Abstract
We identified a gene cluster that is involved in the γ-resorcylate (2,6-dihydroxybenzoate) catabolism of the aerobic bacterium Rhizobium sp. strain MTP-10005. The cluster consists of the graRDAFCBEK genes, and graA, graB, graC, and graD were heterologously expressed in Escherichia coli. Enzymological studies showed that graD, graA, graC, and graB encode the reductase (GraD) and oxygenase (GraA) components of a resorcinol hydroxylase (EC 1.14.13.x), a maleylacetate reductase (GraC) (EC 1.3.1.32), and a hydroxyquinol 1,2-dioxygenase (GraB) (EC 1.13.11.37). Bioinformatic analyses suggested that graE, graR, and graK encode a protein with an unknown function (GraE), a MarR-type transcriptional regulator (GraR), and a benzoate transporter (GraK). Quantitative reverse transcription-PCR of graF, which encodes γ-resorcylate decarboxylase, revealed that the maximum relative mRNA expression level ([5.93 ± 0.82] × 10−4) of graF was detected in the total RNA of the cells after one hour of cultivation when γ-resorcylate was used as the sole carbon source. Reverse transcription-PCR of graDAFCBE showed that these genes are transcribed as a single mRNA and that the transcription of the gene cluster is induced by γ-resorcylate. These results suggested that the graDAFCBE genes are responsible as an operon for the growth of Rhizobium sp. strain MTP-10005 on γ-resorcylate and are probably regulated by GraR at the transcriptional level. This is the first report of the γ-resorcylate catabolic pathway in an aerobic bacterium.
γ-Resorcylate decarboxylase (EC 4.1.1.x) reversibly catalyzes the decarboxylation of γ-resorcylate (2,6-dihydroxybenzoate) to resorcinol (18, 21, 25, 38, 39). The enzyme is expected to be useful for the industrial production of γ-resorcylate, which is an important intermediate in the synthesis of pharmaceuticals and agricultural chemicals (29, 30). Our research group, as well as a few other researchers, have reported the occurrence and biochemical properties of the enzyme (18, 25, 38, 39). We isolated a microorganism, identified as Rhizobium sp. strain MTP-10005, which shows a high level of γ-resorcylate decarboxylase activity (38). We then purified the enzyme and expressed its gene in Escherichia coli for the first time (38). Moreover, we determined the three-dimensional structure of the enzyme and found that one Zn2+ binds to Glu8, His10, His164, and Asp287 in the active center of the enzyme and that the enzyme is a novel Zn2+-dependent decarboxylase (15). During the course of cloning of the γ-resorcylate decarboxylase gene, graF, we found that the homologous genes encoding a hypothetical resorcinol hydroxylase (graA), a hydroxyquinol 1,2-dioxygenase (graB), and a maleylacetate reductase (graC) were localized immediately upstream and downstream of it (38). Recently, we found four additional genes homologous to an NAD(P)H-flavin oxidoreductase (graD), a MarR-type transcriptional regulator (graR), the hypothetical protein Atu2526 (graE), and a benzoate transport protein (graK). Genetic analysis of this γ-resorcylate catabolic gene cluster from aerobic bacteria has not been done up to now (21), but it deserves much attention, especially because of the first γ-resorcylate catabolic pathway in aerobic bacteria and the importance for the regulation of microbial production of γ-resorcylate and development of effective γ-resorcylate production systems in industry.
We here describe the genetic analysis of the γ-resorcylate catabolic pathway in the aerobic bacterium Rhizobium sp. strain MTP-10005, as well as the identification and functional analysis of its gene cluster.
MATERIALS AND METHODS
Materials.
E. coli BL21(DE3), E. coli NovaBlue, a pET3b vector, a pET14b vector, and a pT7Blue T-Vector were purchased from EMD Biosciences, Inc. (San Diego, CA). Ex Taq DNA polymerase, a DNA ligation kit (Mighty Mix), the LA PCR in vitro cloning kit, the chaperone plasmid pG-Tf2, containing groES, groEL, and tig, and the restriction enzymes BamHI, EcoRI, NdeI, PstI, and SalI were purchased from TaKaRa Bio, Inc. (Shiga, Japan). BglII was purchased from Roche Diagnostics (Basel, Switzerland). E. coli BL21 Star (DE3), DNase I (amplification grade), and the SuperScript III Platinum SYBR green one-step quantitative reverse transcription-PCR (qRT-PCR) kit were purchased from Invitrogen Corp. (Carlsbad, CA). The UltraClean15 DNA purification kit was purchased from Mo Bio Laboratories, Inc. (West Carlsbad, CA). Ni-nitrilotriacetic acid (NTA) agarose, the OneStep RT-PCR kit (OneStep RT-PCR buffer and OneStep RT-PCR enzyme mix), and the RNeasy mini kit were purchased from QIAGEN (Hilden, Germany). A DNA ladder was purchased from New England Biolabs, Inc. (Ipswich, MA). Precision Plus Protein all-blue standards (Bio-Rad Laboratories, Inc., Hercules, CA) and protein molecular weight markers (Fermentas International, Inc., Burlington, ON, Canada) were used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). All other reagents were the best grade commercially available and were purchased from Kanto Kagaku Co. (Tokyo, Japan), Kishida Chemical Co. (Osaka, Japan), Sigma-Aldrich Co. (St. Louis, MO), Tokyo Kasei Kogyo Co. (Tokyo, Japan), or Wako Chemical Co. (Osaka, Japan) unless otherwise stated.
Bacterial strains, plasmids, and primers.
The bacterial strains and plasmids used in this study are summarized in Table 1. The primer sequences used for genome-walking PCR, the construction of expression vectors, qRT-PCR, and RT-PCR are summarized in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Rhizobium sp. strain MTP-10005 | γ-Resorcylate decarboxylase-producing bacterium | 38 |
| E. coli BL21 (DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | EMD Biosciences |
| E. coli BL21 Star (DE3) | F−ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Invitrogen |
| E. coli NovaBlue | Tcr; endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 lac F′[proA+B+lacIqZΔM15::Tn10 (Tcr)] | EMD Biosciences |
| Plasmids | ||
| Vector plasmids | ||
| pET3b | Apr; expression vector | EMD Biosciences |
| pET14b | Apr; expression vector | EMD Biosciences |
| pT7Blue T-Vector | Apr; TA cloning vector | EMD Biosciences |
| Plasmids used for cloning of γ-resorcylate degradation genes | ||
| pGWU1 | Apr; pT7Blue T-Vector, a 1.2-kb SalI fragment | This study |
| pGWU2 | Apr; pT7Blue T-Vector, a 0.75-kb PstI fragment | This study |
| pGWU3 | Apr; pT7Blue T-Vector, a 1.0-kb SalI fragment | This study |
| pGWU4 | Apr; pT7Blue T-Vector, a 0.75-kb BamHI fragment | This study |
| pGWD1 | Apr; pT7Blue T-Vector, a 3.0-kb EcoRI fragment | This study |
| pGWD2 | Apr; pT7Blue T-Vector, a 1.5-kb EcoRI fragment | This study |
| pGWD3 | Apr; pT7Blue T-Vector, a 2.0-kb PstI fragment | This study |
| Plasmids used for expression of γ-resorcylate catabolic genes | ||
| pGRA | Apr; pET14b-graA | This study |
| pGRA3 | Apr; pET3b-graA | This study |
| pGRB | Apr; pET14b-graB | This study |
| pGRC | Apr; pET14b-graC | This study |
| pGRD | Apr; pET14b-graD | This study |
| pGRDA | Apr; pET14b-graDA | This study |
| Plasmids used for qRT-PCR | ||
| pTGF | Apr; pT7Blue T-Vector-graF | 38 |
| pTR16 | Apr; pT7Blue T-Vector-16S rRNA | 38 |
TABLE 2.
