Abstract
Two genes encoding transcriptional regulators involved in sulfur assimilation pathways in Burkholderia cenocepacia strain 715j have been identified and characterized functionally. Knockout mutations in each of the B. cenocepacia genes were constructed and introduced into the genome of 715j by allelic replacement. Studies on the utilization of various sulfur sources by 715j and the obtained mutants demonstrated that one of the B. cenocepacia regulators, designated CysB, is preferentially involved in the control of sulfate transport and reduction, while the other, designated SsuR, is required for aliphatic sulfonate utilization. Using transcriptional promoter-lacZ fusions and DNA-binding experiments, we identified several target promoters for positive control by CysB and/or SsuR—sbpp (preceding the sbp cysT cysW cysA ssuR cluster), cysIp (preceding the cysI cysD1 cysN cysH cysG cluster), cysD2p (preceding a separate cluster, cysD2 cysNC), and ssuDp (located upstream of the ssuDCB operon)—and we demonstrated overlapping functions of CysB and SsuR at particular promoters. We also demonstrated that the cysB gene is negatively controlled by both CysB and SsuR but the ssuR gene itself is not significantly regulated as a separate transcription unit. The function of B. cenocepacia CysB (in vivo and in vitro) appeared to be independent of the presence of acetylserine, the indispensable coinducer of the CysB regulators of Escherichia coli and Salmonella. The phylogenetic relationships among members of the “CysB family” in the γ and β subphyla are presented.
The gram-negative genus Burkholderia is a member of the β-proteobacterial subphylum and comprises nutritionally versatile, nonsporulating bacilli that inhabit diverse environments, including freshwater, soil, and plant rhizospheres. Although, Burkholderia isolates were initially identified as plant pathogens causing soft onion rot (6), potentially beneficial properties of members of the genus attracted interest as well, e.g., the ability to utilize groundwater pollutants and chlorinated aromatic compounds as nutrient sources and their antagonistic effects on the growth of soilborne plant pathogens (32, 41). Perhaps the major interest in Burkholderia spp. emerged from their pathogenic traits in humans. Many studies have documented the occurrence of Burkholderia cepacia complex (BCC) infections of the respiratory tracts of immunocompromised individuals, in particular those with cystic fibrosis (CF) and chronic granulomatous disease (see references 35, 36, and 52 for reviews). BCC isolates were formerly classified into nine closely related “genomovars” that have been recently assigned species designations (4, 54). Although essentially all BCC genomovars have been isolated from infected CF patients, Burkholderia multivorans (formerly genomovar II) and Burkholderia cenocepacia (genomovar III) predominate among the isolates, and the latter has been also associated with both increased virulence and epidemic transmission. It is possible that BCC species responsible for infection in humans are genetically indistinguishable from soil isolates (33). The high degree of adaptability of Burkholderia species to different lifestyles correlates with the large sizes of their multireplicon genomes (6 to 9 Mb), as well as the presence of a variety of insertion sequences that promote genomic plasticity (32). Complete genome sequences for Burkholderia pseudomallei and Burkholderia mallei have been published (19, 39), and those of several members of the BCC, including B. cenocepacia strain J2315 (a CF isolate, originally documented as strain CF5610) are publicly available (http://www.sanger.ac.uk/Projects/B_cenocepacia/) but await formal annotation. In the past few years, a number of studies have been devoted to assessing the significance of potential virulence factors of BCC species (35, 36). Some features connected with virulence, e.g., intrinsic resistance to multiple antibiotics, production of catalase and superoxide dismutase, unusual lipopolysaccharide structure, presence of LuxR-type quorum-sensing systems, formation of flagella, ability to form biofilms, and production of siderophores, are hallmarks of all BCC species. Genomewide analyses of BCC bacteria provide a further opportunity to recognize regulatory connections between the production of virulence factors and general anabolic pathways. Recent studies on the codependence of siderophore biosynthesis and sulfate transport by B. cenocepacia (15) prompted us to assess experimentally the regulatory circuits involved in sulfur assimilation by B. cenocepacia as a model for ubiquitous BCC species.
The assimilation of sulfur from inorganic sulfate through the cysteine biosynthetic pathway has been best studied in Salmonella enterica serovar Typhimurium and Escherichia coli (30). Over 20 genes participating in this process form a cysteine (cys) regulon, and most cys genes are coordinately controlled by the LysR-type transcriptional activator CysB. CysB is highly conserved among gram-negative bacteria examined so far, but E. coli and several other species (although not all) also possess another LysR-type regulator, designated Cbl (CysB-like) for its high (60%) similarity to CysB (22). Studies on E. coli Cbl ascribed its regulatory function to the tauABCD and ssuEADCB operons, which encode proteins involved in transport and desulfonation of the organic sulfur sources taurine and aliphatic sulfonates, respectively (58, 59). In E. coli, CysB and Cbl are encoded by loci unlinked to their target genes, and expression of cbl itself is positively controlled by CysB (22).
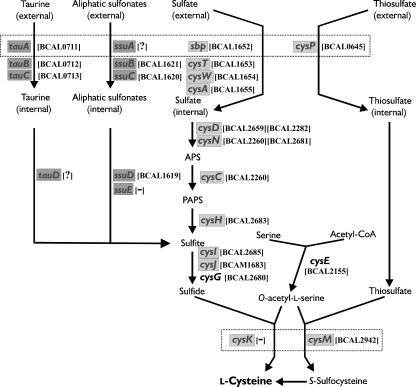
Our inspection of the genomic sequence of the B. cenocepacia J2315 strain allowed us to identify preliminarily several potential counterparts of E. coli genes participating in sulfur flux from substrates to cysteine, as shown in Fig. 1. In addition, we noticed that two open reading frames (ORFs) (BCAL1656 and BCAL2686) present in chromosome 1 are predicted to encode polypeptides exhibiting homology to either of the E. coli regulators CysB and Cbl, as well as high mutual similarity on the amino acid level.
FIG. 1.
Genes involved in the l-cysteine biosynthetic pathway in E. coli and their putative counterparts identified in B. cenocepacia. Genes of E. coli and S. enterica serovar Typhimurium involved in the sulfate/thiosulfate assimilatory pathway are shown according to the scheme of Kredich (30); those activated by CysB (as transcription units: sbp, cysPTWA, cysDNC, cysJIH, cysK, and cysM) are highlighted in light gray. Shaded in dark gray are the genes of E. coli participating in sulfonate-sulfur utilization (transcription units tauABCD and ssuEADCB) and requiring Cbl (as a direct activator) and also CysB (as an activator of the cbl gene) according to a scheme adapted from van der Ploeg et al. (57). The E. coli genes whose products display overlapping functions (Sbp/CysP and CysK/CysM) or partially overlapping functions (TauA/SsuA) are boxed. The putative counterparts of E. coli genes identified by us in the B. cenocepacia genome are indicated in brackets; “−” indicates no counterpart found, and “?” indicates that several ORFs of limited homology to the E. coli counterparts were identified by TBLASTN search. For the deduced functions of B. cenocepacia ORFs, see Discussion.
