Abstract
Epstein–Barr virus (EBV) transforms B lymphocytes into lymphoblastoid cell lines usurping the Notch and tumor necrosis factor receptor pathways to effect transcription including NF-κB activation. To determine whether NF-κB activity is essential in the growth and survival of EBV-transformed lymphoblastoid cell lines, a nondegradable IκBα mutant was expressed under tetracycline regulation. Despite continued Bcl-2 and Bcl-x/L expression, NF-κB inhibition induced apoptosis as evidenced by poly(ADP-ribose) polymerase cleavage, nuclear condensation and fragmentation, and hypodiploid DNA content. Both caspase 3 and 8 activation and loss of mitochondrial membrane potential were observed in apoptotic cells. However, caspase inhibition failed to block apoptosis. These experiments indicate that NF-κB inhibitors may be useful in the therapy of EBV-induced cellular proliferation.
Epstein-Barr virus (EBV) infection induces B lymphocyte proliferation. This and the ensuing T cell response can result in infectious mononucleosis. In the absence of an effective T cell response, EBV-infected B lymphocytes can be malignant. EBV is also implicated in human malignancies that occur long after primary EBV infection, including anaplastic nasopharyngeal carcinoma and Hodgkin's disease (reviewed in refs. 1 and 2).
EBV causes resting B lymphocyte proliferation and growth transformation by encoding nuclear and integral membrane proteins that usurp the Notch and tumor necrosis factor (TNF) receptor signaling pathways, thereby altering transcription (3–20). Unlike some of the other proteins expressed in EBV-transformed B lymphocytes, latent membrane protein 1 (LMP1) has oncogene-like activity in rodent fibroblasts and is expressed in most other malignancies associated with EBV infection (21–25). LMP1 may mediate proliferative and survival effects not only in EBV-transformed B lymphocytes but also in these malignancies that occur long after primary infection. LMP1 engages TNF receptor-associated factors (TRAFs) and TNF receptor-associated death domain protein (TRADD) (17, 26); through these proteins, LMP1 strongly activates NF-κB and stress-activated protein kinases to effect transcription (4, 7, 17, 27–33).
The experiments reported here test the importance of NF-κB in lymphoblastoid cell line (LCL) growth and survival. NF-κB can regulate cell growth and survival (reviewed in ref. 34) through the transcriptional activation of genes such as c-Myc and A20 (reviewed in ref. 35). For example, TNFα induces apoptosis in fibroblasts and LCLs in which NF-κB is inhibited ( refs. 36–38 and reviewed in ref. 34). Inhibition of NF-κB causes apoptosis in normal murine B lymphocytes or in WEHI 231 murine B lymphoma cells (39–41). Gene-targeting studies show a requirement for the NF-κB components, c-Rel or p105, in murine B lymphocyte survival after mitogenic stimulation or at rest (42, 43). However, LCLs express high levels of the anti-apoptotic proteins Bcl-2, Bcl-x/L, and Mcl-1, and the role of NF-κB in the regulation of these proteins in LCLs is uncertain. Thus, LCL survival may or may not be NF-κB dependent.
Materials and Methods
Cell Lines, Plasmids, and Antibodies.
IB4, an EBV-transformed normal human cord blood lymphoblastoid cell line, was cultured in RPMI 1640 supplemented with 10% FBS, l-glutamine, streptomycin, and penicillin. Bfl-1 plasmid was provided by Céline Gélinas of the University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, Piscataway, NJ (44). pJEF3 and pJEF4 plasmids were obtained from M. Rowe, (Univ. of Wales College of Medicine, Cardiff, U.K.) (45). The Flag epitope-tagged amino-terminal truncation of IκBα is a deletion of amino acids 1–36, called here F-ΔN-IκBα, and was a gift from Dean Ballard of Vanderbilt University (Nashville, TN) (46). F-ΔN-IκBα was cloned as a HindIII to Sma fragment and was blunt-end ligated into the EcoRI site of pJEF4 resulting in plasmid no. 406. IB4 was electroporated with pJEF3, plated in 96-well plates at 5000 cells per well, and selected with 0.6 μg/ml hygromycin. Subclones were tested for the presence of the tetracycline (Tc) transactivator using a plasmid-encoding luciferase under the control of the Tc operator sequences. IB4 subclone 1E9 was electroporated with plasmid 406, plated at 5000 cells per well in a 96-well plate, and selected in 0.8 mg/ml G418 and 1 μg/ml Tc. Two subclones, clones 1 and 11, were chosen for further study. Results are shown for clone 1 and are similar to those obtained with clone 11. Cells were maintained in media containing 0.2 μg/ml hygromycin and 0.25 mg/ml G418 and 1 μg/ml Tc. Cell concentration and viability was determined by trypan blue exclusion as visually determined using a hemacytometer and Zeiss compound microscope.
F-ΔN-IκBα Induction.
Cells were washed three times with RPMI and recultured in Tc+ or Tc− media at a concentration of 105 cells per ml in R10 with no selection. Cells were analyzed starting 24 h after the initial cultures were established (day 1) and on 20- to 24-h intervals subsequently.
Cell Cycle, Confocal Microscopy, and FACs Analysis.