Primer sequences used in this study
| Primer | Sequencea |
|---|---|
| Primers used for genome-walking PCR | |
| Sense primers used for PCR and nested PCR | |
| pGWU1 PCR sense primer | 5′-GCA TTC AGC GAC AGG ATC ATG GT-3′ |
| Nested PCR sense primer | 5′-TCC ATC AGC TTC AGG CGC GTA TC-3′ |
| pGWU2 PCR sense primer | 5′-CGG TCA GAA GCG GCA TGT TG-3′ |
| Nested PCR sense primer | 5′-TGG GAA CAA AGA CAT CAT CGA GAA CCA-3′ |
| pGWU3 PCR sense primer | 5′-GGC GCT GGA CCG GAA AAG AT-3′ |
| Nested PCR sense primer | 5′-TGT GGC GCA TGG CTC ATA TCG-3′ |
| pGWU4 PCR sense primer | 5′-GAT CAC CCG TGG CAC AGG C-3′ |
| Nested PCR sense primer | 5′-ATC GTC GTG CGG AAA TTC TGG C-3′ |
| pGWD1 PCR sense primer | 5′-CTC GAA GAG CAT TTC GCA ATC CC-3′ |
| Nested PCR sense primer | 5′-GTG CCC GGT GAT TAC TGG AAG GAA CT-3′ |
| pGWD2 PCR sense primer | 5′-CAG AGG CGC TTT ATG CTC GGG A-3′ |
| Nested PCR sense primer | 5′-GCA CTT TAC GGC GCG TGG C-3′ |
| pGWD3 PCR sense primer | 5′-CTT CAC CAC GCT CGT CAC GC-3′ |
| Nested PCR sense primer | 5′-TGC GGT TTT CGG TGT GAA GGA AAG-3′ |
| Antisense primers used for PCR and nested PCR with pGWU1, pGWU2, pGWU3, pGWU4, pGWD1, pGWD2, and pGWD3 | |
| PCR antisense primer | 5′-GTA CAT ATT GTC GTT AGA ACG CGT AAT ACG ACT CA-3′ |
| Nested PCR antisense primer | 5′-CGT TAG AAC GCG TAA TAC GAC TCA CTA TAG GGA GA-3′ |
| Primers used for construction of expression vectors | |
| graA sense primer | 5′-TCA TAT GAA CGA TAT GAG CCA TGC G-3′ |
| Antisense primer | 5′-AGG ATC CTC AAT ATT GGC CCT TGG-3′ |
| graB sense primer | 5′-CAT ATG GAC ATG AAA ACA ACC GGT GAC GAC G-3′ |
| Antisense primer | 5′-GGA TCC TCA TCG GGC GAG CAC G-3′ |
| graC sense primer | 5′-CGC ATA TGC AGC CGT TCG TCT ATA C-3′ |
| Antisense primer | 5′-GGA TCC TCA CTC CGG CCT TG-3′ |
| graD sense primer | 5′-CCA TAT GAC ATC AGC ACT GTT TGG CCT-3′ |
| Antisense primer | 5′-GGA TCC TCA GGC GGA CAG GC-3′ |
| Primers used for qRT-PCR | |
| graF sense primer | 5′-GGT GGA AAA GCT TGA TGT GC-3′ |
| Antisense primer | 5′-TGG CCA AGA ATG ATG TTG AG-3′ |
| 16S rRNA sense primer | 5′-GAT CCT GGC TCA GAA CGA AC-3′ |
| Antisense primer | 5′-GGC TCA TCA TAC CCC GAT AA-3′ |
| Primer sequences used for RT-PCR | |
| Sense primer for all amplifications | 5′-CTG TTT GGC CTG AAC AAT CTT GC-3′ |
| Antisense primer for graDA region | 5′-CTT CGA CAA GCT CGG CAA ATT C-3′ |
| Antisense primer for graDAF region | 5′-TGG CCA AGA ATG ATG TTG AG-3′ |
| Antisense primer for graDAFC region | 5′-CCA TAG CGT CGA GAT CTT CC-3′ |
| Antisense primer for graDAFCB region | 5′-TGC CGA AAC GAT GTA GTG C-3′ |
| Antisense primer for graDAFCBE region | 5′-AGC CAT GAC GTC CTC GAT AC-3′ |
Sequences shown in boldface type are as follows: CATATG, NdeI site; GGATCC, BamHI site.
Cloning and sequence analysis of graA, graB, graC, graD, graE, graK, and graR.
Genome-walking PCR was done with the LA PCR in vitro cloning kit to analyze the genes upstream and downstream of graF. The chromosomal DNA extracted from Rhizobium sp. strain MTP-10005 cells was digested with BamHI, which produces the same type of sticky end as Sau3AI, EcoRI, PstI, or SalI and is ligated to the Sau3AI, EcoRI, PstI, or SalI cassette, respectively. The DNA fragments obtained were used as the template for PCR amplifications. PCR amplification was carried out in a Gene Amp PCR system 9700 (PE Applied Biosystems, Piscataway, NJ). The PCR fragments obtained were purified from the agarose gel with the UltraClean15 DNA purification kit and ligated into a pT7Blue T-Vector with the DNA ligation kit (Mighty Mix). The plasmids constructed (pGWU1, pGWU2, pGWU3, pGWU4, pGWD1, pGWD2, and pGWD3) were transformed into E. coli NovaBlue. The white ampicillin-resistant colonies were selected and grown in 5 ml of LB medium containing ampicillin (100 μg/ml). The plasmid extracted from the cells was purified and sequenced by means of a DNA sequencer SQ5500E (Hitachi High-Technologies Corp., Tokyo, Japan) with a Thermo Sequenase primer cycle sequencing kit (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). A homology search was done with a BLAST program (http://www.ncbi.nlm.nih. gov/BLAST/).