In this study, we focused on the cloning, expression in E. coli, and functional characterization of the two “cysB/cbl-like” genes of B. cenocepacia 715j. We also identify some target promoters for both gene products, and in consequence, we propose the designations SsuR and CysB for the B. cenocepacia regulators. The specific or overlapping functions of SsuR and CysB at target promoters are discussed.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The strains of E. coli and B. cenocepacia used in this study are listed in Table 1. All E. coli strains were grown at 37°C with aeration either in Luria-Bertani (LB) medium (44) or in modified M9 medium supplemented with 0.2% glucose, thiamine (0.2 mM), and tryptophan (0.2 mM). In the modified M9 medium (55), MgSO4 was replaced by an equimolar concentration of MgCl2, and sulfur sources were provided as sulfate, taurine, ethanesulfonate (sodium salts; 0.25 mM each), l-cysteine (0.1 mM), or l-djenkolic acid (1 mM). l-Djenkolic acid (S,S′-methylene-bis-cysteine) was used routinely as a “derepressing” sulfur source for E. coli and B. cenocepacia that satisfied the cellular requirement for cysteine but obviated the usual repressing effect of l-cysteine itself on the expression of sulfur metabolism-related genes. To monitor the ability of B. cenocepacia to utilize different sulfur sources, overnight cultures grown in brain-heart infusion broth (Difco) were washed once in modified M9 medium containing 0.5% glucose, and either no sulfur source or magnesium sulfate (1 mM), sodium sulfite (1 mM), sodium ethanesulfonate (0.5 mM), or cysteine (0.5 mM) was added. For performing β-galactosidase assays on B. cenocepacia strains harboring promoter-lacZ fusions, overnight cultures were washed twice in modified M9 medium containing glucose (0.5%) plus “18 amino acids” (10 μg/ml each amino acid, except cysteine and methionine), djenkolic acid, and chloramphenicol. Strains containing plasmids were grown with appropriate antibiotics: ampicillin (100 μg/ml), tetracycline (15 μg/ml), chloramphenicol (20 μg/ml for E. coli; 50 μg/ml for B. cenocepacia), kanamycin (25 μg/ml), or trimethoprim (50 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description (genotype or relevant features) | Reference or source |
|---|---|---|
| E. coli K-12 strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| CC118(λpir) | araD139 Δ(ara, leu)7697 ΔlacX74 phoA20 galE galK thi rpsE rpoB argEam recAl λpir | 18 |
| S17-1(λpir) | thi proA hsdR recA RP4-2-tet::Mu-1 kan::Tn7 integrant (Tpr Smr) λpir | 47 |
| MC4100 | araD139 Δ(lac)U169 strA thi | 8 |
| EC1250 | MC4100 trp-1 | Laboratory collection |
| EC2541 | EC1250 Δcbl::kan | 59 |
| EC2549 | EC1250 ΔcysB::kan | 34 |
| EC2625 | SE70 Δcbl::cam | Laboratory collection |
| EC2626 | EC1250 sbppBc-lacZ | This study |
| EC2627a | EC2626 cysB | This study |
| EC2628 | EC1250 cysBpBc-lacZ | This study |
| EC2629a | EC2628 cysB | This study |
| EC2630 | EC1250 cysIpBc-lacZ | This study |
| EC2631a | EC2630 cysB | This study |
| EC2635 | EC1250 ssuDpBc-lacZ | This study |
| EC2636a | EC2635 ΔcysB::kan | This study |
| EC2637 | EC1250 cysD2pBc-lacZ | This study |
| EC2638a | EC2637 cysB | This study |
| EC2655 | EC1250 ssuRpBc-lacZ | This study |
| EC2656 | EC2655 cysB | This study |
| EC2672a | EC2541 ΔcysB::cam | This study |
| EC2673b | EC2626 cysE | This study |
| EC2674b | EC2630 cysE | This study |
| EC2676a | EC2626 ΔcysB::cam | This study |
| EC2677a | EC2673 ΔcysB::cam | This study |
| EC2678a | EC2630 ΔcysB::cam | This study |
| EC2679a | EC2674 ΔcysB::cam | This study |
| SE40 | EC1250 ssuE-lacZ ΔssuADCB | 58 |
| SE45 | SE40 Δcbl::kan | 58 |
| SE47 | SE40 trpB::Tn10 cysB | 58 |
| SE70 | EC1250 tauA-lacZ ΔtauBCD | Laboratory collection |
| SP53 | lacIqrrnB ΔlacZ hsdR514 ΔaraBAD ΔrhaBAD ΔcysB::cam | 40 |
| B. cenocepacia strains | ||
| 715j | CF isolate; prototroph | 10, 15, 37 |
| 715j-ssuR::Tp | 715j with dfrB2 gene inserted in ssuR | This study |
| 715j-cysB::Tp | 715j with dfrB2 gene inserted in cysB | This study |
| Plasmids | ||
| p34E-Tp | Source of trimethoprim resistance cassette dfrB2; Tpr | 12 |
| pACYC184 | Medium-copy-number cloning vector; Cmr Tcr | 9 |
| pBBR1MCS | Broad-host-range cloning vector; Cmr | 29 |
| pGEM T Easy | Vector for direct cloning of PCR fragments; Apr | Promega |
| pKAGd4 | Broad-host-range lacZ transcription fusion vector; Apr Cmr | 2 |
| pRS551 | lacZYA+; used for construction of chromosomal lacZ fusions in E. coli; Apr Kanr | 48 |
| pSHAFT | Mobilizable pir-dependent suicide vector; Apr Cmr | 2 |
| pTrc99A | Expression vector; trc promoter; pBR322 ori; Apr | Pharmacia |
| pMH176 | cblEc ORF under trc promoter control in pTrc99A | 59 |
| pMH199 | cysBEc ORF under trc promoter control in pTrc99A | 34 |
| pMH260c | ssuRBc ORF under trc promoter control in pTrc99A; encodes SsuR with substitution F87S | This study |
| pMH262 | ssuRBc ORF (wt) under trc promoter control in pTrc99A | This study |
| pMH266 | Derivative of pMH262 with dfrB2 cassette inserted into BclI site internal to ssuR; Apr Tpr | This study |
| pMH267 | ssuRBc ORF under trc promoter control cloned in pACYC184 EcoRV-BamHI sites; Cmr | This study |
| pMH269 | Derivative of pSHAFT containing ssuRBc::dfrB2 inserted into NotI site; Apr Cmr Tpr | This study |
| pMH284 | cysBBc ORF (wt) under trc promoter control in pTrc99A | This study |
| pMH289 | Derivative of pMH284 with dfrB2 cassette inserted into NruI site internal to cysBBc; Apr Tpr | This study |
| pMH291 | Derivative of pSHAFT containing cysBBc::dfrB2 inserted into NotI site; Apr Cmr Tpr | This study |
| pMH292 | sbppBc-lacZ fusion in pRS551; Apr Kanr | This study |
| pMH293 | cysBpBc-lacZ fusion in pRS551; Apr Kanr | This study |
| pMH294 | cysIpBc-lacZ fusion in pRS551; Apr Kanr | This study |
| pMH295 | ssuRpBc-lacZ fusion in pRS551; Apr Kanr | This study |
| pMH297 | ssuDpBc-lacZ fusion in pRS551; Apr Kanr | This study |
| pMH299 | cysD2pBc-lacZ fusion in pRS551; Apr Kanr | This study |
| pMH609 | cysIpBc-lacZ fusion in pKAGd4; Apr Cmr | This study |
| pMH610 | sbppBc-lacZ fusion in pKAGd4; Apr Cmr | This study |
| pMH611 | ssuDpBc-lacZ fusion in pKAGd4; Apr Cmr | This study |
| pMH612 | cysD2pBc-lacZ in p KAGd4; Apr Cmr | This study |
| pMH637 | ssuRBc-cysBBc (tandem) under trc promoter control in pTrc99A; Apr | This study |
| pMH648 | ssuRpBc-lacZ fusion in pKAGd4; Apr Cmr | This study |
| pMH651 | cysBBc amplified with primers CB2 and CB3 and cloned in pBBR1MCS BamHI site | This study |
| pMH653 | ssuRBc amplified with primers MX3 and MX12 and cloned in pBBR1MCS BamHI site | This study |
cysB (point mutation; CysB-null phenotype), ΔcysBEc::kan, or ΔcysBEc::cam allele was P1-transduced to relevant recipient strains from SE47, EC2549, or SP53, respectively.
cysE-null mutation was P1-transduced to relevant recipient strains from EC1250 cysE Tn10 zia-207 (laboratory collection).
ssuRBc with asingle nucleotide change resulting in substitution F87S in the gene product, obtained fortuitously during PCR.
DNA manipulations and reagents.
Standard procedures (3) were used for restriction enzyme digestions, ligation, 5′-end labeling of DNA fragments, and transformation of E. coli. Plasmid DNA was isolated by using a Plasmid Midi Kit (QIAGEN). Restriction endonucleases, DNA-modifying enzymes, T4 polynucleotide kinase, and T4 DNA ligase were obtained from MBI Fermentas or Invitrogen. Taq polymerase was from MBI Fermentas, and High Fidelity PCR Master from Roche. [γ-32P]ATP used for 5′-end labeling was from Amersham Pharmacia Biotech, and all other chemicals (of the highest purity grade available) were from Sigma-Aldrich, Fluka, Promega, or Merck. Oligonucleotide synthesis and DNA sequencing (using the dideoxy chain termination method and an ABI Prism 3730 DNA sequencer [Applied Biosystems]) were performed at the Institute of Biochemistry and Biophysics, Warsaw, Poland.
Plasmid constructions.
The plasmids used in this study are listed in Table 1. All the B. cenocepacia 715j sequences used for plasmid constructions were amplified by PCR using SacII-digested total genomic DNA as a template and appropriate oligonucleotide primers listed in Table 2. Routinely, two PCR amplifications of each sequence were performed, the obtained fragments were ligated with pGEM-T-easy vector, and independent isolates of each construct were sequenced to ensure that no undesired mutations had been introduced during PCR. Inserts were recovered from pGEM-T-easy derivatives by restriction enzyme digestions, and they were subsequently cloned into appropriate vectors. The sequences of ssuR and cysB (the ORF plus the upstream intergenic region) were amplified with primers MX3/MX12 or CB2/CB3, respectively, and inserted into the BamHI site of pBBR1MCS. For expression of ssuR and cysB in E. coli, the respective ORFs were amplified with primers SR1/SR3 or CB1/CB2, and the PCR products (containing flanking BspHI and BamHI sites) were inserted into the NcoI/BamHI sites of pTrc99A. In the resultant plasmids, pMH262 and pMH284, wild-type (wt) ssuR and cysB were placed under the control of the trc promoter and an appropriately positioned Shine-Dalgarno (SD) sequence of E. coli. Plasmid pMH637, simultaneously expressing ssuR and cysB, was obtained by PCR amplification of a fragment containing the cysB ORF and SD from a pMH284 template (with primers TRCBam and CB2), digestion of the obtained product with BamHI, and ligation with the BamHI-cleaved pMH262. To obtain derivatives with interruptions of ssuR and cysB, the trimethoprim resistance (Tp) cassette (∼600-bp, containing the dfrB2 gene) was excised from plasmid p34E-Tp with either BamHI or SmaI and inserted into the BclI site of pMH262 (internal to the ssuR ORF) or the NruI site of pMH284 (internal to the cysB ORF), respectively, to give plasmids pMH266 and pMH289. The dfr-interrupted genes were amplified by PCR with primers MX10 and MX11 (for ssuR) or primers CB1 and CB2 (for cysB), and the PCR products were ligated with pGEM-T-easy. The respective sequences were recovered as NotI fragments and subsequently cloned into the NotI site of the suicide plasmid pSHAFT, giving rise to pSHAFT-ssuR::Tp and pSHAFT-cysB::Tp. The resultant plasmids, pMH269 and pMH291, were selected from trimethoprim-resistant transformants of E. coli strain CC118(λpir), and they were exploited further to obtain the respective gene knockouts in the genome of B. cenocepacia 715j by an allele replacement procedure (see below).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Regionb |
|---|---|---|
| CB1 | 5′-GGTTATTCATGAACCTGCACC-3′ | cysB |
| CB2 | 5′-CATACGGATCCGTCAAAGCTC-3′ | cysB |
| CB3 | 5′-TGTAGGATCCGTTCTGCAGA-3′ | cysB |
| CB4 | 5′-GTAGGGATCCTTGGCGGC-3′ | cysB |
| CB5 | 5′-TCGAGGATCCTCTGGTCG-3′ | cysI |
| CB6 | 5′-GGCGATCGCGGCCGGCAG-3′ | cysB |
| CD1 | 5′-CAGAATTCGGCCCAGGTCTG-3′ | cysD2 |
| CD2 | 5′-AGCGGATCCAGGTGGGTCAA-3′ | cysD2 |
| SD1 | 5′-GGAATTCGGATAGGTGCGC-3′ | ssuD |
| SD2 | 5′-AAGGATCCAGAACACATTC-3′ | ssuD |
| SB6 | 5′-GGTCATGGATCCACTGTATGG-3′ | sbp |
| SB7 | 5′-AGTCGGATCCGCCTGGGC-3′ | sbp |
| SR1 | 5′-TACTCATGAATTTTCAGC-3′ | ssuR |
| SR2 | 5′-GCACGTCGACAACGGCTTCG-3′ | ssuR |
| SR3 | 5′-CGGGATCCGGTCAAACGGCTTC-3′ | ssuR |
| SR4 | 5′-GCCGTGCACCAGCTGATCG-3′ | ssuR |
| SR5 | 5′-GTCGTCGCGACGACGAGG-3′ | ssuR |
| MX3 | 5′-CGGGATCCGGTCAAACGGCTTC-3′ | ssuR |
| MX10 | 5′-CACAGCGGCCGCAGACCATGAATTTTCAGC-3′ | pTrc99A |
| MX11 | 5′-CCTGGCGGCCGCCTCTAGAGGATCCGGTCAAA-3′ | pTrc99A |
| MX12 | 5′-AGGCGGATCCCGACCGCGAC-3′ | ssuR |
| MX13 | 5′-GTCAGGATCCTGTTCTGGCGC-5′ | ssuR |
| TRCBam | 5′-GGGATCCAGGAAACAGACCATG-3′ | pTrc99A |
| cysBfor | 5′-CGACAGCCTAAGAGGCAT-3′ | ssuR |
| cysBrev | 5′-GTGCTTCAGCCGTGATGG | ssuR |
| cysBfor2 | 5′-GCTGGATTGCTAATGACG-3′ | cysB |
| cysBrev2 | 5′-ATGCGGGAATCTCCATCT-3′ | cysB |
Nucleotides indicated in italics were changed to create restriction sites (underlined).