Cells were prepared for cell cycle analysis and confocal microscopy by fixation in 75% ethanol in PBS and stored at 4°C. Cells were stained with propidium iodide (Molecular Probes) for 30 min before analysis on a FACScalibur (Becton Dickinson) using the cellquest software. Polyploids and cell conjugates were eliminated by using the double discrimination mode FL2-A vs. FL-2W on linear scales. A portion of the same sample was analyzed by confocal microscopy. Cells were wet mounted in PBS and analyzed on a PCM 2000 (Nikon) coupled to a Zeiss microscope using Simple 32 software (Nikon). For analysis of cell surface expression of intercellular adhesion molecule-1 (ICAM-1), 5 × 105 cells were incubated with anti-human CD54 directly conjugated to phycoerythrin (PharMingen) and analyzed on the FACScalibur. Dead cells were eliminated from the analysis by gating on the forward and side scatter properties of the population. Fluorescence intensity was determined on a log scale in the FL-2 channel.
Electromobility Shift Assays (EMSA).
Cell extracts were generated by lysis in extraction buffer (1% Nonidet P-40/20 mM Hepes, pH 7.9/350 mM NaCl/1 mM MgCl2/0.5 mM EDTA/0.1 mM EGTA/20% glycerol) supplemented before use with 0.5 mM DTT, 0.5 mM PMSF, and 1% aprotinin (Sigma) at a concentration of 108/ml. Protein concentration was determined by using the bicinchoninic acid reagents (Pierce). Three to five micrograms of extract were used per reaction with 0.7 ng of radiolabeled double-stranded NF-κB probe derived from the HIV long terminal repeat (LTR). Competition assays included 75 ng of unlabeled wild-type competitor or mutant HIV NF-κB probe. After incubation at room temperature for 30 min in binding buffer [0.5% Triton X-100/2.5% glycerol/4 mM DTT/10 mM Hepes, pH 7.5/80 μg/ml poly(dI-dC)], complexes were resolved by native PAGE on a 4.5% gel. Antibodies to the specific NF-κB subunits (Santa Cruz Biotechnology) were included during the complex formation incubation at room temperature. Gels were dried and subjected to autoradiography and phosphoimagery (Molecular Dynamics).
Western Blot Analysis.
Western Blot analysis was performed resolving a portion of the extracts prepared for EMSA after normalization by SDS/PAGE, transferred to nitrocellulose, and probed with antibodies at a concentration of 1–5 μg/ml in PBS with 0.05% Tween 20. The exception is that the Western blot to c-Myc was performed on cells directly solubilized in SDS/PAGE sample buffer containing DTT. The antibodies used are to IκBα (c-15), TRAF1 (H-132), Bcl-x/L (S-18), BID (C-20), p53 (Santa Cruz Biotechnology), Bcl-2 (R & D Systems), pro-Caspase-3, and poly(ADP-ribose) polymerase (PARP) (PharMingen), pro-Caspase-8 (Upstate Biotechnology), the FLAG tag M5 (Sigma).
Northern Blot Analysis.
RNA was extracted from cells 2 days after the induction of F-ΔN-IκBα using RNAzol (Tel-Test Inc., Friendswood, TX) as instructed by the manufacturer. Five micrograms of total RNA were resolved on a 1.2% agarose formaldehyde gel and transferred to the membrane ZetaProbe (Bio-Rad) by capillary action. Membranes were probed and washed according to the manufacturer. 32P-labeled probes were generated by the random priming method using RediPrime II (Amersham Pharmacia) and 50 μCi [α-32P]dCTP (3000 Ci/mmol; NEN) according to the manufacturer. Unincorporated nucleotides were eliminated by purification through a G-50 spin column. The results were visualized by autoradiography and phosphoimagery. Quantitation was determined by phosphoimagery.
Cytochrome c Release Assay.
S-100 pellet and nuclear fractions of cells undergoing apoptosis were generated as in ref. 47. Briefly, cells were swollen on ice in 1 ml of hypotonic buffer A [20 mM Hepes, pH 7.5/10 mM KCl/1.5 mM MgCl2/1 mM EDTA/1 mM EGTA/1 mM DTT/0.1 mM PMSF/1:100 dilution of protease inhibitor mixture (Sigma)], homogenized by douncing, and fractionated at 100,000 × g. The pellet was resuspended in buffer A in 50 μl. After protein quantitation, extracts were visualized by Western blot analysis using antibodies to cytochrome c (Cyt c) (Santa Cruz Biotechnology).
Mitochondrial Potential Determination.
Cells were incubated in complete media with 65 nM of DiOC6 (Molecular Probes) for 30 min at 37°C. After two washes in PBS, cells were resuspended in propidium iodide. DiOC6 was measured in cells that were negative for propidium iodide staining. BJAB cells were treated with 20 ng/ml of anti-Fas Ab (Becton Dickinson) and used as a positive control.
Caspase Enzyme Assay.
The colorimetric caspase-3, -8, and -9 enzyme assay kits were performed according to the manufacturer (R & D Systems) in a microtiter plate. Optical densities were determined at 2 and 4 h after the assay was initiated on a Microtiter Plate Reader (Bio-Rad) at 405 nm. z-VAD.FMK (Calbiochem) was added to cultures at the initiation of some experiments at a concentration of 50 μM. Cells were washed extensively to assure that no z-VAD.FMK carried over into the enzyme assay.
Results
Tc-Regulated F-ΔN-IκBα Expression Inhibits NF-κB Activity in an LCL.