Construction of expression vectors of graA, graB, graC, and graD.
graA, graB, graC, and graD were amplified from Rhizobium sp. strain MTP-10005 genomic DNA (253 ng) with the primers (10 pmol) by PCR. PCR amplification was carried out with Ex Taq DNA polymerase in a Gene Amp PCR system 9700. The PCR fragments obtained were purified from the agarose gel with an UltraClean15 DNA purification kit and subcloned into a pT7Blue T-Vector. After the DNA sequence was confirmed, the subcloned gene was digested with NdeI/BamHI and cloned into a pET14b vector digested with the same restriction enzymes to form pGRA, pGRB, pGRC, or pGRD. The graDA coexpression vector, pGRDA, was constructed as follows: graA was ligated into the NdeI/BamHI sites of pET3b vector; the plasmid obtained, pGRA3, was digested with BglII/EcoRI; and the fragment containing graA was ligated into pGRD digested with BamHI/EcoRI to form pGRDA.
Expression of graA, graB, graC, and graD in E. coli BL21 Star (DE3).
pGRA, pGRB, pGRC, pGRD, or pGRDA was transformed into E. coli BL21 Star (DE3). The clones were grown in a Sakaguchi flask (500 ml) containing 200 ml of LB medium supplemented with ampicillin (100 μg/ml). After cultivation at 37°C with shaking (130 rpm) until the optical density at 600 nm (OD600) reached approximately 0.6, the cultures were placed at 15°C for at least 30 min and cultivated again at 15°C for 24 h. pGRD was transformed into E. coli BL21(DE3) harboring a chaperone plasmid, pG-Tf2, containing groES, groEL, and tig. The cells were grown in a Sakaguchi flask (500 ml) containing 200 ml of LB medium supplemented with ampicillin (50 μg/ml) and chloramphenicol (20 μg/ml). The cells were grown at 37°C with shaking (130 rpm), and tetracycline was added to the medium (final concentration, 10 ng/ml) when the OD600 reached approximately 0.3. The cultures were then placed at 15°C for at least 30 min when the OD600 reached approximately 0.6 and cultivated again at 15°C for 24 h.
Preparation of cell extracts.
After cultivation, the cells were collected by centrifugation (6,500 × g, 15 min) and washed in a 20 mM Tris-HCl buffer (pH 8.0). The washed cells were suspended in a 20 mM Tris-HCl buffer (pH 8.0) and disrupted with an ultrasonic disintegrator, model UD-201 (Tomy Seiko Co., Tokyo, Japan). The disruption was done in an ice bath for 3 min and repeated twice after the output and duty cycle were adjusted to 6 and 30, respectively. The cell debris was removed by centrifugation (16,000 × g, 15 min), and the supernatant solution was used as a cell extract.
Enzyme assays.
In all enzyme assays described below, a blank assay against the enzyme reaction was taken with a cell extract of the host cells harboring the corresponding empty vector, and the value obtained in the blank assay was subtracted as the background activity. We also subtracted the blank activities of the oxidation of NADH. One unit of enzyme was defined as the amount of enzyme that transforms 1 μmol of NADH, resorcinol, hydroxyquinol, or γ-resorcylate per min at 30°C. The molar extinction coefficients (M−1·cm−1) were as follows: 6,220 for NADH at 340 nm and 42,000 for maleylacetate at 243 nm.
The flavin reductase (EC 1.5.1.30) activity was assayed by monitoring the oxidation rate of NADH spectrophotometrically at 340 nm. The reaction mixture (final volume, 3 ml) contained 0.1 mM FAD, 0.1 mM NADH, a 0.1 M potassium phosphate buffer (pH 7.0), and the cell extract. The reaction was started by the addition of NADH and was carried out at 30°C.
The resorcinol hydroxylase (EC 1.14.13.x) activity was assayed by measuring the remaining resorcinol by high-performace liquid chromatography (HPLC), according to the modified method of Otto et al. (26). The reaction mixture (final volume, 1 ml) contained 2 mM resorcinol, 50 mM NADH, a 50 mM Tris-HCl buffer (pH 8.0), 150 mM potassium formate, 0.5 U/ml formate dehydrogenase (Roche Diagnostics), 650 U/ml catalase (Wako Chemical Co.), and the cell extract. The reaction was started by the addition of NADH and was carried out at 30°C with shaking (150 rpm). After 0, 30, 60, 120, 180, 240, or 300 min, the reactions were stopped with 1 ml of acetonitrile containing 10% (vol/vol) acetic acid. The mixture (10 μl) was then subjected to HPLC analysis.
The hydroxyquinol 1,2-dioxygenase (EC 1.13.11.37) activity was assayed by measuring the produced maleylacetate spectrophotometrically at 243 nm (9, 12, 35). The reaction mixture (final volume, 3 ml) contained 0.5 mM hydroxyquinol, a 50 mM 2-morpholinoethanesulfonate-NaOH buffer (pH 6.0), and the cell extract. The reaction was started by the addition of hydroxyquinol and was carried out at 30°C.
The maleylacetate reductase (EC 1.3.1.32) activity was assayed by measurement of the oxidation rate of NADH spectrophotometrically at 340 nm. The reaction mixture (final volume, 3 ml) contained 0.5 mM hydroxyquinol, 0.2 mM NADH, a 50 mM 2-morpholinoethanesulfonate-NaOH buffer (pH 6.5), and the cell extract. The reaction was started by the addition of hydroxyquinol and was carried out at 30°C.
The γ-resorcylate decarboxylase (EC 4.1.1.x) activity was assayed according to a previously described method (38), except that the reaction time was changed to 10 min.
qRT-PCR of graF.
Rhizobium sp. strain MTP-10005 was cultivated in an Erlenmeyer flask (100 ml) containing 20 ml of LB medium on a rotary shaker (150 rpm) at 30°C. After 21 h, 2 ml of the culture was transferred into a Sakaguchi flask (500 ml) containing 200 ml of LB medium and cultivated on a reciprocal shaker (130 rpm) at 30°C for 7 h. The cells were harvested by centrifugation (7,500 × g, 20 min), suspended in a Sakaguchi flask (500 ml) containing 200 ml of a minimum medium supplemented with 0.3% (wt/vol) γ-resorcylate or glycerol, and cultivated on a reciprocal shaker (130 rpm) at 30°C. After 0, 0.5, 1, 3, 6, or 9 h, 15 ml of the culture was taken from the flask and the cells were collected by centrifugation (7,500 × g, 20 min) and washed twice with a 10 mM potassium phosphate buffer (pH 7.0) containing 0.75% (wt/vol) NaCl. The washed cells were suspended in 750 μl of a 10 mM potassium phosphate buffer (pH 7.0) containing 0.02% (wt/vol) 2-mercaptoethanol, and 500 μl of the cell suspension obtained was subjected to an ultrasonic disintegrator, model UD-201 (Tomy Seiko Co.), at about 4°C (two times for 9 s; output, 6; duty cycle, 30). The cell debris was removed by centrifugation (16,000 × g, 15 min), and the supernatant solution was used as a cell extract. The total RNA was extracted from the remaining 250 μl of cell suspension obtained above with the RNeasy mini kit according to the manufacturer's instructions and was treated with DNase I. qRT-PCR was performed with an Mx3000P Real-Time PCR system (Stratagene Corp., La Jolla, CA). The reaction mixture for qRT-PCR consisted of SuperScript III RT/Platinum Taq mix, 1× SYBR green reaction mix, 1 mM MgSO4, 0.5 μM ROX reference dye, 0.2 μM sense and antisense primers, and 100 ng of total RNA. The housekeeping gene used as a control was 16S rRNA, and the mRNA expression level was normalized in such a way that the 16S rRNA expression level was equal to 1 and was averaged with three independent measurements.
