detected the insertion of a MITE related to ISC1057 and ISC1058 and homologous to an element in the Sulfolobus solfataricus P2 genome classified as SM3A (13) in a Sulfolobus strain typed as conspecific to Sulfolobus islandicus (23).
Besides mobile elements that carry inverted repeats and are thus more easily identified by a bioinformatic approach, there are other types of mobile elements that do not exhibit terminal inverted repeats. Elements with subterminal inverted repeats have been reported (22), as have some classes of IS elements that do not contain inverted repeats at all and that transpose not by a cut-and-paste mechanism but by a mechanism similar to the rolling-circle (RC) replication of some bacterial plasmids, such as the members of the IS91 and IS605/IS200 families (10). Nonautonomous mobile elements that lack terminal inverted repeats are very difficult to detect by bioinformatic approaches, and so far none has been described for Sulfolobus.
Sulfolobus solfataricus strains P1 and P2 and Sulfolobus islandicus strains REN1H1, REN2H1, and HVE10/4 (23), and S1 (a lacS mutant strain derived from REN1H1) were grown in Brock's basal salts medium at pH 3.5 (4) supplemented with 0.2% arabinose (Fluka) and 0.1% tryptone (BD Biosciences) for liquid medium and with 0.2% dextrin 10 (Fluka) and 0.1% tryptone for plates. For growth of pyrEF mutant strains PH1-16 (11), R20, R21, R22, S1R1, and R1 (derived from REN1H1), 20 μg ml−1 of uracil was added to the medium. Plates were solidified by addition of 0.6% Gelrite (Sigma) and 10 mM CaCl2.
Mobile elements were detected by screening for insertions into the pyrEF genes, coding for orotidine phosphoribosyltransferase and orotidine-5′-monophosphatedecarboxylase, after plating of Sulfolobus cells on Gelrite plates containing 50 μg ml−1 5′-fluoroorotic acid (FOA) (Apollo Scientific) and 20 μg ml−1 uracil. Colonies obtained after 5 to 7 days of growth at 75°C were cultivated in 200 μl of nonselective medium for 2 to 3 days before genomic DNA was prepared by phenol-chloroform extraction and ethanol precipitation. pyrEF genes were amplified by PCR using primers 5′-TTCACCTTTTGCTATCGAAG and 5′-GTTTATAAAGACCGGCTATT, resulting in a fragment of 1,584 bp containing the promoter region of pyrB and the pyrE and pyrF genes. PCR products differing in length from the wild-type PCR fragment were cloned and sequenced.
Genomic DNA for Southern blots was prepared from 25 ml of exponentially growing Sulfolobus cells. Cell pellets were resuspended in 1 ml Tris-EDTA buffer-0.06% N-laurylsarcosine-0.8% Triton X-100 and incubated at room temperature for 30 min (18). Cell debris was removed by centrifugation at 15,000 × g for 30 min at 4°C. The supernatant was treated with RNase for 60 min at room temperature, and then the preparation was extracted once with phenol, with phenol-chloroform-isoamyl alcohol (25:24:1), and finally with chloroform. After isopropanol precipitation, the DNA was resuspended in 100 μl of Tris-EDTA buffer. Five micrograms of HindIII-digested DNA was separated on a 0.8% agarose gel. The gel was stained with ethidium bromide before denaturation and capillary transfer to a Hybond N membrane as described by Sambrook and Russel (16). Probes spanning 319 bp of the 321 bp of the insertion sequence were generated using a digoxigenin PCR labeling kit (Roche). Hybridization (50% formamide) and stringency washes (0.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) were carried out either at 42°C and 60°C or at 53°C and 68°C. UV cross-linking hybridization was carried out overnight. Detection was done using antidigoxigenin antibodies coupled to alkaline phosphatase with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (Roche) as the substrate. Color development was stopped after 1 to 5 h.
Detection of insertion sequences.