Regions are indicated according to the current gene predictions in B. cenocepacia (Fig. 3).
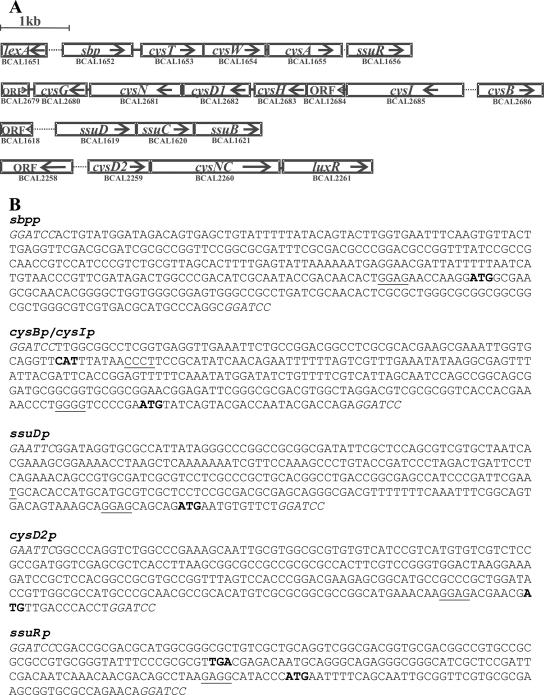
Plasmids carrying promoter-reporter (lacZ) fusions were constructed with the aid of E. coli plasmid pRS551 and with pKAGd4, which replicates in both E. coli and B. cenocepacia. The putative regulatory regions of selected B. cenocepacia genes were chosen as sequences encompassing small portions of downstream open reading frames (including the ATG start codon) and extending upstream at least 200 bp (see Fig. 4B). These sequences were amplified from 715j genomic DNA by PCR as follows: sbpp fragment with primers SB6 and SB7, the intergenic fragment between cysB and cysI with primers CB4 and CB5, the ssuDp fragment with primers SD1 and SD2, the cysD2p fragment with primers CD1 and CD2, and the ssuRp fragment with primers MX12 and MX13. Each of the fragments obtained was sequenced, cleaved with the corresponding restriction enzymes, and inserted into the BamHI or EcoRI/BamHI sites of pRS551, in front of the promoterless lacZ gene; the intergenic fragment between cysB and cysI was inserted in both orientations. The pRS551 derivatives, carrying transcriptional fusions sbpp-lacZ (pMH292), cysIp-lacZ (pMH293), cysBp-lacZ (pMH294), ssuDp-lacZ (pMH297), cysD2p-lacZ (pMH299), and ssuRp-lacZ (pMH295), were subsequently exploited to transfer the respective fusions into the chromosome of E. coli. In parallel, each of the promoter fragments described above was inserted into pKAGd4 MCS upstream of the lacZ gene. The pKAGd4 derivatives carrying the analogous fusions sbpp-lacZ (pMH610), cysIp-lacZ (pMH609), ssuDp-lacZ (pMH611), cysD2p-lacZ (pMH612), and ssuRp-lacZ (pMH648) were used as multicopy promoter-reporter systems in B. cenocepacia.
FIG. 4.
Organization of chromosomal regions encoding putative enzymes of sulfate and sulfonate assimilatory pathways in B. cenocepacia J2315. (A) The ORF numbers are shown according to current gene predictions in B. cenocepacia J2315 (http://www.sanger.ac.uk./projects/B_cenocepacia); the gene designations are proposed on the basis of homology of the predicted products to their counterparts in E. coli (or other bacteria) (see Discussion); “ORF” denotes a gene of unknown function; transcription directions are indicated by arrows. Intergenic regions indicated by dots were analyzed in this study. (B) Sequences of promoter fragments (isolated from the strain 715j) with indicated ATG start codons (boldface) and predicted SD elements (underlined); the restriction sites (introduced by primers) used for cloning in lacZ vectors are shown in italics.
Construction of single-copy B. cenocepacia promoter-lacZ fusions in E. coli.
The transcriptional fusions of B. cenocepacia promoter regions with lacZ, encoded by derivatives of pRS551, were transferred to the chromosome of E. coli strain EC1250 (Δlac recA+) by the method of Simons et al. (48) utilizing the transducing phage λRS45. The transductants, containing single-copy chromosomal fusions, were selected on LB agar containing kanamycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside), streak purified, screened for loss of ampicillin resistance, PCR verified, and then used for β-galactosidase assays.
β-Galactosidase assays.
β-Galactosidase activities were assayed by the method of Miller (38), with ONPG (o-nitrophenyl-β-galactopyranoside) as a substrate, in cells taken from mid-log-phase cultures of either E. coli or B. cenocepacia. Some assays with E. coli were performed with 4-methylumbelliferyl-β-d-galactopyranoside as a substrate, and methylumbelliferon released from the substrate was assayed fluorometrically as previously described (55). Routinely, triplicate cultures were grown for each assay, and the assays were repeated at least twice.
Construction of B. cenocepacia ssuR and cysB mutants by allelic replacement.
Plasmids pMH269 (pSHAFT-ssuR::Tp) and pMH291 (pSHAFT-cysB::Tp) were mobilized from E. coli S17-1(λpir) into B. cenocepacia 715j as previously described (11, 18), and recombinants were selected on M9-glucose agar containing Casamino Acids (0.5%), cysteine (40 μg/ml), trimethoprim (50 μg/ml), and kanamycin (50 μg/ml). Candidate double-crossover recombinants, in which vector sequences and the wild-type copy of ssuR or cysB were lost, were identified by virtue of their sensitivity to chloramphenicol (50 μg/ml). The presence of the desired genomic insertion mutation in the candidate null mutants was confirmed by performing PCRs on boiled lysates using primers cysBfor and cysBrev (for candidate ssuR::Tp knockouts) and cysBfor2 and cysBrev2 (for candidate cysB::Tp knockouts).
Protein preparations.
A procedure described earlier for partial purification of the E. coli CysB and Cbl regulators (34, 58) was followed to obtain preparations of B. cenocepacia SsuR and CysB. Briefly, E. coli strain EC2672 (ΔcysB Δcbl) was transformed with pMH262 or pMH284 (expressing the ssuR and cysB genes from the IPTG [isopropyl-thiogalactopyranoside]-inducible trc promoter), the transformed cells were grown in LB-ampicillin to early exponential phase (A600 = 0.15), IPTG was added to a final concentration of 0.1 mM, and growth was continued for a further 2 to 3 h. Cells were collected by centrifugation, resuspended in buffer A (50 mM Tris-Cl, pH 7.5, 1 mM Na2EDTA, 1 mM phenylmethylsulfonyl fluoride), and disrupted by sonication. The cellular debris was removed by centrifugation, and clear extracts were fractionated by ammonium sulfate precipitation. The protein fractions precipitated with 229 mg/ml of ammonium sulfate were collected, suspended in buffer A at a total protein concentration of 1 to 2 mg/ml, and stored in aliquots at −70°C. The amounts of both B. cenocepacia proteins in the preparations obtained were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining to be not less than 50% of the total protein. Protein extracts prepared in an analogous way from the same host cells but transformed with an empty vector (pTrc99A) served as a control in DNA-binding studies utilizing partially purified B. cenocepacia proteins.
DNA-binding assays.