RT-PCR of the graDAFCBE genes.
Total RNA was extracted from Rhizobium sp. strain MTP-10005 cells that were cultivated as described above in a minimal medium supplemented with γ-resorcylate or glycerol at 30°C for 1 h. RT-PCR was performed with an iCycler thermal cycler (Bio-Rad Laboratories, Inc.). The reaction mixture for RT-PCR consisted of a 1× OneStep RT-PCR buffer, a 400 μM deoxynucleoside triphosphate mix, 0.6 μM sense and antisense primers, the OneStep RT-PCR enzyme mix, and 100 ng of total RNA.
Analytical methods.
Spectrophotometric measurements were done with a UV/visual spectrophotometer U-3210 (Hitachi High-Technologies Corp.). The HPLC analysis was performed with HPLC system LC-10A (Shimadzu Corp., Kyoto, Japan). The column used was an Inertsil ODS-2 column (0.46 by 25 cm; GL Sciences, Inc., Tokyo, Japan), and resorcinol was detected at 280 nm. The mobile phase used was 15% (vol/vol) acetonitrile in water containing 0.1% (vol/vol) trifluoroacetic acid, and the flow rate was 0.6 ml/min. SDS-PAGE was performed by the method of Laemmli (22). The proteins in the gels were stained with Coomassie brilliant blue R-250. The protein concentration was measured by the method of Bradford (7), with bovine serum albumin (Wako Chemical Co.) used as the standard.
Nucleotide sequence accession numbers.
The DNA sequences of graA, graB, graC, graD, graE, graK, and graR of Rhizobium sp. strain MTP-10005 were submitted to the GenBank database under accession numbers AB266210, AB266211, AB266212, AB266213, AB266214, AB266215, and AB266216, respectively.
RESULTS AND DISCUSSION
Cloning and sequencing of the γ-resorcylate catabolic gene cluster and analysis of its gene organization.
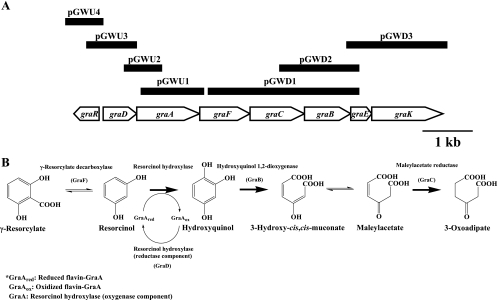
The organization of the γ-resorcylate catabolic gene cluster, based on genome-walking PCR, is summarized in Fig. 1A. The typical −35/−10 promoter sequences were identified only upstream of the genes graD, graR, and graK. The distances between graD and graA, graA and graF, graF and graC, and graC and graB are 36, 47, 3, and 39 bp, respectively. These results suggest that graDAFCBE forms an operon and that graR and graK may be independently transcribed. The putative Shine-Dalgarno sequences precede the start codons of graR, graD, graA, graF, graB, and graK, but such sequences do not exist in front of the start codons of graC and graE. Additionally, the start codon of graE overlaps the stop codon of graB by 4 bp, indicating that graF and graC, as well as graB and graE, are translationally coupled (19). The characteristics of the γ-resorcylate catabolic gene cluster are summarized in Table 3. The bioinformatics analysis suggests that graR, graC, and graB encode a MarR-type transcriptional regulator, a maleylacetate reductase, and a hydroxyquinol 1,2-dioxygenase, respectively. GraK shows similarity to a benzoate transport protein and is probably involved in the transport of γ-resorcylate. GraA shows similarity to the oxygenase component of p-hydroxyphenylacetate hydroxylase, suggesting that the hydroxylation of resorcinol may be catalyzed by the coupled reaction of GraD and GraA. The function of GraE cannot be predicted, because there are no functionally characterized homologues in the data bank.
FIG. 1.
Organization of the genes related to γ-resorcylate catabolism (A) and the γ-resorcylate catabolic pathway in Rhizobium sp. strain MTP-10005 (B).
TABLE 3.
Characteristics of the γ-resorcylate catabolic gene cluster
| ORFa | Gene | Length (aa) | Molecular wt | Identical protein(s) | Identity (%) | Accession no. |
|---|---|---|---|---|---|---|
| 1 | graR | 183 | 20,868 | Transcriptional regulator (MarR family) from Azoarcus sp. EbN1 | 26 | CR555306 |
| Putative transcriptional regulator (MarR family) from Agrobacterium tumefaciens strain C58 | 91 | AE009200 | ||||
| 2 | graD | 179 | 19,429 | NAD(P)H-flavin oxidoreductase from Pseudomonas syringae pv. tomato strain DC3000 | 38 | AE016853 |
| Putative actinorhodin polyketide dimerase from Agrobacterium tumefaciens strain C58 | 94 | AE009199 | ||||
| 3 | graA | 409 | 43,305 | p-Hydroxyphenylacetate hydroxylase C2, the oxygenase component from Acinetobacter baumannii | 27 | AY566612 |
| Putative oxidoreductase from Agrobacterium tumefaciens strain C58 | 92 | AE009199 | ||||
| 4 | graF | 327 | 37,422 | 2,3-Dihydroxybenzoate decarboxylase from Aspergillus oryzae | 43 | AP007151 |
| Hypothetical protein Atu2529 from Agrobacterium tumefaciens strain C58 | 96 | AE009199 | ||||
| 5 | graC | 351 | 36,405 | Maleylacetate reductase from Ralstonia eutropha JMP134 | 46 | U16782 |
| Putative maleylacetate reductase from Agrobacterium tumefaciens strain C58 | 90 | AE009199 | ||||
| 6 | graB | 295 | 33,349 | Hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E | 46 | AY822041 |
| Putative dioxygenase from Agrobacterium tumefaciens strain C58 | 94 | AE009199 | ||||
| 7 | graE | 112 | 12,195 | Hypothetical protein Atu2526 from Agrobacterium tumefaciens strain C58 | 75 | AE009199 |
| 8 | graK | 428 | 45,002 | Benzoate transport protein from Acinetobacter baylyi ADP1 | 35 | AF009224 |
| Putative MFS permease from Agrobacterium tumefaciens strain C58 | 94 | AE009199 |
ORF, open reading frame.