Two hundred thirty FOA-resistant colonies of Sulfolobus islandicus REN1H1 were screened by PCR. In nine cases, PCR products longer than expected for the wild-type pyrEF sequence were observed. All other observed FOA-resistant colonies did not show any difference in PCR product length and were assumed to carry point mutations. Three of the nine insertions showed a length of about 0.3 kb. Sequencing revealed that they are all identical and have a length of 321 bp. This element was named SMN1 (for Sulfolobus miniature noninverted repeat transposable element). Two of the other six insertions correspond to an insertion sequence related to ISC796, and the other four correspond to an insertion sequence related to ISC735 (94%/91% transposase sequence identity).
The 321-bp insertions are thought to be nonautonomous, as they are too short to contain a transposase-encoding open reading frame (ORF). The shortest transposases so far described (e.g., TnpA from ISHp608) comprise around 150 amino acid residues (15), which is much longer than a putative open reading frame corresponding to 59 amino acids found in SMN1. Another remarkable feature is the high GC content of SMN1 (54%).
Related sequences.
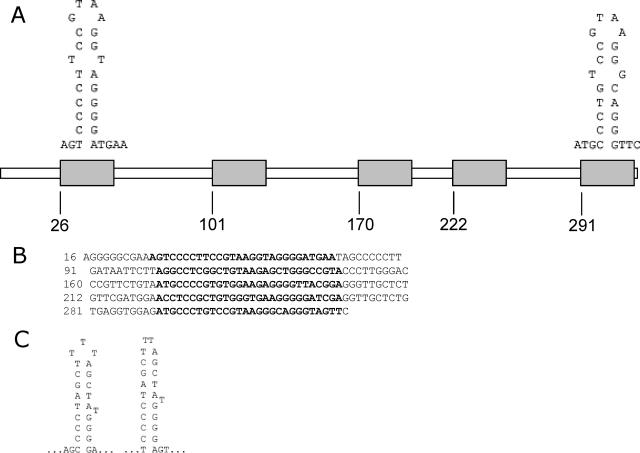
We were not able to identify related characterized IS elements by a standard nucleotide BLAST search or protein BLAST search (1) using only the putative ORF59 sequence (nucleotides [nt] 104 to 280). However, by this database search we found two highly similar sequences in the Sulfolobus islandicus plasmids pARN4 and pHVE14 (8) (only one nucleotide mismatch each). In pHVE14 (accession number AJ748324), SMN1 is inserted after nucleotide 31457. It overlaps with the annotated ORF46. All ORFs surrounding SMN1 are annotated as encoding hypothetical proteins. In pARN4 (accession number AJ748323) it is found inserted after nucleotide 25272 (see Fig. 2).
FIG. 2.
Alignment of sequences adjacent to the insertion sites of SMN1 in the three identical insertions reported here and in the plasmids pHVE14 and pARN4. For SMN1 inserted into the pyrE promoter region, the BRE and TATA boxes are underlined and the start codon of pyrE is indicated by an arrow and boldface. For both plasmid sequences the starts of putative open reading frames downstream and upstream of the insertion site are also indicated by arrows and boldface. The consensus sequence of the insertion site (TTT/A) is also shown in boldface.
In the Sulfolobus solfataricus P2 genome, several similar sequences were detected, but they never spanned the full length of SMN1 and had a lower degree of similarity (five hits spanning more than 50 nt with 81% to 89% similarity). These hits did not map to annotated open reading frames, nor were any ORFs found repeatedly adjacent to the sequences similar to SMN1. No similar sequences of more than 50 nt in length were found in the other sequenced Sulfolobus genomes.
Presence in other species and strains.