The abilities of B. cenocepacia proteins to bind DNA at putative target promoter regions were tested by the electrophoretic mobility shift assay (EMSA) (16). DNA fragments containing promoter regions of interest were the same as those exploited to construct the respective promoter-reporter fusions. PCR-amplified promoter fragments were labeled at the 5′ ends with [γ-32P]ATP and polynucleotide kinase. Reaction mixtures (20 μl) contained approximately 10 ng of labeled DNA fragment and 2 μg of sonicated calf thymus DNA per ml (as a nonspecific competitor) in a buffer consisting of 40 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 100 mM KCl, 1 mM dithiothreitol, and 100 μg bovine serum albumin per ml. The protein preparations were added at various final concentrations (typically 1 to 20 μg/ml), and the reaction mixtures were incubated at 37°C for 5 min. Some samples also contained 10 mM O-acetyl-l-serine (OAS), which was tested as a potential cofactor of the B. cenocepacia regulatory proteins. After incubation, the reaction mixtures were separated in a 5% acrylamide-bisacrylamide (82:1) nondenaturing gel in 0.05 M Tris-borate-EDTA buffer (pH 8.3) for 1.5 h at 10 V/cm. Radiolabeled bands were visualized by autoradiography.
Sequence analysis.
For phylogenetic analysis, the amino acid sequences of ORFs similar to CysB and Cbl were retrieved from the GenBank Sequence Database. The sequences were aligned with the Clustal W program (http://www.ebi.ac.uk/clustalw/index.html) (53) using the default parameters. The alignments were edited with the BioEdit software package (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). A phylogenetic tree was constructed by the neighbor-joining and minimal evolution methods implemented in MEGA 3.1 (http://www.megasoftware.net) (31), using distance matrix calculations for all pairs from the sequence alignments and the p distance and Poisson correction substitution models. The bootstrap values for confidence limits of branch points were estimated from 10,000 replicates.
RESULTS
Two genes in chromosome 1 of the B. cenocepacia genome encode proteins similar to the E. coli CysB and Cbl regulators.
Examination of the genomic sequence of B. cenocepacia J2315 (http://www.sanger.ac.uk/Projects/B_cenocepacia/) (not formally annotated) revealed that chromosome 1 contains two open reading frames, BCAL1656 and BCAL2686, whose deduced products share homology with both E. coli CysB and Cbl transcriptional regulators. It has been previously reported that one of “cysB/cbl-like” sequences in B. cenocepacia strain 715j is adjacent to the locus encoding a putative sulfate transporter (homologs of E. coli Sbp, CysT, CysW, and CysA), and it was tentatively designated “cysB” (15). The second “cysB-like” gene, BCAL2686, present in the J2315 genome, is oriented divergently to a cluster of genes encoding putative enzymes of sulfate activation and reduction (homologs of E. coli CysI, CysH, CysD, CysN, and CysG, as inferred from TBLASTN analysis) (Fig. 1) (see Discussion). After inspection of available genomic sequences of other members of the genus Burkholderia, B. pseudomallei and B. mallei, we noticed the presence of potential orthologs of BCAL1656 and BCAL2686 in genomic contexts analogous to those present in B. cenocepacia J2315.
Cloning of BCAL1656 and BCAL2686 from B. cenocepacia and complementation studies in E. coli.
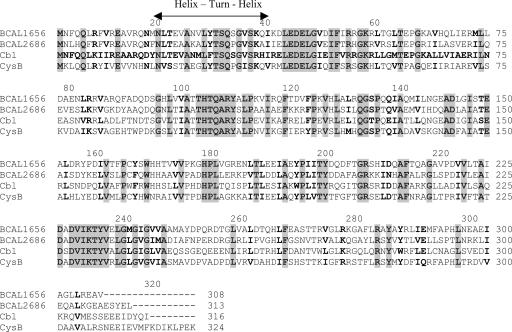
The two “cysB-like” ORFs of B. cenocepacia were amplified from genomic DNA of strain 715j and sequenced, which revealed 100% conservation of deduced products between strains 715j and J2315. As can be seen in Fig. 2, the mutual similarity of these products is higher than that of E. coli CysB and Cbl, and their functional analogy to E. coli regulators cannot be simply deduced. The BCAL1656 and BCAL2686 sequences were cloned in the vector pTrc99A under the control of the trc promoter and an E. coli Shine-Dalgarno sequence to give plasmids pMH262 and pMH284, respectively. Both gene products could be effectively overproduced from these plasmids in E. coli strain DH5α, as well as in a ΔcysB Δcbl derivative of strain MC4100 (EC2672). The latter strain was exploited to obtain cellular extracts enriched with each of the B. cenocepacia proteins, which were subsequently used in DNA-binding experiments.
FIG. 2.
Amino acid sequence alignment of the E. coli CysB and Cbl proteins and deduced products of B. cenocepacia BCAL1656 and BCAL2686, designated ssuR and cysB, respectively. Identical amino acid residues at the same relative position in all four proteins are shaded, and similar residues are shown in boldface. The helix-turn-helix motif typical of LysR family members (45) is shown by arrows.
To investigate whether any of these proteins replace the functions of the E. coli CysB and Cbl regulators in vivo, we tested the abilities of plasmids pMH262 and pMH284 to complement the growth of E. coli cysB and cbl mutants in the presence of selected sulfur sources. The E. coli cysB-null mutant requires cysteine for growth and is unable to utilize sulfate as a sulfur source (due to a lack of expression of nearly all the cys genes involved in the sulfate assimilatory pathway); this mutant is also unable to utilize taurine and aliphatic sulfonates, since sulfite released from these substrates must be further metabolized by the cys pathway, which is dependent on genes controlled by CysB (Fig. 1). In contrast, the E. coli cbl-null mutant grows well on sulfate but does not utilize taurine and aliphatic sulfonates, since expression from the tau and ssu promoters is strictly dependent on Cbl. Since expression of E. coli cbl itself is dependent on CysB (22), the cysB-null mutant is practically devoid of both CysB and Cbl functions. We found that neither of the B. cenocepacia genes could complement the cysteine deficiency of the E. coli cysB-null mutant. In contrast, utilization of ethanesulfonate (but not taurine) by the E. coli cbl-null strain SE45 was supported in the presence of pMH262 containing BCAL1656. To examine whether the partial cross-activity of the BCAL1656 product in E. coli correlates specifically with expression of the ssuEADCB operon, we assayed the β-galactosidase levels in strains with chromosomal ssuE-lacZ and tauA-lacZ fusions in the presence of plasmid pMH262. The results (Table 3) confirmed that the BCAL1656 product is able to activate expression from the E. coli ssuE promoter (although to a lesser extent than the cognate Cbl regulator), but it is inactive in upregulation of the E. coli tauA promoter (consistent with the inability of strain Δcbl/pMH262 to utilize taurine as a sole sulfur source).
TABLE 3.
Effects of BCAL1656 product on the activities of the E. coli ssu and tau promoters
| Straina | Plasmidb | β-Galactosidase activity (Miller units)c |
|---|---|---|
| SE40 (ssuE-lacZ) | None | 888 ± 84 |
| SE45 (ssuE-lacZ Δcbl) | None | 28 ± 2 |
| SE45 | pMH176 (cblEc) | 2,676 ± 252 |
| SE45 | pMH260 (BCAL1656*) | 139 ± 13 |
| SE45 | pMH262 (BCAL1656) | 1,139 ± 18 |
| SE70 (tauA-lacZ) | None | 1,037 ± 108 |
| EC2625 (tauA-lacZ Δcbl) | None | 34 ± 4 |
| EC2625 | pMH176 | 958 ± 78 |
| EC2625 | pMH260 | 70 ± 5 |
| EC2625 | pMH262 | 55 ± 7 |
E. coli strains harboring single-copy chromosomal ssuE-lacZ or tauA-lacZ fusions.
Plasmids were derivatives of pTrc99A; BCAL1656* contained on pMH260 encodes a mutant protein with a single amino acid substitution, F87S.
β-Galactosidase was assayed in cells from mid-log-phase cultures grown in minimal medium with l-djenkolic acid (1 mM) as a sulfur source. Triplicate assays were performed for each culture of at least three independent transformants.
The above-mentioned results provided a first indication that the BCAL1656 product may be a functional analogue of the E. coli Cbl regulator rather than the CysB regulator. Since the designation “cbl” in Burkholderia species has already been exploited for genes involved in the formation of cell surface structures termed “cable pili” (43), we propose to designate BCAL1656 “ssuR.” For BCAL2686, we propose the designation “cysB” on the basis of further characterization of its function in B. cenocepacia (see below). These designations are used throughout the following text and figures.
Construction and phenotypes of cysB and ssuR mutants of B. cenocepacia.
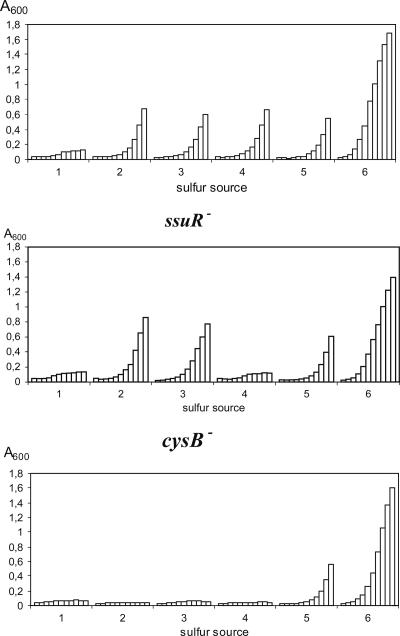
The B. cenocepacia mutants devoid of CysB or SsuR function were constructed by an allele replacement technique using plasmids carrying corresponding ORFs interrupted with a Tp cassette. The growth of both mutant strains 715j-ssuR::Tp and 715j-cysB::Tp on various sulfur sources was compared with that of the parental strain 715j. As shown in Fig. 3, the cysB mutant was not able to utilize any of the tested potential substrates (sulfate, sulfite, and ethanesulfonate) for cysteine biosynthesis, and hence, its phenotype is analogous to that of an E. coli cysB mutant. In contrast, the ssuR mutant was able grow well with sulfate but was unable to utilize ethanesulfonate as a sulfur source, a hallmark phenotype of an E. coli cbl mutant. The growth of the cysB mutant on sulfate and ethanesulfonate was restored to the wild-type level in the presence of plasmid pMH651 carrying wt B. cenocepacia cysB (cysBBc), and growth of the ssuR mutant on ethanesulfonate was restored by plasmid pMH653, carrying wt ssuRBc (not shown). It has also been noted that l-cysteine as a sole sulfur source does not support full growth of strain 715j even over a 24-h period. As shown in Fig. 3, supplementation with 18 amino acids significantly increased growth with l-cysteine. This effect is consistent with earlier observations in E. coli of transient amino acid starvation (especially for threonine and branched-chain amino acids) caused by cysteine excess (17, 50, 51).