Expression of graA, graB, graC, and graD in E. coli BL21 Star (DE3).
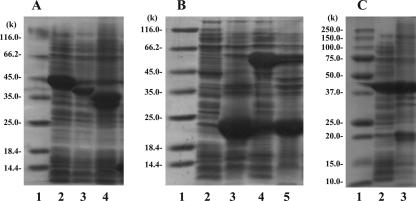
The plasmids pGRD, pGRA, pGRC, pGRB, and pGRDA were constructed with a high-expression vector (pET14b) and graD, graA, graC, graB, and both graD and graA, respectively (Table 1), and each plasmid was introduced into E. coli BL21 Star (DE3) by transformation. The cell extracts of these clones were subjected to SDS-PAGE analysis (Fig. 2). Overexpressed proteins with molecular weights of approximately 22,100 (22.1K), 45.2K, 38.2K, 35.2K, and 21.2K or 43.2K were observed in E. coli BL21 Star (DE3) harboring pGRD, pGRA, pGRC, pGRB, and pGRDA, respectively (Fig. 2). The molecular weights of these overexpressed proteins agreed well with the expected values of the N-terminal His-tagged gene products of graD (21.6K), graA (45.5K), graC (38.6K), and graB (35.5K) and the gene product of graA (43.3K). GraD expressed in E. coli BL21 Star (DE3) harboring pGRD was present mostly in the insoluble fractions. However, GraD was overexpressed in the soluble fractions of E. coli BL21(DE3) harboring pGRD when the chaperone plasmid pG-Tf2 was coexpressed (Fig. 2B). The chaperon probably helps in the correct folding of GraD.
FIG. 2.
SDS-PAGE of cell extracts from E. coli harboring γ-resorcylate catabolic genes. (A) Lane 1, molecular weight marker (Fermentas International, Inc.); lane 2, cell extract of E. coli BL21 Star (DE3) harboring pGRA (38.1 μg); lane 3, cell extract of E. coli BL21 Star (DE3) harboring pGRC (21.7 μg); lane 4, cell extract of E. coli BL21 Star (DE3) harboring pGRB (39.2 μg). (B) Lane 1, molecular weight marker (Fermentas International, Inc.); lane 2, cell extract of E. coli BL21 Star (DE3) harboring pGRD (16.6 μg); lane 3, insoluble fraction of E. coli BL21 Star (DE3) harboring pGRD; lane 4, cell extract of E. coli BL21 (DE3) harboring pGRD and the chaperone plasmid pG-Tf2 (32.8 μg); lane 5, insoluble fraction of E. coli BL21(DE3) harboring pGRD and the chaperone plasmid pG-Tf2. (C) Lane 1, molecular weight marker (Bio-Rad Laboratories, Inc.); lane 2, cell extract of E. coli BL21 Star (DE3) harboring pGRDA (24.6 μg); lane 3, insoluble fraction of E. coli BL21 Star (DE3) harboring pGRDA.
Identification of GraA, GraB, GraC, and GraD.
The enzyme activities of the expressed proteins were measured as described in Materials and Methods to clarify their functions. The experimental data of the enzyme activities are summarized in Table 4. The enzyme activities detected for GraA, GraB, GraC, and GraD agreed well with the expected functions based on the bioinformatic analysis.
TABLE 4.
Experimental data for measurement of enzyme activitiesa
| Enzymeb | Change in valuec/min | Total activity (U) | Total protein (mg) | Sp act (U/mg) |
|---|---|---|---|---|
| Flavin reductase | ||||
| GraD* | 0.271 ± 0.021 | 65.4 ± 5.0 | 16.6 ± 0.1 | 3.94 ± 0.30 |
| (GraD + chaperone)† | 0.332 ± 0.018 | 801 ± 44 | 32.8 ± 0.2 | 24.4 ± 1.3 |
| Resorcinol hydroxylase | ||||
| GraA* | 0 | 0 | 68.3 ± 0.4 | 0 |
| (GraD + chaperone)† + GraA* | 1,390 ± 140 | (9.63 ± 0.94) × 10−2 | (58.6 ± 0.3) + (68.3 ± 0.4) | (1.41 ± 0.14) × 10−3 |
| GraDA* | 2,960 ± 220 | 0.103 ± 0.008 | 48.9 ± 0.1 | (2.11 ± 0.15) × 10−3 |
| Hydroxyquinol 1,2-dioxygenase | ||||
| GraB* | 0.165 ± 0.004 | 23.6 ± 0.6 | 39.2 ± 0.6 | 0.602 ± 0.016 |
| Maleylacetate reductase | ||||
| GraC* | 0.304 ± 0.005 | 293 ± 5 | 21.7 ± 0.5 | 13.5 ± 0.2 |
Cell extract was used for all enzyme assays. Values are means ± standard deviations.
Symbols: *, expressed in E. coli BL21 Star (DE3); †, GroES + GroEL + Tig, expressed in E. coli BL21 (DE3).
Units: flavin reductase, ΔA340/min; resorcinol hydroxylase, change in peak area of resorcinol per min; hydroxyquinol 1,2-dioxegenase, ΔA243/min; maleylacetate reductase, ΔA340/min. Hydroxyquinol and resorcinol were eluted at retention times of 5.06 and 10.1 min, respectively.