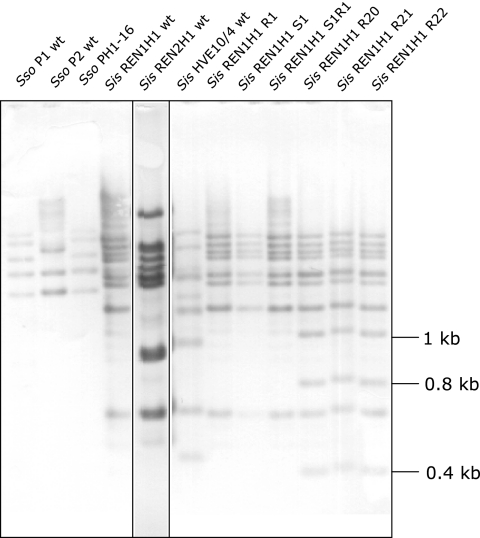
The occurrence of the insertion sequence in other, not-yet-sequenced S. islandicus and S. solfataricus strains was examined by Southern blotting (Fig. 1). At least eight copies of the element could be identified in the Sulfolobus islandicus REN1H1 wild-type strain and a variety of mutant strains. In the three mutant strains showing the insertion of SMN1 in the pyrEF genes (R20, R21, and R22), three additional bands in comparison to those in the wild-type strain were detected. The presence of the same three additional bands in the three different isolates makes it very probable that they all originate from one transposition event. As none of the bands present in the wild type disappeared, the element probably moves not by a cut-and-paste mechanism but by some process involving the replication of the element. One of the additional bands has a predicted length of about 1.1 kb, corresponding to the HindIII fragment of the pyrB and pyrE genes (755 bp) plus the insertion sequence (321 bp). The other observed restriction fragments are 0.8 and 0.4 kb long and correspond to insertions of the element in other unidentified genomic regions. Other pyrEF or lacS mutants of strain REN1H1 were also examined by Southern blotting but showed the same pattern as the wild type, indicating that the element had not been active. In S. islandicus strains REN2H1 and HVE10/4, eight copies of similar sequences were identified by the blot. For S. solfataricus, strain P1 and the derived pyrEF/lacS mutant PH1-16 as well as strain P2 were examined. Both P1 strains produced the same five bands, whereas for P2 three similar sequences were detected. For P2 the genomic sequence and the size of the restriction fragments on the blot were compared and found to correspond to the best BLAST hits. Hybridization and stringency washes were carried out under standard conditions (42°C/60°C). Additional experiments were done using more stringent conditions yielding only a positive signal at about 90% similarity. Under the higher-stringency conditions all S. islandicus REN1H1 strains, as well as REN2H1 and HVE10/4, showed the same bands as in Fig. 1, while no bands were detected for S. solfataricus P1, P2, or PH1-16.
FIG. 1.
Southern blotting using a probe spanning nucleotides 2 to 321 of the SMN1 sequence with several different Sulfolobus species, strains, and mutants. Genomic DNA was digested with HindIII. PH1-16 has been described by Martusewitsch et al. (11). R1 is a pyrEF mutant, S1 is a lacS mutant, and S1R1 is a pyrEF lacS double mutant (unpublished).
Insertion sites and target site duplication.
The three different insertion sites are shown in Fig. 2. The consensus for insertion of SMN1 shown by all three target sites is TTT/A. In the case of the pyrB and pyrEF genes, the promoter region has been studied in detail and BRE and TATA boxes have been identified (20). SMN1 has inserted into the TATA box of the pyrEF genes and probably blocks the transcription of the genes causing the FOA-resistant phenotype. In plasmids pARN4 and pHVE14 the elements are also found in intergenic regions, and putative open reading frames start 40 bp downstream of the insertion in pARN4 and 28 bp upstream in pHVE14. Thus, the element likely also inserted into the promoter regions of these putative open reading frames.
As all known right-end flanking sequences of SMN1 show an A (Fig. 2) and the sequence of the element also starts with an A, it is impossible to distinguish whether a single base is duplicated or no target site duplication occurs at all. So far, no insertion sequence that produces a target site duplication of one nucleotide has been described (10; IS Finder [http://www-is.biotoul.fr/]).
Similarities to known elements.
As no terminal inverted repeats are present in the element and no characterized sequences showed similarity to SMN1, we tried to find some characteristics that would allow classification of the element. The lack of terminal inverted repeats narrowed down the number of IS families under consideration to the IS91 family of RC transposons, which have as eukaryotic counterparts the helitrons (9), and the IS605/IS200 family.