To determine the role of NF-κB in LCL growth and survival, we established clones of an EBV-transformed B lymphocyte LCL (IB4 line) in which expression of a degradation-resistant mutant IκBα, F-ΔN-IκBα, was regulated by Tc. F-ΔN-IκBα has a FLAG epitope tag and is deleted for residues 1–36 that include phosphorylation and ubiquitination sites required for proteosome-mediated degradation. Twenty four hours after culture in media lacking Tc (Tc− media), F-ΔN-IκBα was induced to high level (Fig. 1A).
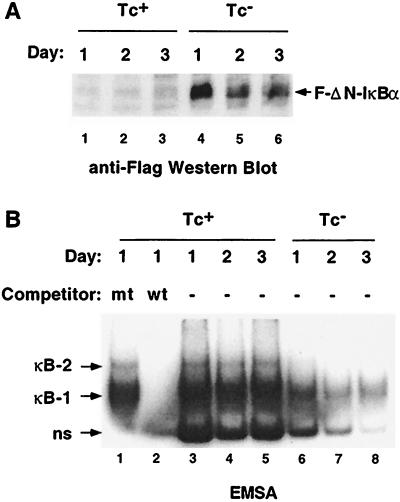
Figure 1.
Growth in Tc− media induced Flag-ΔN-IκBα expression and inhibited NF-κB activity. (A) F-ΔN-IκBα expression was induced within 1 day after Tc withdrawal. Western blot analysis using M5 mAb. Lanes 1–3, extracts from cells grown in Tc+ media days 1–3; lanes 4–6, extracts from cells grown in Tc− media, days 1–3. (B) EMSA indicated that F-ΔN-IκBα expression inhibited NF-κB activity. Two specific complexes were formed after incubating the extracts from A with a NF-κB probe from the HIV LTR marked κB-1 and κB-2. Lane 1, 100-fold mutant cold competitor. Lane 2, 100-fold excess wild-type cold competitor. Lanes 3–5, extracts from cells grown in Tc+ media, days 1–3; lanes 6–8, extracts from cells grown in Tc− media, days 1–3
F-ΔN-IκBα expression had a significant effect on NF-κB activity, as determined by EMSA. The HIV promoter NF-κB oligonucleotide formed two specific major protein complexes with nuclear extracts from cells grown in Tc+ media (Fig. 1B, lane 3, indicated as κb-1 and κb-2). Complex formation was inhibited by excess wild-type oligonucleotide but not with an oligonucleotide mutated in the NF-κB site (Fig. 1B, lanes 1 and 2). Antibodies to Rel A, Rel B, c-Rel, and p50 induced equally significant supershifts of complex 1 (data not shown). Antibodies to Rel B and p50, but not Rel A or c-Rel, induced a partial supershift of complex 2 (data not shown). Cells grown in Tc+ media had similar NF-κB-specific complex formation over 3 days (Fig. 1B, lanes 3–5). In contrast, cells grown in Tc− media had substantially reduced NF-κB activity by day 1 (Fig. 1B, lane 6). As early as 24 h after F-ΔN-IκBα expression, complex 2 formation was undetectable. On days 1, 2, and 3, complex 1 formation was diminished by 71, 84, and 82%, respectively (Fig. 1B, lanes 6, 7, and 8). Thus, F-ΔN-IκBα was effective at inhibiting NF-κB activity in this LCL.
NF-κB Inhibition Down-Regulated Gene Expression.
An early effect of growth in Tc− media was that the cells no longer grew in aggregates. Homotypic adhesion of LCLs is because of lymphocyte function-associated antigen 1 and ICAM-1 (48, 49), and ICAM-1 expression can be regulated by NF-κB (50). FACS analysis revealed a 2-fold decrease in ICAM-1 expression on cells grown in Tc− media (Fig. 2A).
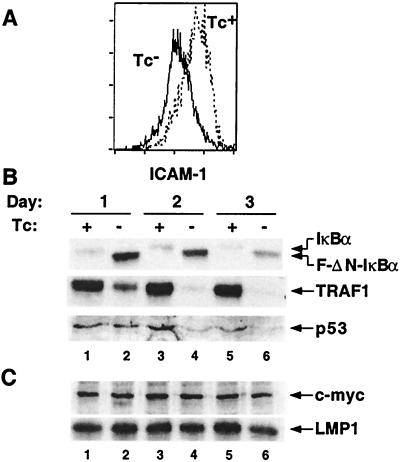
Figure 2.
NF-κB inhibition down-regulated specific gene expression. (A) FACs analysis of ICAM-1 on cells grown in Tc+ (dashed line) or Tc- (solid line) media for 3 days. (B) Western blot analysis of extracts were examined for endogenous IκBα, TRAF1, and p53 expression. Lanes 1, 3, and 5 are extracts from cells grown in Tc+ media; lanes 2, 4, and 6 are from cells grown in Tc− media. Lanes 1 and 2, day 1; lanes 3 and 4, day 2; and lanes 5 and 6, day 3. (C) c-Myc and LMP-1 expression was unaffected by inhibition of NF-κB. Lanes are as noted in B.
Numerous proteins known to be regulated by NF-κB (reviewed in ref. 35) were no longer expressed in cells grown in Tc− media. For example, endogenous IκBα was no longer detectable after 24 h of growth in Tc− media (Fig. 2B, lane 2). Similarly, TRAF1 was substantially diminished at day 1 and almost undetectable at day 2 (Fig. 2B, lanes 2 and 4). p53 expression was decreased at day 2 and was diminished further at day 3 (Fig. 2B, lanes 4 and 6). In contrast to earlier findings (41, 51–53), c-Myc expression was unaffected by NF-κB inhibition (Fig. 2C). Importantly, EBV LMP1 and latent gene expression were unaffected in these experiments (Fig. 2C, and data not shown).