Flavin reductase activity was detected when the cell extract of E. coli BL21 Star (DE3) harboring pGRD or E. coli BL21(DE3) harboring pGRD and the chaperone plasmid, pG-Tf2, were used. The specific activity for FAD was determined to be 3.94 ± 0.30 U/mg for E. coli BL21 Star (DE3) harboring pGRD. About six-times-higher specific activity (24.4 ± 1.3 U/mg) was observed when the chaperone plasmid pG-Tf2 was coexpressed. However, the cell extract of E. coli BL21 Star (DE3) harboring pGRA did not show resorcinol hydroxylase activity by itself. The resorcinol hydroxylase activity was detected only when the cell extract of E. coli BL21(DE3) harboring pGRD and pG-Tf2 was mixed with that of E. coli BL21 Star (DE3) harboring pGRA. The specific activity was calculated to be (1.41 ± 0.14) × 10−3 U/mg. In addition, when GraD and GraA were coexpressed in E. coli BL21 Star (DE3) harboring pGRDA, resorcinol hydroxylase activity was also detected, and about 1.5-times-higher specific activity was observed ([2.11 ± 0.15] × 10−3 U/mg). Accordingly, the chaperone plasmid pG-Tf2 was effective for the expression of GraD in the soluble fractions of E. coli BL21(DE3). GraD and GraA were eluted separately when the cell extract of E. coli BL21 Star (DE3) harboring pGRDA was applied to a column of Ni-NTA agarose (data not shown). These results suggest that GraD and GraA are not subunits but rather two essential components of resorcinol hydroxylase. By considering the sequence similarities (Table 3), GraA and GraD are identified as the oxygenase and reductase components of resorcinol hydroxylase, respectively. Two types of flavin-dependent aromatic hydroxylases, i.e., single-component and two-component enzymes, have been reported previously. The single-component enzyme contains one subunit with oxygenase and reductase functions, whereas the two-component enzyme has two components that catalyze the oxygenation of the substrate and transfer a reduced form of flavin to the oxygenase component separately. The single-component NADPH-dependent resorcinol hydroxylase from Corynebacterium glutamicum has recently been described (17), and the two-component p-hydroxyphenylacetate hydroxylases from Acinetobacter baumannii (6), Pseudomonas aeruginosa (6), and E. coli W ATCC 11105 (23) are well characterized. The recombinant resorcinol hydroxylase from Rhizobium sp. strain MTP-10005 showed the enzyme activity only when both GraA (the oxygenase component) and GraD (the reductase component) existed in the assay mixture and therefore is classified as a two-component flavin-dependent aromatic hydroxylase. This is the first example of a two-component resorcinol hydroxylase. In contrast to the resorcinol hydroxylase from C. glutamicum, the reductase component of the resorcinol hydroxylase from Rhizobium sp. strain MTP-10005 requires NADH as a coenzyme. The sequence similarities between C. glutamicum resorcinol hydroxylase and GraA or GraD are 2.9 and 4.5%, respectively.
Hydroxyquinol 1,2-dioxygenase activity was detected in the cell extract of E. coli BL21 Star (DE3) harboring pGRB (specific activity, 0.602 ± 0.016 U/mg). Accordingly, GraB is identified as a hydroxyquinol 1,2-dioxygenase. In the hydroxyquinol 1,2-dioxygenase reaction, 3-hydroxy-cis,cis-muconate produced from hydroxyquinol is spontaneously and reversibly converted to its tautomer, maleylacetate. The three-dimensional structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E has been solved, and the essential amino acid residues for the enzyme activity have been determined (13). Tyr164, Tyr197, His221, and His223 form a polyhedron structure with Fe(III) that is essential for the catalytic activity, and a similar structure has been observed for the catechol 1,2-dioxygenase from Acinetobacter baylyi ADP1 (37). These amino acid residues are well conserved as Tyr165, Tyr200, His224, and His226 in the primary structure of the hydroxyquinol 1,2-dioxygenase from Rhizobium sp. strain MTP-10005 and could be the probable iron ligands.
Maleylacetate reductase activity was detected in the cell extract of E. coli BL21 Star (DE3) harboring pGRC in the presence of the cell extract of E. coli BL21 Star (DE3) harboring pGRB and hydroxyquinol to supply maleylacetate as a substrate. The specific activity was determined to be 13.5 ± 0.2 U/mg. Accordingly, GraC is identified as a maleylacetate reductase. We found that Cys242 in the primary structure of the maleylacetate reductase from Rhizobium sp. strain MTP-10005 is conserved in the enzyme from Ralstonia eutropha JMP134. The R. eutropha JMP134 enzyme was inhibited by p-chloromercuribenzoate and stabilized by dithiothreitol (31). Accordingly, Cys242 is probably necessary for the catalytic activity of the maleylacetate reductase from Rhizobium sp. strain MTP-10005, too. Although the maleylacetate reductase gene from Ralstonia eutropha 335T was expressed in E. coli (32) and the enzymological characteristics of the R. eutropha JMP134 enzyme were well characterized (31, 36), the three-dimensional structures of the enzymes have not been determined.
Based on these enzymological studies, the γ-resorcylate catabolic pathway of Rhizobium sp. strain MTP-10005 is proposed in Fig. 1B.
Regulation of the graDAFCBE operon.
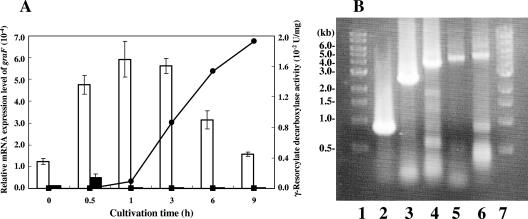
The MarR-type transcriptional regulator homolog gene graR (Table 3) exists upstream of the graDAFCBE genes (Fig. 1A). The MarR transcriptional regulator from E. coli negatively regulates the expression of the antibiotic resistance genes, marAB (1, 4, 10, 24, 33). Inducers of these genes, such as 2-hydroxybenzoate and 2,4-dinitrophenol, have been shown to bind to the marAB operator region and allow transcription of the marAB operon. The relative mRNA expression level of graF was measured by qRT-PCR in the presence and absence of γ-resorcylate (Fig. 3A). The total RNA from cells grown on γ-resorcylate or glycerol was extracted and used. The relative mRNA expression level of graF increased significantly in the total RNA of the cells when γ-resorcylate was used as the sole carbon growth source, and the maximum relative value ([5.93 ± 0.82] × 10−4) was detected after 1 h of cultivation. Moreover, γ-resorcylate decarboxylase activity was detected after 1 h of cultivation ([8.97 ± 0.08] × 10−4 U/mg) and increased with the cultivation time up to 9 h (Fig. 3A). The results of RT-PCR against graDAFCBE genes showed that the amplified fragments with molecular sizes of approximately 796, 2,494, 3,780, 4,616, and 5,041 bp agree well with the expected values of those for graDA (790 bp), graDAF (2,453 bp), graDAFC (3,677 bp), graDAFCB (4,687 bp), and graDAFCBE (5,082 bp) (Fig. 3B), respectively, and that the graDAFCBE genes are transcribed as a single mRNA and the transcription of the gene cluster is induced in the presence of γ-resorcylate. These results suggest that the graDAFCBE genes are expressed as an operon for the growth of Rhizobium sp. strain MTP-10005 on γ-resorcylate and are probably regulated by GraR at the transcriptional level. The homologous benzoate transport gene of GraK (Table 3) existed downstream of the graDAFCBE operon (Fig. 1A). BenK, the benzoate transport protein that is involved in the benzoate degradation of Acinetobacter baylyi ADP1, is induced by a benzoate metabolite, cis,cis-muconate, and transports benzoate and benzaldehyde into the cytoplasm (11). Judging from the sequence similarity, GraK is probably involved in transporting γ-resorcylate.