FIG. 3.
Growth of B. cenocepacia 715j and isogenic ssuR and cysB mutants in the presence of various sulfur sources. Growth was monitored by A600 measurements over 11 h after inoculation of cells taken from LB cultures and washed in modified (i.e., sulfate-free) M9 minimal medium supplemented with (1) no sulfur source, (2) sulfate (1 mM), (3) sulfite (1 mM), (4) ethanesulfonate (0.5 mM), (5) l-cysteine (0.5 mM), or (vi) l-cysteine (0.5 mM) plus 18 amino acids.
Identification of B. cenocepacia genes controlled by SsuR and CysB.
Following the TBLASTN analysis of the genomic sequence of B. cenocepacia J2315 for the presence and organization of ORFs encoding putative enzymes of the sulfate and sulfonate assimilatory pathways (Fig. 1) (see Discussion), we chose four regions, shown in Fig. 4, for further analysis. We noted that the organizations of these regions are similar (but not strictly identical) to that found in genomic sequences of other Burkholderia spp. published to date. In all of these genomic sequences, ORFs encoding the same pair of potential regulators, here referred to as ssuR and cysB, are clustered with the “cysA locus” and the “cysI locus,” respectively.
To investigate whether the intergenic regions shown in Fig. 4A contain target sequences for CysB or/and SsuR, appropriate “promoter fragments” (the sequences are shown in Fig. 4B) were amplified from genomic DNA of B. cenocepacia 715j and used to construct transcriptional fusions with the promoterless lacZ gene contained on plasmid pKAGd4. Plasmids carrying promoter-reporter fusions were introduced into the B. cenocepacia wt strain 715j and its derivative cysB-null or ssuR-null. Table 4 shows the results of β-galactosidase assays, reflecting transcription initiating from within the five intergenic regions (cloned in pKAGd4) in the three B. cenocepacia genetic backgrounds. High promoter activity above the background level (afforded by pKAGd4) was detected in sbpp-, cysIp-, and ssuDp-lacZ fusions in wt 715j. In the absence of functional cysB, expression from cysIp was moderately decreased (2.3-fold), while the absence of functional ssuR strongly affected expression from ssuDp (170-fold) and also that from sbpp, albeit less severely (2.4-fold). It was noted that β-galactosidase activity measured with a plasmid containing ssuDp-lacZ in an ssuR-null strain was lower than background levels afforded by pKAGd4, which might suggest the presence of a strong transcription termination signal residing in a cloned ssuDp fragment upstream of the SsuR-dependent promoter. The expression levels of lacZ from cysD2p and ssuRp were lower by an order of magnitude than those of other fusions, indicating lower promoter activities, but in both cases, some negative effect of either a cysB or ssuR knockout was detectable.
TABLE 4.
Effects of mutant cysB and ssuR alleles on the activities of promoters of sulfur metabolism genes in B. cenocepacia
| Plasmid (fusion) | β-Galactosidase activity (Miller units)a in B. cenocepacia 715j strain:
|
||
|---|---|---|---|
| Wild type | cysB-null | ssuR-null | |
| pKAGd4 (none) | 136 ± 5 | 157 ± 16 | 126 ± 4 |
| pMH610 (sbpp-lacZ) | 13,462 ± 1,159 | 15,906 ± 450 | 5,731 ± 423 |
| pMH609 (cysIp-lacZ) | 4,509 ± 351 | 2,003 ± 198 | 4,451 ± 196 |
| pMH611 (ssuDp-lacZ) | 5,726 ± 530 | 5,390 ± 1,056 | 31 ± 1 |
| pMH612 (cysD2p-lacZ) | 568 ± 25 | 433 ± 18 | 258 ± 10 |
| pMH648 (ssuRp-lacZ) | 456 ± 12 | 336 ± 13 | 292 ± 11 |
β-Galactosidase was assayed in cells from mid-log-phase cultures grown M9 with 18 amino acids (minus cysteine and methionine) and l-djenkolic acid (1 mM) as a sulfur source. Triplicate cultures were grown for each transformant, and each culture was assayed in duplicate for enzyme activity.
Suspecting that the roles of B. cenocepacia CysB and SsuR at particular targets may partially overlap, we decided to test the individual contribution of each regulator to expression from promoter regions of interest in a heterologous system, using E. coli as a host. Transcriptional fusions of the respective B. cenocepacia promoter regions (the same as those present in the pKAGd4 derivatives) with promoterless lacZ were constructed in the vector pRS551, and they were subsequently recombined into the E. coli chromosome. To avoid possible cross talk between B. cenocepacia promoters and resident E. coli regulators (CysBEc and CblEc), all of the strains obtained with chromosomal B. cenocepacia promoter-lacZ fusions were made cysBEc null. As the expression of cblEc is under the control of CysBEc, this background indicated the absence of both CysB and Cbl activities. β-Galactosidase activity was assayed in the resultant strains in the absence and in the presence of B. cenocepacia CysB and SsuR expressed from plasmids (Table 5). Consistent with the results obtained with the B. cenocepacia host strains (compare Table 4), the ssuDp-lacZ fusion was highly and specifically upregulated by SsuR in E. coli. Expression of two other fusions, sbpp-lacZ and cysIp-lacZ, was upregulated by SsuR and CysB; however, cysIp was much more sensitive to the stimulatory activity of CysB, whereas sbpp appeared more sensitive to that of SsuR. The activity of β-galactosidase in E. coli harboring the cysD2p-lacZ fusion was barely detectable in the presence of SsuR or CysB in a standard Miller assay, but a more sensitive method revealed elevation of cysD2p activity when B. cenocepacia SsuR and CysB were delivered jointly (expressed from two plasmids present in the same cell). Using the same system, we observed that expression of lacZ from cysBpBc was negatively affected by the presence of either SsuR (3.8-fold) or CysBBc (2.8-fold), while expression of lacZ from “ssuRp” (taken as a cysA-ssuR intergenic region) was barely detectable.
TABLE 5.
Effect of SsuR and CysB on the activity of B. cenocepacia promoters of sulfur metabolism genes measured in a heterologous system (E. coli)
| E. coli strain | Chromosomal fusion (B. cenocepacia promoter-lacZ) | Plasmidb (relevant B. cenocepacia gene) | β-Galactosidase activitya
|
|
|---|---|---|---|---|
| Miller unitsc | MUF unitsd | |||
| EC2627 | sbpp-lacZ | pTrc99A | 12 ± 2 | ND |
| EC2627 | sbpp-lacZ | pMH262 (ssuR) | 7,600 ± 491 | ND |
| EC2627 | sbpp-lacZ | pMH284 (cysB) | 4,560 ± 366 | ND |
| EC2631 | cysIp-lacZ | pTrc99A | 42 ± 2 | ND |
| EC2631 | cysIp-lacZ | pMH262 (ssuR) | 950 ± 99 | ND |
| EC2631 | cysIp-lacZ | pMH284 (cysB) | 4,370 ± 260 | ND |
| EC2638 | cysD2p-lacZ | pTrc99A | ND | 33 ± 5 |
| EC2638 | cysD2p-lacZ | pMH262 (ssuR) | ND | 61 ± 7 |
| EC2638 | cysD2p-lacZ | pMH284 (cysB) | ND | 68 ± 6 |
| EC2638 | cysD2p-lacZ | pMH267 (ssuR) | ND | 61 ± 7 |
| EC2638 | cysD2p-lacZ | pMH284 + pMH267 | ND | 257 ± 20 |
| EC2636 | ssuDp-lacZ | pTrc99A | 10 ± 1 | ND |
| EC2636 | ssuDp-lacZ | pMH262 (ssuR) | 3,480 ± 360 | ND |
| EC2636 | ssuDp-lacZ | pMH284 (cysB) | 40 ± 7 | ND |
| EC2629 | cysBp-lacZ | pTrc99A | 116 ± 7 | ND |
| EC2629 | cysBp-lacZ | pMH262 (ssuR) | 31 ± 3 | ND |
| EC2629 | cysBp-lacZ | pMH284 (cysB) | 42 ± 2 | ND |
| EC2656 | ssuRp-lacZ | pTrc99A | ND | 37 ± 3 |
| EC2656 | ssuRp-lacZ | pMH262 (ssuR) | ND | 65 ± 2 |
| EC2656 | ssuRp-lacZ | pMH284 (cysB) | ND | 60 ± 2 |
In all EC strains, the wild-type cysBEc was replaced with a cysB-null allele from SE47. ND, not determined.
Plasmids were derivatives of pTrc99A (Apr), with the exception of pMH267(Cmr), which contains ssuR under the trc promoter in pACYC184.
Assays were performed under the same conditions as indicated in Table 4.
MUF (4-methylumbelliferone) units are shown as pmol MUF/ml/min/A600 released from 4-methylumbelliferyl-β-d-galactopyranoside. In our hands, 100 MUF units correspond approximately to 1 Miller unit of β-galactosidase activity.