NF-κB Inhibition Induced Apoptosis.
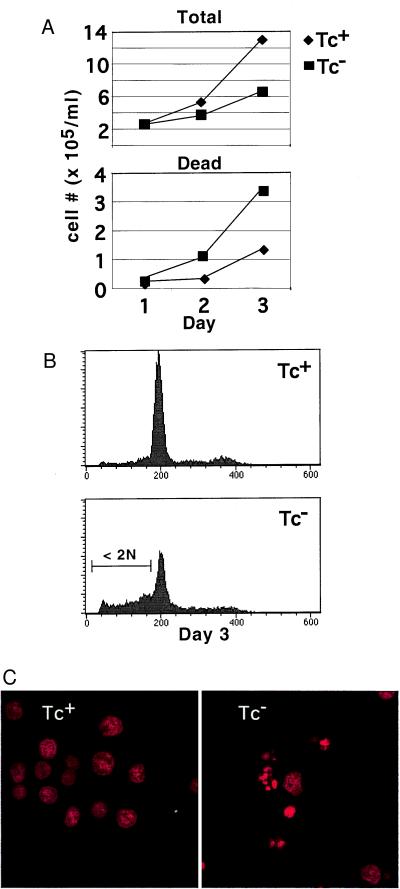
To determine the effect of F-ΔN-IκBα expression on cell growth and survival, the concentration of viable cells was monitored by trypan blue dye exclusion. In Tc+ or Tc− media, cells doubled in the first 24 h. Thereafter, cells grown in Tc+ media continued to double every 24 h, achieving a total (live and dead) cell concentration of 5.33 × 105 and 12.93 × 105 on days 2 and 3 (Fig. 3A, Top). Cells grown in Tc− media exhibited a modest decrease in cell growth, achieving a cell concentration of 3.67 × 105 and 6.60 × 105 on days 2 and 3 (Fig. 3A, Top). The number of dead cells defined by trypan blue uptake grown in Tc+ media was 0.33 × 105 (6%) and 1.33 × 105 (10%) on days 2 and 3. In contrast, dead cells grown in Tc− media increased to 1 × 105 (33%) and 3.2 × 105 (50%) on days 2 and 3. (Fig. 3A, Bottom). Dead cells in Tc− media were hypodipliod, whereas those in Tc+ media were not. The number of hypodiploid cells by propidium iodide staining and FACS analysis (Fig. 3B) was 70–80% of the number of dead cells, as assessed by trypan blue uptake. Consistent with the modest affect on cell growth, the number of cells in S phase was unaffected, and the number in G2/M slightly reduced over the course of these experiments (Fig. 3B, and data not shown). Cells grown in Tc− media had condensed and fragmented nuclei indicative of apoptosis (Fig. 3C). Thus, NF-κB activity is necessary for wild-type cell growth and cell survival. Interestingly, when cells were grown at 10-fold lower concentration, hypodiploid cells accumulated more rapidly and to a greater extent, suggesting that autocrine or paracrine growth factors may partially protect cells from NF-κB inactivation (data not shown).
Figure 3.
NF-κB inhibition caused apoptosis. (A) Cell growth was determined by counting on a hemacytometer with the inclusion of trypan blue. (Top) Total cell number—positive and negative for trypan blue. (Bottom) Cell number positive for trypan blue uptake. ⧫, Tc+ media; ■, Tc− media. Cultures were initially seeded at 105/ml and counted at 24-h intervals thereafter. (B) Examination of DNA content on day 3 showed the accumulation of hypodiploid (<2 N) cells. After ethanol fixation, cells were stained with propidium iodide and examined by FACS. (Top) Tc+ media. (Bottom) Tc− media. (C) Confocal microscopy showing apoptotic bodies of cells in B. (Left) Tc+. (Right) Tc−.
Apoptosis-Induced NF-κB Inhibition Had Unique Biochemical Properties.
Apoptosis can be initiated by multiple pathways and is frequently accompanied by a reduction in cell size, movement of phosphatidyl choline from the inner to the outer aspect of the plasma membrane, loss of mitochondrial membrane potential, and cleavage of DNA between nucleosomes. Apoptosis induced by F-ΔN-IκBα had some, but not all, of these features. Cell size decreased as cells underwent F-ΔN-IκBα-mediated apoptosis (data not shown). However, annexin V staining for phosphatidyl choline on the cell surface was negative in cells grown in Tc− media (data not shown). Furthermore, electrophoresis of soluble DNA from cells grown in Tc− media for 3 days yielded DNA fragments of 2 kb or larger but not the 200-bp ladder characteristic of DNA cleavage between nucleosomes (data not shown).