Characteristics of the members of IS family IS91 (10, 12, 14, 19) are relatively constant 3′ ends (right ends in Fig. 2 and 3) and variations in 5′ ends except for a well-conserved 20- to 25-bp stretch. The IS inserts behind a specific tetranucleotide sequence identical to the last four nucleotides found on the other end of the element (19). No target site duplication occurs upon insertion. RC transposons also have been described for eukaryotes (7, 9). Common features are the target site 5′ A/T, no target site duplication, and no terminal inverted repeats. They begin with the sequence 5′-TC and end with CTRR-3′. Palindromes of 16 to 20 nt separated by 10 to 12 nt from the 3′ ends are found. The majority of family members are nonautonomous internal deletion variants of small size (0.5 to 3 kb), whereas intact variants are large (5.5 to 15 kb).
FIG. 3.
(A) Distribution of the 28-bp repeats identified by MEME search (gray boxes) within SMN1. For the first and the last repeats, possible hairpin structures are shown above. (B) Alignment of the sequences of the repeats (boldface) from panel A. (C) Hairpin structures from ISHp608 (15).
When comparing SMN1 to the above-described mobile elements one can conclude that SMN1 shows features of both prokaryotic and eukaryotic RC transposons. The target site (TT)T/A is similar to eukaryotic RC transposons. Probably no target site duplication occurs, and no terminal inverted repeats are found, like for IS91 and eukaryotic RC transposons. The right-end sequence (GTTC) of SMN1 is identical to the sequence found in the IS91 family (IS1294) (19); the left-end sequence does not fit any known IS. Subterminal hairpin structures can be found on the right end, as has been described for the IS91 family (e.g., for nucleotides 316 to 296 a 6-bp stem and 9-nt loop or for nucleotides 315 to 292 a 10-bp stem [two imperfect] and 4-nt loop). Also on the left end subterminal hairpin structures can be found, reminiscent of helitrons, which have palindromic sequences on their 3′ ends (e.g., for nucleotides 13 to 40 an 11-bp stem [two imperfect] and 6-nt loop, which matches the given numbers for helitrons).
Another possibility for classifying SMN1 is to put it into the IS605/IS200 group, the other IS family that does not show terminal inverted repeats and has no target site duplication. By use of a MEME motif search (2), subterminal imperfect direct repeats of 28 bp in length were identified, which are able to form hairpin structures. These subterminal hairpin structures show some similarity to IS605 family transposons from Helicobacter pylori (ISHp608) (21) (Fig. 3). The transposase of ISHp608 has been shown to bind to these hairpin structures (21) and catalyze single-strand breaks at each hairpin. The mechanism of replication of ISHp608 is not identical to the rolling-circle mode of replication, but they might have some steps in common (15).
Excision of SMN1.
The excision of SMN1 appears to be a very rare event. We were not able to obtain any revertants of the pyrEF mutants by plating on selective medium without uracil. However, when we used the mutant strain with SMN1 inserted into the pyrEF genes as the recipient strain for transformation by electroporation, several colonies were obtained on selective medium. The length of the pyrEF PCR product had decreased to the wild-type pyrEF length. Sequencing of this PCR product showed that SMN1 had been completely removed, restoring the original pyrEF sequence.
There are two possible ways for a miniature transposable element to be created. One possible way is the loss of internal sequence parts and thus the sequence coding for the transposase gene. The other possibility is the acquisition of the flanking regions necessary as recognition sites for transposition by an internal unrelated sequence region, which can then be mobilized (6). Since database searches of the P2 genome comprising the whole 321-bp sequence of SMN1 reveal only sequences that do not comprise the terminal parts of the element and since searches conducted only with the terminal sequences yield different hits, the latter possibility might be true for SMN1. The availability of the genome sequence of an S. islandicus strain will probably help to unravel the element responsible for mobilization of SMN1.
Nucleotide sequence accession number.
The sequence of SMN1 has been deposited in GenBank under accession number EF127252.
Acknowledgments
This work was supported by the DFG (grant Li913/3 to G.L.).