Binding of SsuR and CysB proteins to DNA at target promoter regions.
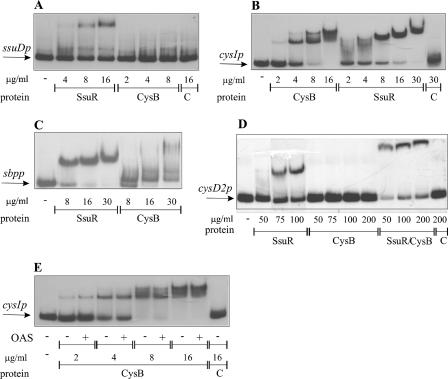
The promoter regions of B. cenocepacia identified as targets for CysB- and SsuR-mediated regulation in vivo were subsequently tested by EMSA for the ability to bind each regulator in vitro. The DNA fragments used as promoter probes (ssuDp, cysIp/cysBp, sbpp, and cysD2p) (Fig. 3B) encompassed the same DNA regions as those used to construct the respective promoter-lacZ fusions. As a source of B. cenocepacia regulators, we used protein extracts of a ΔcysB Δcbl E. coli strain enriched with either CysBBc or SsuRBc (following their overproduction from the corresponding plasmids, pMH262 and pMH284). As shown in Fig. 5A, the ssuDp promoter fragment was able to bind SsuR only, giving a single shifted band. Further increase of the SsuR concentration (data not shown) did not result in the appearance of additional bands, suggesting that a single binding site for this regulator may exist in the ssuDp region. The intergenic region separating cysI and cysB (cysIp/cysBp) (Fig. 5B) gave rise to shifted bands with both SsuR and CysB. It is also evident that CysB was able to produce more than one distinct complex with the cysIp/cysBp probe, suggesting the presence of at least two target sequences (binding sites) for CysB in this region. SsuR produced only one shifted band with cysIp/cysBp, but the electrophoretic mobility of the DNA-SsuR complex decreased with the protein concentration; this effect might also suggest some changes in the DNA-binding stoichiometry. The sbpp probe was up-shifted by both SsuR and CysB (Fig. 5C); however, apparently weaker and diffuse shifted bands observed with CysB might reflect lower affinity of CysB for the target site(s) in the sbpp region than that of SsuR.
FIG. 5.
Binding of SsuR and CysB to DNA at target promoter regions. EMSA was performed with radiolabeled B. cenocepacia promoter probes (PCR-amplified fragments of 715j DNA; sequences are shown in Fig. 4B). (A) ssuDp. (B and E) cysBp/cysIp. (C) sbpp. (D) cysD2p. Protein extracts enriched with SsuR, CysB, or control extract (in panel C, prepared from cells containing vector pTrc99A) were added to the final protein concentrations as indicated (in μg/ml). OAS (10 mM) was included where indicated by + in the reactions shown in panel E. Free probes (unbound DNA) are indicated by arrows.
Using the cysD2 promoter region as a probe, a shifted band was detected after incubation with SsuR, but not with a CysB preparation (Fig. 5D). The same result (a single shifted band corresponding to the presumed SsuR-DNA complex) was obtained when both SsuR and CysB (individually overproduced from plasmids pMH262 and pMH284, respectively) were added together to the reaction mixture (data not shown). However, a distinct retarded band (a very slowly migrating complex) was clearly seen after incubation of the cysD2p probe with protein extract containing SsuR and CysB overproduced simultaneously in the same cell (from plasmid pMH637; trcp-ssuR-cysB). This suggests that cysD2p may be regulated by some cooperation of SsuR-CysB. No complex of either CysB or SsuR was detectable by EMSA using the intergenic region cysA-ssuR (ssuRp) as a probe (not shown).
A hallmark feature of the CysB proteins from E. coli and S. enterica serovar Typhimurium (CysBSt) is either qualitative or quantitative modulation of their DNA-binding mode in the presence of a cognate inducer, acetylserine (30, 34). Using the B. cenocepacia promoter probes sbpp and cysD2p (data not shown) and cysIp/cysBp (Fig. 5E), we found no effect of O-acetylserine on either the abundance or the relative mobilities of complexes formed by SsuR and/or CysBBc. The possibility that CysBBc may function without a coinducer (in contrast to CysBEc and CysBSt) was further tested in vivo (see below).
The function of CysBBc in vivo is independent of acetylserine.
In S. enterica serovar Typhimurium and E. coli, OAS is synthesized in the reaction catalyzed by serine acetyltransferase (the product of the cysE gene) and serves as an acceptor of the sulfide moiety in the final reaction of cysteine biosynthesis. In addition, OAS, and also the product of its nonenzymatic conversion, N-acetyl-l-serine, serve as coinducers of CysB-activated transcription in both these species (30, 34). Thus, the phenotypes of cysB and cysE null mutants are very similar in terms of the lack of expression from CysB-dependent promoters. In order to test if CysBBc-mediated activation of B. cenocepacia promoters requires OAS as an inducer, expression of chromosomal sbppBc-lacZ and cysIpBc-lacZ fusions was measured in E. coli strains cysBEc-null and cysEEc-null in the presence of CysBBc or CysBEc expressed from corresponding plasmids (Table 6). The CysBBc-activated expression from sbppBc and cysIpBc was maintained at the same level, irrespective of the presence of CysE (an enzyme producing OAS). In addition, expression of the sbppBc-lacZ and cysIpBc-lacZ fusions was unchanged by growth of the corresponding strains (EC2676/pMH284 and EC2678/pMH284) with cysteine, which acts as an inhibitor of serine acetyltransferase (CysE) activity (data not shown). It can be noted that a low level of expression from sbppBc was observed in the presence of CysBEc, which may reflect a regulatory “cross talk” between the B. cenocepacia target and the E. coli regulator. Significantly, upregulation of sbppBc by CysBEc was entirely abolished in the absence of functional cysE. These pieces of evidence strongly suggest that B. cenocepacia CysB may not require acetylserine as a coinducer to act as a transcriptional activator.
TABLE 6.
Expression of lacZ from B. cenocepacia promoters in E. coli in the presence/absence of functional serine transacetylase (CysE)
| Strain | Fusion | Background | Plasmid (relevant gene) | β-Galactosidase activity (Miller units) |
|---|---|---|---|---|
| EC2676 | sbppBc-lacZ | ΔcysB | pMH289 (cysBBc::Tp)b | <5 |
| EC2676 | sbppBc-lacZ | ΔcysB | pMH284 (cysBBc) | 4,769 ± 325 |
| EC2677 | sbppBc-lacZ | ΔcysB ΔcysE | pMH284 (cysBBc) | 4,361 ± 134 |
| EC2676 | sbppBc-lacZ | ΔcysB | pMH199 (cysBEc)c | 152 ± 3 |
| EC2677 | sbppBc-lacZ | ΔcysB ΔcysE | pMH199 (cysBEc) | <5 |
| EC2678 | cysIpBc-lacZ | ΔcysB | pMH289 (cysBBc::Tp) | <5 |
| EC2678 | cysIpBc-lacZ | ΔcysB | pMH284 (cysBBc) | 2,860 ± 260 |
| EC2679 | cysIpBc-lacZ | ΔcysB ΔcysE | pMH284 (cysBBc) | 2,727 ± 209 |
| EC2678 | cysIpBc-lacZ | ΔcysB | pMH199 (cysBEc) | <5 |
| EC2679 | cysIpBc-lacZ | ΔcysB ΔcysE | pMH199 (cysBEc) | <5 |
Assays were performed under the same conditions as indicated in Table 5.
Plasmid carrying cysBBc disrupted by a dfrB2 cassette.
Plasmid encoding wt E. coli CysB.
DISCUSSION
Relatively little is known about sulfur assimilation pathways and their regulation in ubiquitous bacteria belonging to the genus Burkholderia, and the presence of relevant traits can be only tentatively predicted from analysis of available genomic sequences. As a starting point to assess experimentally the selectivity of sulfur source utilization by these bacteria, we focused here on transcriptional regulation of some “signature” genes for sulfur metabolism in B. cenocepacia 715j. The TBLASTN search of the B. cenocepacia J2315 genome with the E. coli proteins involved in sulfur flux from external substrates to l-cysteine (Fig. 1) as queries allowed us to recognize their candidate counterparts in B. cenocepacia. Notably, no putative ortholog of the E. coli major O-acetylserine sulfhydrylase A (CysK) involved in the final reaction of cysteine biosynthesis has been found in B. cenocepacia. Perhaps this reaction relies solely on the O-acetylserine sulfhydrylase B-like enzyme (CysM), the product of BCAL2942, sharing 60% identity and 73% similarity with E. coli CysM.
SsuRBc is a specific activator of genes involved in aliphatic sulfonate transport and desulfonation.