FIG. 3.
qRT-PCR and RT-PCR analyses. (A) Relative mRNA expression levels of graF and γ-resorcylate decarboxylase activity versus cultivation time. White bars show the relative mRNA expression levels of graF in total RNA prepared from Rhizobium sp. strain MTP-10005 cells cultivated in a minimal medium supplemented with 0.3% (wt/vol) γ-resorcylate. Black bars show the relative mRNA expression levels of graF in the total RNA prepared from Rhizobium sp. strain MTP-10005 cells cultivated in a minimal medium supplemented with 0.3% (wt/vol) glycerol. •, γ-resorcylate decarboxylase activity of cell extract prepared from Rhizobium sp. strain MTP-10005 cells cultivated in a minimal medium supplemented with 0.3% (wt/vol) γ-resorcylate; ▪, γ-resorcylate decarboxylase activity of cell extract prepared from Rhizobium sp. strain MTP-10005 cells cultivated in a minimal medium supplemented with 0.3% (wt/vol) glycerol. The temperature profile used for qRT-PCR was as follows: RT step (one cycle, 50°C, 30 min), initial PCR activation step (one cycle, 95°C, 15 min), PCR step (50 cycles, 94°C, 15 s; 55°C for 30 s; 72°C for 1 min), final annealing step (one cycle, 55°C, 30 s), and final denaturation step (one cycle, 95°C, 30 s). The sequence-specific standard curves were described with 1 ng, 10 pg, 100 fg, and 1 fg of pTGF or pTR16. (B) RT-PCR analysis of the graDAFCBE operon. Lanes 1 and 7, 1-kb DNA ladder; lane 2, internal 796 bp of graDA region; lane 3, internal 2,494 bp of graDAF region; lane 4, internal 3,780 bp of graDAFC region; lane 5, internal 4,616 bp of graDAFCB region; lane 6, internal 5,041 bp of graDAFCBE region. The temperature profile used for RT-PCR was as follows: RT step (one cycle, 45°C, 30 min), initial PCR activation step (one cycle, 95°C for 15 min), PCR step (forty cycles, 94°C, 10 s; 55°C, 30 s; 68°C, 5 min), and final extension step (one cycle, 68°C, 10 min).
A novel pathway for γ-resorcylate catabolism in Rhizobium sp. strain MTP-10005.
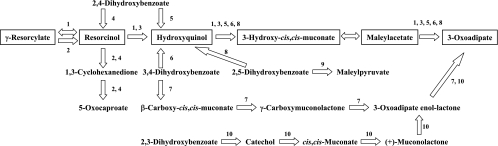
The biodegradation of various dihydroxybenzoates has been reported only in microorganisms, and their pathways are summarized in Fig. 4. The γ-resorcylate pathway has been reported previously only for an anaerobic bacterium, Clostridium sp. KN 245 (21), in which γ-resorcylate is converted into 5-oxocaproate via resorcinol and 1,3-cyclohexanedione (Fig. 4, pathway 2), and is quite different from the γ-resorcylate pathway of the aerobic bacterium Rhizobium sp. strain MTP-10005 (Fig. 4, pathway 1). The present study is the first report of the γ-resorcylate catabolic pathway in an aerobic bacterium. In the γ-resorcylate pathway of Rhizobium sp. strain MTP-10005 (Fig. 4, pathway 1), the oxygenase component of the resorcinol hydroxylase requires an oxygen molecule to produce hydroxyquinol, whereas in that of Clostridium sp. KN 245 (Fig. 4, pathway 2), resorcinol produced from γ-resorcylate is converted to 1,3-cyclohexanedione by the resorcinol reductase without the participation of oxygen. Accordingly, we can assume that these γ-resorcylate catabolic pathways have evolved independently in bacteria adapted to oxic and anoxic environments. We also investigated whether the γ-resorcylate catabolic pathway exists in microorganisms whose genomes have been sequenced. The results of BLAST analyses indicated that homologous genes for the γ-resorcylate catabolic pathway of Rhizobium sp. strain MTP-10005 are present only in the genome of Agrobacterium tumefaciens strain C58 (Table 3). The genes are clustered in a manner similar to that of the graRDAFCBEK genes and are likely to be functional, since γ-resorcylate decarboxylase was purified from the other strain of A. tumefaciens (39).
FIG. 4.
Various dihydroxybenzoate catabolic pathways in microorganisms. 1, γ-resorcylate pathway of Rhizobium sp. strain MTP-10005; 2, γ-resorcylate pathway of Clostridium sp. KN 245 (21); 3, resorcinol pathway of Corynebacterium glutamicum (17), Pseudomonas putida (9), and Trichosporon cutaneum (14); 4, 2,4-dihydroxybenzoate pathway of Clostridium sp. KN 245 (21); 5, 2,4-dihydroxybenzoate pathway of Pseudomonas sp. BN9 (34) and Sphingomonas sp. strain RW1 (5); 6, 3,4-dihydroxybenzoate pathway of Trichosporon cutaneum (2); 7, 3,4-dihydroxybenzoate 3,4-cleavage pathway of Agrobacterium sp. (27, 28), Bradyrhizobium sp. (27), Rhizobium sp. (27), Roseobacter sp. (8), and Streptomyces sp. strain 2065 (19); 8, 2,5-dihydroxybenzoate pathway of Trichosporon cutaneum (2); 9, 2,5-dihydroxybenzoate 1,2-cleavage pathway of Pseudomonas acidovorans (16) and Pseudomonas testosteroni (16); 10, 2,3-dihydroxybenzoate pathway of Aspergillus niger (20) and Trichosporon cutaneum (2, 3).
We are currently studying the enzymological properties of GraD, GraA, GraC, and GraB in detail and trying to clarify the function of GraE.
Acknowledgments
This research was partially supported by the High-Tech Research Center Project for Private Universities, with a matching-fund subsidy from MEXT (2002-2006).