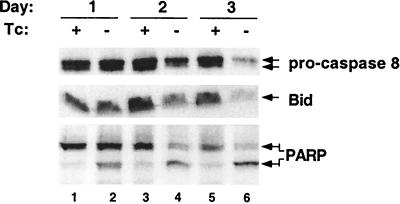
Apoptosis is frequently mediated by the activation of aspartyl proteases. These caspases exist as pro-forms that are activated by cleavage by upstream caspases or are autocatalytically cleaved. TNFα and Fas-mediated apoptosis is initiated by the aggregation by TNFR death domain-containing proteins that activate caspase 8 (reviewed in refs. 54–57). Although Western blot analysis with antibodies to the pro-form of caspase 8 revealed lower levels at day 2 and much lower levels at day 3 (Fig. 4A, lanes 4 and 6), active subunits could not be detected. Thus, the decreased pro-caspase 8 levels could reflect transcriptional control of caspase 8 by NF-κB or caspase activation. Enzymatic assays showed a low level of cleavage activity for the caspase 8 substrate, IETD-pNA favoring the former hypothesis over the later (data not shown). Levels of Bid, a pro-apoptotic Bcl-2 family member that is cleaved by caspase-8, decreased in parallel to pro-caspase 8 in one of two clones (Fig. 4A, lanes 4 and 6) consistent with caspase 8 activation in some cells undergoing F-ΔN-IκBα-mediated apoptosis. Growth factor withdrawal-mediated apoptosis is initiated by mitochondrial damage and couples to the activation of caspases 9 and 3 (review in ref. 58). As early as 24 h after the induction of F-ΔN-IκBα, caspase-3 was activated, before caspase-8, as is evident from the cleavage of PARP from 116 kDa to 86 kDa. (Fig. 4, lane 2). Enzymatic assays for caspase-3 using DEVD-pNA as a substrate confirm caspase-3 activation at 24 h (data not shown).
Figure 4.
PARP cleavage occurred before detectable activation of caspase-8 or Bid cleavage. Western blot analysis of extracts for pro-caspase 8, Bid, and PARP. Lanes 1, 3, and 5 are extracts from cells grown in Tc+ media. Lanes 2, 4, and 6 are from cells grown in Tc− media. Lanes 1 and 2, day 1; lanes 3 and 4, day 2; and lanes 5 and 6, day 3.
Caspase Activity Was Not Essential for F-ΔN-IκBα-Mediated Apoptosis.
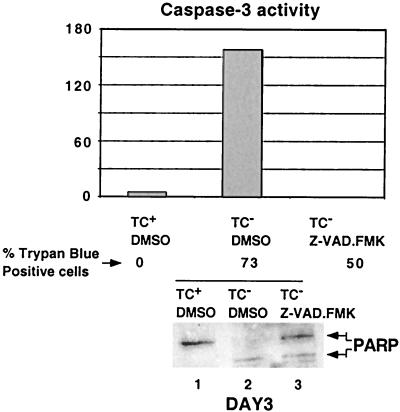
To ascertain the role of caspases in apoptosis mediated by NF-κB inactivation, cells were treated with the general caspase inhibitor z-VAD.FMK in DMSO or with DMSO alone. The addition of 50 μM z-VAD.FMK reduced the percentage of dead cells at day 3 from 73% to 50%, indicating that caspases activity was not essential for F-ΔN-IκBα-mediated cell death. To be certain that the inhibitor was functional over the time course of these experiments, caspase 3 activity was measured on day 3. In F-ΔN-IκBα-expressing cells, caspase-3 activity was readily detectable and was absent in cell extracts from cells cultured in z-VAD.FMK (Fig. 5A, lanes 2 and 3). Furthermore, PARP cleavage was largely inhibited (Fig. 5B, lanes 2 and 3). Thus, in contrast to TNFα- or Fas-mediated apoptosis (reviewed in refs. 54–57), caspase activity was not essential for F-ΔN-IκBα-mediated apoptosis.
Figure 5.
Caspase activation was not required for F-ΔN-IκBα-mediated apoptosis. A total of 50 μM Z.VAD-FMK or 0.1% DMSO, the carrier, were added at day 0. Caspase 3 activity was determined by using the colorimetric substrate DEVD-pNA at Day 3. Cell viability was determined as in Fig. 3. PARP cleavage was determined as in Fig. 4. Lane 1, Tc+ media plus DMSO. Lane 2, Tc− media plus DMSO. Lane 3, Tc− media with 50 μM z-VAD.FMK.
Expression of the Bcl-2 Family Member Bfl-1 Was Regulated by F-ΔN-IκBα in IB4 Cells.
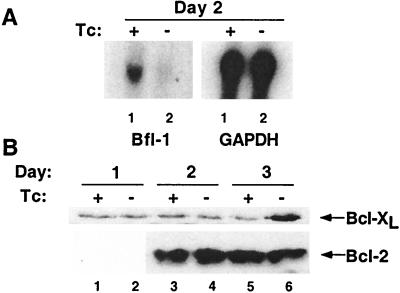
Caspase-independent cell death has been attributed to mitochondrial damage that can be regulated by Bcl-2 family members. The anti-apoptotic Bcl-2 family member Bfl-1 has recently been shown to be regulated by NF-κB (44, 59, 60), and Bfl-1 expression was affected by F-ΔN-IκBα expression. By Northern blot, Bfl-1 mRNA was decreased 50% on day 2, whereas glyceraldehyde-3-phosphate dehydrogenase RNA levels were unaffected (Fig. 6A). In contrast, F-ΔN-IκBα expression had no effect on Bcl-2 nor Bcl-x/L levels. In one experiment, Bcl-x/L expression increased (Fig. 6B).
Figure 6.