The organization of a gene cluster of B. cenocepacia designated by us ssuD ssuC ssuB (Fig. 4A) is essentially identical to that found in other Burkholderia genomes annotated to date (19, 39). The translated gene products are likely to constitute two components of the transporter for aliphatic sulfonates (SsuB and SsuC) and an FMNH2-dependent monooxygenase-type enzyme (SsuD) involved in desulfonation of substrates. The components of homologous bacterial systems devoted to alkanesulfonate transport and desulfonation (Fig. 1 shows those in E. coli) typically include the periplasmic substrate-binding protein (SsuA), in addition to SsuB and SsuC, and the NAD(P)H-dependent flavin mononucleotide reductase (SsuE) acting in complex with SsuD; all of these proteins are usually encoded within a single operon (13, 14, 23, 24, 25, 56, 58). In B. cenocepacia, the ssu gene cluster does not include an ssuE-like ORF, as in B. subtilis (56), and the SsuE ortholog is apparently not encoded by any other genomic locus. Possibly, the SsuD monooxygenase in Burkholderia may interact with a different type of flavin reductase, as suggested for B. subtilis (24). Our search for a gene encoding a potential counterpart of SsuA revealed the presence of several ORFs (e.g., BCAL1552, BCAM1118, and BCAS0769) sharing limited similarities (44 to 59% over the parts of the deduced protein sequences) with SsuA from E. coli or Pseudomonas putida. The genomic contexts of these ORFs do not encourage speculation on their relevance to sulfonate-assimilation processes, and the regulation of their expression remains to be investigated. However, we have shown here that expression of contiguous ssuDCB genes (presumably an operon), controlled by a promoter preceding ssuD, is strictly dependent on the SsuR regulator (the product of BCAL1656) in B. cenocepacia. In E. coli, the ssuEADCB operon is under the positive control of the Cbl regulator (58), and some cross-activation of the ssuEEc promoter by SsuR was demonstrated in our study. B. cenocepacia also possesses the putative orthologs of the tauABCD genes (Fig. 1), whose expression in E. coli is strictly dependent on Cbl (7, 59). However, we were not able to detect regulation of “tauApBc” by SsuRBc (data not shown), and no upregulation of tauApEc by SsuRBc was detected in our assays. Nevertheless, we believe that SsuRBc is most likely an ortholog of E. coli Cbl. It should be stressed, however, that SsuRBc is not an ortholog of the recently described “SsuR” activator of ssu genes in Corynebacterium japonicum (27); the latter belongs to the ROK family of proteins, and it was discussed as a functional (but not structural) counterpart of E. coli Cbl.
SsuRBc and CysBBc control the steps of sulfate transport, activation, and reduction.
The region encoding the “sulfate transporter” of B. cenocepacia 715j has been identified (15) as a cluster, sbp cysT cysW cysA, where Sbp is an ortholog of the periplasmic sulfate-binding protein and CysT, CysW, and CysA represent orthologs of the ABC-type sulfate transporter components of several bacteria (25). The TBLASTN analysis revealed that in B. cenocepacia, B. mallei, and B. pseudomallei, the product of the first ORF in the locus is more similar to Sbp than to CysP of E. coli (despite overall similarities between the proteins). In all these species, cysA is followed by a gene encoding a transcriptional regulator, designated “cysB” in a previous study on B. cenocepacia (15) and here renamed “ssuR.” E. coli and Salmonella possess the sulfate/thiosulfate transporter locus organized as the cysPTWA operon, transcribed from the cysP promoter, where CysP is a thiosulfate-binding protein (20) and Sbp is encoded by an unlinked gene. We demonstrated that the B. cenocepacia region preceding the sbp gene contains a functional promoter whose full activity requires SsuR in vivo. However, since both SsuR and CysB were active in upregulation of sbpp in a heterologous system and complexes of sbpp probe with both these proteins were detectable in vitro by EMSA, the functions of SsuR and CysB at sbpp can apparently replace each other.
It seems that the ssuR gene of B. cenocepacia, which is clustered with sbp cysTWA and separated by 75 bp from the cysA ORF, may be expressed at a low level as a separate transcription unit. Our probing of the promoter activity within the cysA-ssuR region (as a plasmid-encoded ssuRp-lacZ fusion including 172 bp upstream of the ssuR start codon, ATG) revealed only a weak decrease in ssuRp-lacZ expression in B. cenocepacia mutants devoid of CysB or SsuR function. Since we have not detected binding of CysB and SsuR in this “ssuRp region,” regulation of the ssuR gene itself may be not of physiological relevance. This is in contrast to the E. coli paradigm, where expression of the cbl gene from its own promoter is strongly activated by CysB (22). At the moment, we can speculate that in Burkholderia the expression of ssuR is regulated in parallel with either the sbp cysTWA transcription unit (from sbpp) or the cysTWA transcription unit, if the latter is preceded by a functional promoter.
Apart from the sbp cysTWA ssuR locus, two other gene clusters of B. cenocepacia have been recognized by homology searches and designated here as loci devoted to sulfate metabolism: the cluster cysI cysH orf cysD1 cysN cysG and the separate cluster cysD2 cysNC (Fig. 4). The proteins encoded in the former locus, CysD1 (a putative ATP sulfurylase) and CysN (a putative GTPase coupling GTP hydrolysis to the sulfurylation of ATP) are likely to be associated with sulfate “activation” to the adenosine 5′-phosphosulfate (APS), while CysH, CysI, and CysG are likely to participate in subsequent reduction steps of S6+ to S2−. Notably, a gene encoding an ortholog of CysC (an APS kinase producing 3′ phosphoadenosine 5′ phosphosulfate from APS) is absent from this gene cluster. However, there is an ORF encoding a putative ortholog of bacterial “CysH”-type reductases that utilizes APS as a substrate (1, 5, 26, 28). The cluster also encodes a putative sulfite reductase component (CysI), which is similar (43%) to the hemoprotein subunit of E. coli sulfite reductase and 78% similar to that of P. aeruginosa (21), and a putative ortholog of uroporphyrinogen III methylase (CysG), an enzyme involved in synthesis of the siroheme cofactor of sulfite reductase (60). The arrangement of ORFs in the cysI-cysG locus of B. cenocepacia is identical to that in two other Burkholderia genomes (19, 39), including the presence of an ORF of unknown function located between cysI and cysH and an ORF encoding a CysG-type protein (annotated as cobA in the B. mallei and B. psedomallei genomes). The deduced functions of proteins encoded in this gene cluster are consistent with a view that in Burkholderia they may be sufficient to perform conversion of inorganic sulfate to sulfide.
The gene designated by us cysB (BCAL2686) is oriented divergently from cysI, and its location is conserved in the B. mallei and B. pseudomallei genomes. We demonstrated that the intergenic region between cysI and cysB contains targets for binding of both SsuR and CysB and that expression from cysIp is elevated by both CysB and SsuR (albeit to a lesser extent by the latter). This suggests that these regulators may replace each other in positive control of cysIp, similarly to sbpp. An analogous interpretation of overlapping functions of B. cenocepacia regulators may be applied to the observed negative control of the divergently oriented cysB gene by either CysB or SsuR.
In addition to cysD1 cysN genes contained within the cysI-cysG cluster, the B. cenocepacia genome contains another “sulfate activation locus,” cysD2 cysNC (Fig. 4). The translated product, CysD2, shares 44% identity and 66% similarity with CysD1, and it is also similar (∼70%) to the NodP-type proteins of Rhizobiaceae. The product of cysNC is likely to be a fusion protein with ATP sulfurylase and APS kinase activities, as judged by comparisons of the CysNCBc and CysNC proteins of P. aeruginosa and Mycobacterium tuberculosis (42) and the NodQ-type proteins of Rhizobiaceae (49) and RaxQ of Xanthomonas oryzae (46). It seems, therefore, that Burkholderia species, like Rhizobiaceae and Mycobacteriaceae but unlike Enterobacteriaceae, can channel the intracellular sulfate either to the reductive pathway (functions encoded in the cluster cysI cysH orf cysD1 cysN cysG) or to sulfatation processes that require phosphoadenosine 5′ phosphosulfate as a sulfate donor (the latter being produced by the sulfate-activating complex CysD2/CysNC). In our assays, activity of the cysD2p promoter appeared weak (compared with those of other promoters tested), but it showed measurable upregulation by SsuR and CysB acting in concert in vivo. Also, an EMSA using a cysD2p probe detected a high-order complex with protein extract containing SsuR and CysB overproduced jointly. Although elucidation of the nature of this “supercomplex” requires further studies, we think that cysD2p is regulated by some cooperation of SsuR-CysB, possibly via formation of mixed hetero-oligomers.
Phylogeny of the “CysB family” of transcriptional regulators.
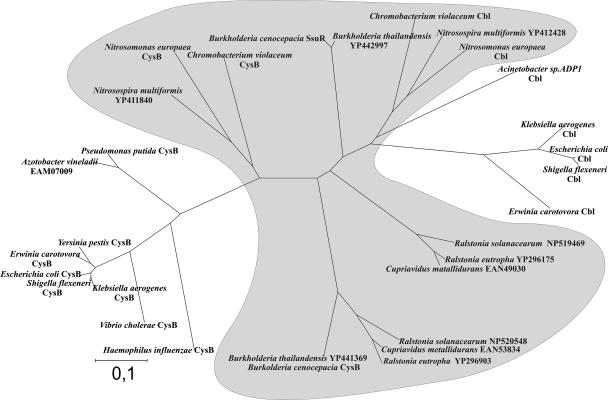
The “cysB-like” genes can be identified in silico in many genomes of Proteobacteria belonging to the β and γ classes, either as single-copy ORFs (e.g., in Haemophilus, Vibrio, Salmonella, and Acinetobacter) or as two ORFs sharing substantial similarity at the amino acid level. It seems that in genomes containing a pair of “cysB-like” sequences, they arose from duplication of a single ancestral gene and subsequent divergence of daughter genes. The functional significance of such divergence has so far been experimentally assessed only in E. coli, where the cysB and cbl (cysB-like) gene products activate expression of different target genes and respond to different metabolic signals. The constructed phylogenetic tree of “cysB-like” gene products (Fig. 6) illustrates various degrees of their relatedness in representative Proteobacteria from the β and γ subphyla. In Enterobacteriales (e.g., Escherichia, Erwinia, Klebsiella, and Shigella), CysB and Cbl appear to represent a pair of the most divergent paralogs in the “CysB family,” while the CysBBc and SsuRBc proteins of B. cenocepacia and their putative counterparts in other Burkholderiaceae (e.g., Ralstonia and Cupriavidus) appear to be the most closely related paralogous proteins. Nevertheless, it has been demonstrated by this study that the two regulatory proteins of B. cenocepacia display distinct preferences for some target genes that justifies a proposed annotation of their genes as cysBBc (encoding an ortholog of E. coli CysB) and ssuRBc (encoding an ortholog of E. coli Cbl). However, our results have highlighted some functional differences between the regulators CysBBc/CysBEc and SsuRBc/CblEc. In contrast to CysBEc, which activates the target cys promoters only in the presence of the inducer acetylserine, the function of CysBBc (at least at the cysIBc promoter) seems to be independent of this cofactor. The Cbl regulator of E. coli activates the target promoters (taup and ssup) without any inducing ligand, but its function is negatively affected by APS, the first intermediate of the sulfate assimilatory pathway (7). This explains the strong inhibition of expression from Cbl-dependent promoters in vivo in the presence of inorganic sulfate in the medium (55, 58, 59). In the case of SsuRBc, no decrease in expression of the responsive fusion ssuDBc-lacZ was observed in the presence of sulfate in the growth medium (measured in E. coli) (data not shown). The intriguing possibility that the function of SsuRBc is independent of any cofactor is being investigated.