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. J., and S. Dagley. 1980. Catabolism of aromatic acids in Trichosporon cutaneum. J. Bacteriol. 141:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. J., and S. Dagley. 1981. Catabolism of tryptophan, anthranilate, and 2,3-dihydroxybenzoate in Trichosporon cutaneum. J. Bacteriol. 146:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariza, R. R., S. P. Cohen, N. Bachhawat, S. B. Levy, and B. Demple. 1994. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 176:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armengaud, J., K. N. Timmis, and R.-M. Wittich. 1999. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J. Bacteriol. 181:3452-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballou, D. P., B. Entsch, and L. J. Cole. 2005. Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases. Biochem. Biophys. Res. Commun. 338:590-598. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Buchan, A., E. L. Neidle, and M. A. Moran. 2004. Diverse organization of genes of the β-ketoadipate pathway in members of the marine Roseobacter lineage. Appl. Environ. Microbiol. 70:1658-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, P. J., and D. W. Ribbons. 1976. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J. Bacteriol. 125:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, S. P., H. Hächler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collier, L. S., N. N. Nichols, and E. L. Neidle. 1997. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J. Bacteriol. 179:5943-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daubaras, D. L., K. Saido, and A. M. Chakrabarty. 1996. Purification of hydroxyquinol 1,2-dioxygenase and maleylacetate reductase: the lower pathway of 2,4,5-trichlorophenoxyacetic acid metabolism by Burkholderia cepacia AC1100. Appl. Environ. Microbiol. 62:4276-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferraroni, M., J. Seifert, V. M. Travkin, M. Thiel, S. Kaschabek, A. Scozzafava, L. Golovleva, M. Schlömann, and F. Briganti. 2005. Crystal structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E, a key enzyme involved in polychlorinated aromatics biodegradation. J. Biol. Chem. 280:21144-21154. [DOI] [PubMed] [Google Scholar]
- 14.Gaal, A., and H. Y. Neujahr. 1979. Metabolism of phenol and resorcinol in Trichosporon cutaneum. J. Bacteriol. 137:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto, M., H. Hayashi, I. Miyahara, K. Hirotsu, M. Yoshida, and T. Oikawa. 2006. Crystal structures of nonoxidative zinc-dependent 2,6-dihydroxybenzoate (γ-resorcylate) decarboxylase from Rhizobium sp. strain MTP-10005. J. Biol. Chem. 281:34365-34373. [DOI] [PubMed] [Google Scholar]
- 16.Harpel, M. R., and J. D. Lipscomb. 1990. Gentisate 1,2-dioxygenase from Pseudomonas. Purification, characterization, and comparison of the enzymes from Pseudomonas testosteroni and Pseudomonas acidovorans. J. Biol. Chem. 265:6301-6311. [PubMed] [Google Scholar]
- 17.Huang, Y., K. X. Zhao, X. H. Shen, M. T. Chaudhry, C. Y. Jiang, and S. J. Liu. 2006. Genetic characterization of the resorcinol catabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72:7238-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii, Y., Y. Narimatsu, Y. Iwasaki, N. Arai, K. Kino, and K. Kirimura. 2004. Reversible and nonoxidative gamma-resorcylic acid decarboxylase: characterization and gene cloning of a novel enzyme catalyzing carboxylation of resorcinol, 1,3-dihydroxybenzene, from Rhizobium radiobacter. Biochem. Biophys. Res. Commun. 324:611-620. [DOI] [PubMed] [Google Scholar]
- 19.Iwagami, S. G., K. Yang, and J. Davies. 2000. Characterization of the protocatechuic acid catabolic gene cluster from Streptomyces sp. strain 2065. Appl. Environ. Microbiol. 66:1499-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamath, A. V., and C. S. Vaidyanathan. 1990. New pathway for the biodegradation of indole in Aspergillus niger. Appl. Environ. Microbiol. 56:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluge, C., A. Tschech, and G. Fuchs. 1990. Anaerobic metabolism of resorcylic acids (m-dihydroxybenzoic acids) and resorcinol (1,3-benzenediol) in a fermenting and in a denitrifying bacterium. Arch. Microbiol. 155:68-74. [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Louie, T. M., X. S. Xie, and L. Xun. 2003. Coordinated production and utilization of FADH2 by NAD(P)H-flavin oxidoreductase and 4-hydroxyphenylacetate 3-monooxygenase. Biochemistry 42:7509-7517. [DOI] [PubMed] [Google Scholar]
- 24.Martin, R. G., and J. L. Rosner. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 92:5456-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsui, T., T. Yoshida, T. Yoshimura, and T. Nagasawa. 2006. Regioselective carboxylation of 1,3-dihydroxybenzene by 2,6-dihydroxybenzoate decarboxylase of Pandoraea sp. 12B-2. Appl. Microbiol. Biotechnol. 73:95-102. [DOI] [PubMed] [Google Scholar]
- 26.Otto, K., K. Hofstetter, M. Röthlisberger, B. Witholt, and A. Schmid. 2004. Biochemical characterization of StyAB from Pseudomonas sp. strain VLB120 as a two-component flavin-diffusible monooxygenase. J. Bacteriol. 186:5292-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parke, D., and L. N. Ornston. 1986. Enzymes of the β-ketoadipate pathway are inducible in Rhizobium and Agrobacterium spp. and constitutive in Bradyrhizobium spp. J. Bacteriol. 165:288-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parke, D. 1995. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J. Bacteriol. 177:3808-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid, J., R. D. Watson, J. B. Cochran, D. H. Sproull, B. E. Clayton, and F. T. Prunty. 1951. Sodium γ-resorcylate in rheumatic fever. Br. Med. J. 4727:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruschig, H., J. Konig, D. Duwel, and H. Loewe. 1973. 2,6-Dihydroxybenzoic acid derivatives as anthelmintics. Arzneimittelforschung 23:1745-1785. [PubMed] [Google Scholar]
- 31.Seibert, V., K. Stadler-Fritzsche, and M. Schlömann. 1993. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 175:6745-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seibert, V., M. Thiel, I. S. Hinner, and M. Schlömann. 2004. Characterization of a gene cluster encoding the maleylacetate reductase from Ralstonia eutropha 335T, an enzyme recruited for growth with 4-fluorobenzoate. Microbiology 150:463-472. [DOI] [PubMed] [Google Scholar]
- 33.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stolz, A., and H.-J. Knackmuss. 1993. Degradation of 2,4-dihydroxybenzoate by Pseudomonas sp. BN9. FEMS Microbiol. Lett. 108:219-224. [DOI] [PubMed] [Google Scholar]
- 35.Tiedje, J. M., J. M. Duxbury, M. Alexander, and J. E. Dawson. 1969. 2,4-d metabolism: pathway of degradation of chlorophenols by Arthrobacter sp. J. Agric. Food Chem. 17:1021-1026. [DOI] [PubMed] [Google Scholar]
- 36.Trefault, N., R. de la Iglesia, A. M. Molina, M. Manzano, T. Ledger, D. Perez-Pantoja, M. A. Sanchez, M. Stuardo, and B. Gonzalez. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6:655-668. [DOI] [PubMed] [Google Scholar]
- 37.Vetting, M. W., and D. H. Ohlendorf. 2000. The 1.8 Å crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure 8:429-440. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida, M., N. Fukuhara, and T. Oikawa. 2004. Thermophilic, reversible γ-resorcylate decarboxylase from Rhizobium sp. strain MTP-10005: purification, molecular characterization, and expression. J. Bacteriol. 186:6855-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida, T., Y. Hayakawa, T. Matsui, and T. Nagasawa. 2004. Purification and characterization of 2,6-dihydroxybenzoate decarboxylase reversibly catalyzing nonoxidative decarboxylation. Arch. Microbiol. 181:391-397. [DOI] [PubMed] [Google Scholar]