NF-κB inhibition reduced A1/Bfl-1 expression, but not Bcl-x/L or Bcl-2 expression. (A) Western blot analysis of extracts for Bcl-x/L and Bcl-2. Lanes 1, 3, and 5 are extracts from cells grown in Tc+ media. lanes 2, 4, and 6 are from cells grown in Tc− media. Lanes 1 and 2, day 1; lanes 3 and 4, day 2; and lanes 5 and 6, day 3. (B) Northern blot analysis. Five micrograms of total RNA generated on day 2 was analyzed for Bfl-1 (Left) and glyceraldehyde-3-phosphate dehydrogenase, (Right). Lane 1, Tc+ media. Lane 2, Tc− media.
F-ΔN-IκBα Expression Resulted in a Loss in Mitochondrial Potential but Not in Cyt c Release.
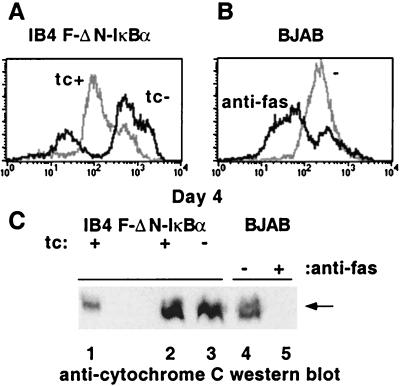
The effect of F-ΔN-IκBα expression on mitochondrial membrane potential was monitored daily using DiOC6. DiOC6 staining decrease in a fraction of cells concomitant with the appearance of hypodiploid cells. In this experiment, 25% of the F-ΔN-IκBα-expressing cells were hypodiploid, and 30% of the cells had low mitochondrial potential (Fig. 7A). In contrast, 70% of BJAB cells treated with anti-Fas antibody showed a decrease in mitochondrial potential and served as a control in these experiments (Fig. 7B). Similar levels of Cyt c were evident in the mitochondrial fraction of LCLs grown in either Tc+ or Tc− media, whereas Cyt c was undetectable in mitochondrial fraction of BJAB cells treated with anti-Fas antibody (Fig. 7C). Furthermore, preliminary experiments indicated that total cellular ATP content was unchanged in cells grown in Tc− media (data not shown). Thus, the mitochondrial membrane potential was diminished in cells expressing F-ΔN-IκBα, without detectable changes in Cyt c release or ATP concentration.
Figure 7.
NF-κB inhibition caused a loss of mitochondrial potential but not Cyt c release. (A and B) DiOC6 fluorescence. (A) IB4 F-ΔN-IκBα cells grown in Tc+ media, gray line, in Tc− media, black line. (B) BJAB, gray line, and BJAB treated with anti-Fas antibody, black line. (C) Cyt c release from the mitochondria. Western blots of S-100 pellets were analyzed for Cyt c. Lane 1, total cell extracts; lane 2, IB4 F-ΔN-IκBα cells, Tc+ media; lane 3, IB4 F-ΔN-IκBα cells, Tc− media; lane 4, BJAB; lane 5, BJAB treated with anti-Fas.
Discussion
These experiments indicated that NF-κB is critical for the survival of EBV-transformed B lymphocytes and has a supportive role in cell growth. NF-κB inhibition caused apoptosis despite Bcl-2 and Bcl-x/L expression, and external inducers of apoptosis were not required for this effect.
The mechanisms by which NF-κB inactivation induces apoptosis differ from those involved in TNFα or Fas. The data presented here indicate there are two phases to apoptosis caused by NF-κB inhibition. The first phase is evidenced by caspase 3 activation and PARP cleavage, the second by caspase 8 activation, Bid cleavage, loss of mitochondrial potential, and nuclear condensation and fragmentation. Altogether, blockade of caspase activity only slightly reduced apoptosis induced by NF-κB inhibition, suggesting that caspase-independent pathways predominate in NF-κB inhibition induced apoptosis.
This NF-κB inactivation-mediated apoptosis shares some features with growth factor withdrawal- and Bax-mediated apoptosis (reviewed in ref. 58). First, apoptosis occurs over days rather than hours. Second, mitochondrial dysfunction may contribute to NF-κB inactivation-mediated apoptosis as it does with growth factor withdrawal. The steady state mRNA level of the anti-apoptotic Bcl-2 family member A1/Bfl-1 is down-regulated by NF-κB inhibition, and this may be the initial insult to these cells. Subsequently, mitochondrial membrane potential changed without other mitochondrial effects. Forced dimerization of Bax causes a similar caspase-independent apoptosis with loss of mitochondrial membrane potential and no Cyt c release (61). After the initial insult of NF-κB inhibition, Bax may be activated by either a change in the stoichometery of Bax to Bfl-1 or, as has been seen in HeLa cells (62), by Bid translocation to the mitochondria. Both Bid and Bax are expressed in LCLs. Bax can cause the release of apoptosis initiating factor from the mitochondria, causing DNA cleavage in large fragments similar to what is observed here (63). Studies are ongoing to more precisely define the potential role of Bfl-1 and Bax in LCL survival in this apoptotic process following NF-κB inhibition. Our preliminary data indicate that Bax is associated with the mitochondria in LCLs.
It is interesting to note that although there is a loss of mitochondrial membrane potential, neither Cyt c was released nor was total ATP concentration changed. Sustained expression of Bcl-2 and Bcl-x/L may have prevented additional mitochondrial effects.