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36. In R. Altman, D. Brutlag, P. Karp, R. Lathrop, and D. Searls (ed.), Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA. [PubMed]
- 3.Blount, Z. D., and D. W. Grogan. 2005. New insertion sequences of Sulfolobus: functional properties and implications for genome evolution in hyperthermophilic archaea. Mol. Microbiol. 551:312-325. [DOI] [PubMed] [Google Scholar]
- 4.Brock, T. D., K. M. Brock, R. T Belly, and R. L. Weiss. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 84:54-68. [DOI] [PubMed] [Google Scholar]
- 5.Brügger, K., P. Redder, Q. She, F. Confalonieri, Y. Zivancovic, and R. A. Garrett. 2002. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 206:131-141. [DOI] [PubMed] [Google Scholar]
- 6.Brügger, K., E. Torarinsson, P. Redder, L. Chen, and R. A. Garrett. 2004. Shuffling of Sulfolobus genomes by autonomous and non-autonomous mobile elements. Biochem. Soc. Trans. 32:179-183. [DOI] [PubMed] [Google Scholar]
- 7.Feschotte, C., and S. R. Wessler. 2001. Treasures in the attic: rolling circle transposons discovered in eucaryotic genomes. Proc. Natl. Acad. Sci. USA 98:8923-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greve, B., S. Jensen, K. Brügger, W. Zillig, and R. A. Garrett. 2004. Genomic comparision of archaeal conjugative plasmids from Sulfolobus. Archaea 1:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapitonov, V., and J. Jurka. 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98:8714-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martusewitsch, E., C. W. Sensen, and C. Schleper. 2000. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J. Bacteriol. 182:2574-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendiola, M. V., I. Bernales, and F. de la Cruz. 1994. Differential roles of the transposon termini in IS91 transposition. Proc. Natl. Acad. Sci. USA 91:1922-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redder, P., Q. She, and R. A. Garrett. 2001. Non-autonomous mobile elements in the crenarchaeon Sulfolobus solfataricus. J. Mol. Biol. 306:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Richter, G. Y., K. Bjöklof, M. Romantschuk, and D. Mills. 1998. Insertion specificity and trans-activation of IS801. Mol. Gen. Genet. 260:381-387. [DOI] [PubMed] [Google Scholar]
- 15.Ronning, D. R., C. Guynet, B. Ton-Hoang, Z. N. Perez, R. Ghirlando, M. Chandler, and F. Dyda. 2005. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol. Cell 20:143-154. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16a.Schleper, C., R. Roder, T. Singer, and W. Zillig. 1994. An insertion element of the extremely thermophilic archaeon Sulfolobus solfataricus transposes into the endogenous beta-galactosidase gene. Mol. Gen. Genet. 243:91-96. [DOI] [PubMed]
- 17.She, Q., X. Peng, W. Zillig, and R. A. Garrett. 2001. Gene capture in archael chromosomes. Nature 409:478. [DOI] [PubMed] [Google Scholar]
- 18.Stedman, K. M., C. Schleper, E. Rumpf, and W. Zillig. 1999. Genetic requierements for the function of the archeal virus SSV1 in Sulfolobus solfataricus: construction and testing of a viral shuttle vector. Genetics 152:1397-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tavakoli, N., A. Comanducci, H. M. Dodd, M.-C. Lett, B. Albiger, and P. Bennett. 2000. IS1294, a DNA element that transposes by RC transposition. Plasmid 44:66-84. [DOI] [PubMed] [Google Scholar]
- 20.Thia-Toong, T.-L., M. Roovers, V. Durbecq, D. Gigot, N. Glansdorf, and D. Charlier. 2002. Genes of de novo pyrimidine biosynthesis from the hyper-thermophilic crenarchaeote Sulfolobus acidocaldarius: novel organization in a bipolar operon. J. Bacteriol. 184:4430-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ton-Hoang, B., C. Guynet, D. R. Ronning, B. Cointin-Marty, F. Dyda, and M. Chandler. 2005. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 24:3325-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu, Z., and S. P. Orphanides. 2001. Microuli, a family of miniature subterminal inverted-repeat transposable elements (MSITES): transposition without terminal inverted repeats. Mol. Biol. Evol. 18:893-895. [DOI] [PubMed] [Google Scholar]
- 23.Zillig, W., A. Kletzin, C. Schleper, I. Holz, D. Janekovic, J. Hain, M. Lanzendörfer, and J. K. Kristiansson. 1994. Screening for Sulfolobales, their plasmids, and their viruses in Islandic solfataras. Syst. Appl. Microbiol. 16:606-628. [Google Scholar]