Overall, these experiments indicate that LCL survival is more precarious than previously appreciated. Through the induction of survival genes such as Bfl-1, TRAF1, A20, and perhaps others, NF-κB likely down-regulates the pro-apoptotic effect of proteins such as Bax and Bak expressed in LCLs (64). Decreased expression of anti-apoptotic proteins following NF-κB inhibition tilts the balance and the cells die. Thus, inhibitors of NF-κB activation may have therapeutic benefit in EBV-infected cells in which NF-κB activation is at a sustained high level in response to LMP1 expression. This would include B lymphocytes proliferating in response to primary EBV infection, EBV-infected B lymphocytes proliferating in response to immune suppression, Hodgkin's disease, or nasopharyngeal carcinoma (21–25). Current knowledge of the signaling pathways downstream of LMP1 indicate that signaling could be inhibited by blockade of TRAF domain interactions with LMP1 (65), TRADD (10, 66, 67), or NIK (68), or by inhibition of the intermolecular interactions or enzymatic activities of NIK, IKKα, or IKKβ (ref. 69, and reviewed in ref. 70). Further proof of this concept will require identification of small molecule inhibitors and testing in cells in vitro, in experimental models, and in patients.
Acknowledgments
This manuscript is dedicated to the memory of Matthew Lee Thomas. We thank Dean Ballard and Céline Gélinas for the generous gifts of plasmids. We thank the other members of the lab for their helpful discussions. This work was supported by Public Health Service (PHS) Grant CA47006. E.D.C.M. is supported by PHS Fellowship CA76727. S.S. is supported by PHS Training Grant AI07245.
Abbreviations
- EBV
Epstein–Barr virus
- LMP1
latent membrane protein 1
- Tc
tetracycline
- LCL
lymphoblastoid cell line
- TNFα
tumor necrosis factor α
- TRAF
TNF-receptor associated factor
- F-ΔN-IκBα
Flag-tagged deletion mutant of IκBα
- ICAM
intracellular adhesion molecule
- LTR
long terminal repeat
- EMSA
electromobility shift assay
- Cyt c
cytochrome c
- PARP
poly(ADP-ribose) polymerase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100119497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100119497
References
- 1.Rickinson A B, Kieff E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 2397–2446. [Google Scholar]
- 2.Kieff E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 2343–2396. [Google Scholar]
- 3.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Izumi K M, Kieff E D. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johannsen E, Miller C L, Grossman S R, Kieff E. J Virol. 1996;70:4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaye K M, Devergne O, Harada J N, Izumi K M, Yalamanchili R, Kieff E, Mosialos G. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson E S, Lin J, Kieff E. J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izumi K M, McFarland E C, Ting A T, Riley E A, Seed B, Kieff E D. Mol Cell Biol. 1999;19:5759–5767. doi: 10.1128/mcb.19.8.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krauer K G, Kienzle N, Young D B, Sculley T B. Virology. 1996;226:346–353. doi: 10.1006/viro.1996.0662. [DOI] [PubMed] [Google Scholar]
- 12.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strobl L J, Hofelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm G W, Zimber-Strobl U. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 14.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waltzer L, Perricaudet M, Sergeant A, Manet E. J Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 18.Harada S, Kieff E. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen J I, Kieff E. J Virol. 1991;65:5880–5885. doi: 10.1128/jvi.65.11.5880-5885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nitsche F, Bell A, Rickinson A. J Virol. 1997;71:6619–6628. doi: 10.1128/jvi.71.9.6619-6628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pallesen G, Hamilton-Dutoit S J, Rowe M, Young L S. Lancet. 1991;337:320–322. doi: 10.1016/0140-6736(91)90943-j. [DOI] [PubMed] [Google Scholar]
- 22.Hitt M M, Allday M J, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbst H, Dallenbach F, Hummel M, Niedobitek G, Pileri S, Muller-Lantzsch N, Stein H. Proc Natl Acad Sci USA. 1991;88:4766–4770. doi: 10.1073/pnas.88.11.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niedobitek G, Young L S, Sam C K, Brooks L, Prasad U, Rickinson A B. Am J Pathol. 1992;140:879–887. [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks L, Yao Q Y, Rickinson A B, Young L S. J Virol. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumi K M, Kaye K M, Kieff E D. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatzivassiliou E, Miller W E, Raab-Traub N, Kieff E, Mosialos G. J Immunol. 1998;160:1116–1121. [PubMed] [Google Scholar]
- 28.Kilger E, Kieser A, Baumann M, Hammerschmidt W. EMBO J. 1998;17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huen D S, Grand R J, Young L S. Oncogene. 1988;3:729–730. [PubMed] [Google Scholar]
- 30.Mitchell T, Sugden B. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eliopoulos A G, Young L S. Oncogene. 1998;16:1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 32.Eliopoulos A G, Gallagher N J, Blake S M, Dawson C W, Young L S. J Biol Chem. 1999;274:16085–16096. doi: 10.1074/jbc.274.23.16085. [DOI] [PubMed] [Google Scholar]
- 33.Eliopoulos A G, Blake S M, Floettmann J E, Rowe M, Young L S. J Virol. 1999;73:1023–1035. doi: 10.1128/jvi.73.2.1023-1035.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barkett M, Gilmore T D. Oncogene. 1999;18:6910–6924. doi: 10.1038/sj.onc.1203238. [DOI] [PubMed] [Google Scholar]
- 35.Pahl H L. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 36.Wang C Y, Mayo M W, Baldwin A S J. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 37.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 38.Asso-Bonnet M, Feuillard J, Ferreira V, Bissieres P, Tarantino N, Korner M, Raphael M. Oncogene. 1998;17:1607–1615. doi: 10.1038/sj.onc.1202365. [DOI] [PubMed] [Google Scholar]
- 39.Arsura M, Wu M, Sonenshein G E. Immunity. 1996;5:31–40. doi: 10.1016/s1074-7613(00)80307-6. [DOI] [PubMed] [Google Scholar]
- 40.Wu M, Lee H, Bellas R E, Schauer S L, Arsura M, Katz D, FitzGerald M J, Rothstein T L, Sherr D H, Sonenshein G E. EMBO J. 1996;15:4682–4690. [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H, Arsura M, Wu M, Duyao M, Buckler A J, Sonenshein G E. J Exp Med. 1995;181:1169–1177. doi: 10.1084/jem.181.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grumont R J, Rourke I J, O'Reilly L A, Strasser A, Miyake K, Sha W, Gerondakis S. J Exp Med. 1998;187:663–674. doi: 10.1084/jem.187.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tumang J R, Owyang A, Andjelic S, Jin Z, Hardy R R, Liou M L, Liou H C. Eur J Immunol. 1998;28:4299–4312. doi: 10.1002/(SICI)1521-4141(199812)28:12<4299::AID-IMMU4299>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 44.Zong W X, Edelstein L C, Chen C, Bash J, Gelinas C. Genes Dev. 1999;13:382–387. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Floettmann J E, Ward K, Rickinson A B, Rowe M. Virology. 1996;223:29–40. doi: 10.1006/viro.1996.0452. [DOI] [PubMed] [Google Scholar]
- 46.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 48.Rothlein R, Springer T A. J Exp Med. 1986;163:1132–1149. doi: 10.1084/jem.163.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Liebowitz D, Wang F, Gregory C, Rickinson A, Larson R, Springer T, Kieff E. J Virol. 1988;62:4173–4184. doi: 10.1128/jvi.62.11.4173-4184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van de Stolpe A, Caldenhoven E, Stade B G, Koenderman L, Raaijmakers J A, Johnson J P, van der Saag P T. J Biol Chem. 1994;269:6185–6192. [PubMed] [Google Scholar]
- 51.Duyao M P, Buckler A J, Sonenshein G E. Proc Natl Acad Sci USA. 1990;87:4727–4731. doi: 10.1073/pnas.87.12.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duyao M P, Kessler D J, Spicer D B, Bartholomew C, Cleveland J L, Siekevitz M, Sonenshein G E. J Biol Chem. 1992;267:16288–16291. [PubMed] [Google Scholar]
- 53.Duyao M P, Kessler D J, Spicer D B, Sonenshein G E. Curr Top Microbiol Immunol. 1990;166:211–220. doi: 10.1007/978-3-642-75889-8_27. [DOI] [PubMed] [Google Scholar]
- 54.Ashkenazi A, Dixit V M. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 55.Wallach D, Varfolomeev E E, Malinin N L, Goltsev Y V, Kovalenko A V, Boldin M P. Annu Rev Immunol. 1999;17:331–367. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 56.Krammer P H. Adv Immunol. 1999;71:163–210. doi: 10.1016/s0065-2776(08)60402-2. [DOI] [PubMed] [Google Scholar]
- 57.Lenardo M, Chan K M, Hornung F, McFarland H, Siegel R, Wang J, Zheng L. Annu Rev Immunol. 1999;17:221–253. doi: 10.1146/annurev.immunol.17.1.221. [DOI] [PubMed] [Google Scholar]
- 58.Gross A, McDonnell J M, Korsmeyer S J. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 59.Lee H H, Dadgostar H, Cheng Q, Shu J, Cheng G. Proc Natl Acad Sci USA. 1999;96:9136–9141. doi: 10.1073/pnas.96.16.9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang C Y, Guttridge D C, Mayo M W, Baldwin A S., Jr Mol Cell Biol. 1999;19:5923–5929. doi: 10.1128/mcb.19.9.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gross A, Jockel J, Wei M C, Korsmeyer S J. EMBO J. 1998;17:3878–3885. doi: 10.1093/emboj/17.14.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonsson B, Martinou J C. J Cell Biol. 1999;144:891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Susin S A, Lorenzo H K, Zamzami N, Marzo I, Snow B E, Brothers G M, Mangion J, Jacotot E, Costantini P, Loeffler M, et al. Nature (London) 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 64.Spender L C, Cannell E J, Hollyoake M, Wensing B, Gawn J M, Brimmell M, Packham G, Farrell P J. J Virol. 1999;73:4678–4688. doi: 10.1128/jvi.73.6.4678-4688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Devergne O, McFarland E C, Mosialos G, Izumi K M, Ware C F, Kieff E. J Virol. 1998;72:7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu H, Xiong J, Goeddel D V. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 67.Hsu H, Shu H B, Pan M G, Goeddel D V. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 68.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 69.Sylla B S, Hung S C, Davidson D M, Hatzivassiliou E, Malinin N L, Wallach D, Gilmore T D, Kieff E, Mosialos G. Proc Natl Acad Sci USA. 1998;95:10106–10111. doi: 10.1073/pnas.95.17.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karin M. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]