FIG. 6.
Dendrogram showing the relationship between “CysB-like” proteins in bacteria. The protein sequences were retrieved from GenBank and analyzed with the programs Clustal and MEGA as described in Materials and Methods. The “CysB” and “Cbl” symbols are assigned to the proteins whose functions were either verified experimentally or annotated as CysB or Cbl in published genomic sequences; the putative members are indicated by accession numbers. The species belonging to the γ and β subphyla are unshaded and shaded, respectively. The scale bar represents the number of amino acid substitutions per site.
Acknowledgments
We are grateful to Beowulf Genomics and the Sanger Institute Pathogen Sequencing Unit for nucleotide sequence determination of the B. cenocepacia strain J2315. We thank J. Zaim for help in identification of the “cysB/cbl-like” ORF2 (BCAL2686) and the ssuD promoter region in this (nonannotated) genomic sequence. We also thank A. Imlay for the strain ΔcysB::cam (SP53).
The work of R.I.-N., A.Z., and M.M.H. was supported in part by Ministry of Science and Higher Education grant no. N303 074 32/2454.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Abola, A. P., M. G. Willits, R. C. Wang, and S. R. Long. 1999. Reduction of adenosine-5′-phosphosulfate instead of 3′-phosphoadenosine-5′-phosphosulfate in cysteine biosynthesis by Rhizobium meliloti and other members of the family Rhizobiaceae. J. Bacteriol. 181:5280-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnoli, K., C. A. Lowe, K. L. Farmer, I. Husnain, and M. S. Thomas. 2006. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an ECF σ factor which is a part of the Fur regulon. J. Bacteriol. 188:3631-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Baldwin, A., E. Mahenthiralingam, K. M. Thickett, D. Honeybourne, M. C. J. Maiden, J. R. Govan, D. P. Speert, J. J. LiPuma, P. Vandamme, and C. G. Dowson. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bick, J. A., J. J. Dennis, G. J. Zylstra, J. Nowack, and T. Leustek. 2000. Identification of a new class of 5′-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J. Bacteriol. 182:135-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 7.Bykowski, T., J. R. van der Ploeg, R. Iwanicka-Nowicka, and M. M. Hryniewicz. 2002. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5′-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol. Microbiol. 43:1347-1358. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 9.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 12.DeShazer, D., and D. E. Woods. 1996. Broad-host-range cloning and cassette vectors based on R388 trimethoprim resistance gene. BioTechniques 20:762-764. [DOI] [PubMed] [Google Scholar]
- 13.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 1999. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274:26639-26646. [DOI] [PubMed] [Google Scholar]
- 14.Eichhorn, E., J. R., van der Ploeg, and T. Leisinger. 2000. Deletion analysis of the Escherichia coli taurine and alkanesulfonate transport systems. J. Bacteriol. 182:2687-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farmer, K. L., and M. S. Thomas. 2004. Isolation and characterization of Burkholderia cenocepacia mutants deficient in pyochelin production: pyochelin biosynthesis is sensitive to sulfur availability. J. Bacteriol. 186:270-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner, M. M., and A. Revzin. 1981. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 9:3047-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, C. L. 1981. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as a target enzyme. J. Bacteriol. 145:1031-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holden, M. T. G., R. W. Titball, S. J. Peacock, A. M. Cerdeño-Tárraga, T. Atkins, L. C. Crossman, T. Pitt, C. Churcher, K. Mungall, S. D. Bentley, M. Sebaihia, N. R. Thomson, N. Bason, I. R. Beacham, K. Brooks, K. A. Brown, N. F. Brown, G. L. Challis, I. Cherevach, T. Chillingworth, A. Cronin, B. Crossett, P. Davis, D. DeShazer, T. Feltwell, A. Fraser, Z. Hance, H. Hauser, S. Holroyd, K. Jagels, K. E. Keith, M. Maddison, S. Moule, C. Price, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, M. Simmonds, S. Songsivilai, K. Stevens, S. Tumapa, M. Vesaratchavest, S. Whitehead, C. Yeats, B. G. Barrell, P. C. F. Oyston, and J. Parkhill. 2004. Genomic plasticity of the causative agent for melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA 101:14240-14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hryniewicz, M. M., A. Sirko, A. Pałucha, A. Böck, and D. Hulanicka. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: identification of a gene encoding a novel protein involved in thiosulfate binding. J. Bacteriol. 172:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hummerjohann, J., E. Kuttel, M. Quadroni, J. Ragaller, T. Leisinger, and M. A. Kertesz. 1998. Regulation of the sulfate starvation response in Pseudomonas aeruginosa: role of cysteine biosynthetic intermediates. Microbiology 144:1375-1386. [DOI] [PubMed] [Google Scholar]
- 22.Iwanicka-Nowicka, R., and M. M. Hryniewicz. 1995. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators belongs to Escherichia coli cys regulon. Gene 166:11-17. [DOI] [PubMed] [Google Scholar]
- 23.Kahnert, A., P. Vermeij, C. Wietek, P. James, T. Leisinger, and M. A. Kertesz. 2000. The ssu locus plays a key role in organosulfur metabolism in Pseudomonas putida S-313. J. Bacteriol. 182:2869-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kertesz, M. A. 1999. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 25.Kertesz, M. A. 2001. Bacterial transporters for sulfate and organosulfur compounds. Res. Microbiol. 152:279-290. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S. K., A. Rahman, J. A. Bick, R. C. Conover, M. K. Johnson, J. T. Mason, M. Hirasawa, T. Leustek, and D. B. Knaff. 2004. Properties of the cysteine residues and iron-sulfur cluster of the assimilatory 5′-adenylyl sulfate reductase from Pseudomonas aeruginosa. Biochemistry 43:13478-13486. [DOI] [PubMed] [Google Scholar]
- 27.Koch, D. J., C. Rückert, A. Albersmeier, A. T. Hüser, A. Tauch, A. Pühler, and J. Kalinowski. 2005. The transcriptional regulator SsuR activates expression of the Corynebacterium glutamicum sulphonate utilization genes in the absence of sulphate. Mol. Microbiol. 58:480-494. [DOI] [PubMed] [Google Scholar]
- 28.Kopriva, S., T. Büchert, G. Fritz, M. Suter, R. Benda, V. Schünemann, A. Koprivova, P. Schürmann, A. X. Trautwein, P. M. H. Kroneck, and C. Brunold. 2002. The presence of an iron-sulfur cluster in adenosine 5′-phosphosulfate reductase separates organisms utilizing adenosine 5′-phosphosulfate and phosphoadenosine 5′-phosphosulfate for sulfate assimilation. J. Biol. Chem. 277:21786-21791. [DOI] [PubMed] [Google Scholar]
- 29.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 30.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 31.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 32.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 33.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzales. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 34.Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz. 2001. Functional dissection of the LysR-type CysB transcriptional regulator: regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276:2098-2107. [DOI] [PubMed] [Google Scholar]
- 35.Mahenthiralingam, E. T., A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 36.Mahenthiralingam, E. T., and P. Vandamme. 2005. Taxonomy and pathogenesis of the Burkholderia cepacia complex. Chron. Respir. Dis. 2:209-217. [DOI] [PubMed] [Google Scholar]
- 37.McKevitt, A. I., S. Bajaksouzian, J. D. Klinger, and D. E. Woods. 1989. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect. Immun. 57:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Nieman, W. C., D. DeShazer, H. S. Kim, H. Tettelin, K. E. Nelson, T. Feldblyum, R. L. Ulrich, C. M. Ronning, L. M. Brinkac, S. C. Daugherty, T. D. Davidsen, R. T. Deboy, G. Dimitrov, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, H. Khouri, J. F. Kolonay, R. Madupu, Y. Mohammoud, W. C. Nelson, D. Radune, C. M. Romero, S. Sarria, J. Selengut, C. Shamblin, S. A. Sullivan, O. White, Y. Yu, N. Zafar, L. Zhou, and C. M. Fraser. 2004. Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. USA 101:14246-14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, S., and J. A. Imlay. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 185:1942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 42.Pinto, R., Q. X. Tang, W. J. Britton, T. S. Leyh, and J. A. Triccas. 2004. The Mycobacterium tuberculosis cysD and cysNC genes form a stress-induced operon that encodes a tri-functional sulfate-activating complex. Microbiology 150:1681-1686. [DOI] [PubMed] [Google Scholar]
- 43.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 46.Shen, Y., P. Sharma, F. G. da Silva, and P. Ronald. 2002. The Xanthomonas oryzae pv. lozengeoryzae raxP and raxQ genes encode an ATP sulphurylase and adenosine-5′-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 44:37-48. [DOI] [PubMed] [Google Scholar]
- 47.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 48.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 49.Snoeck, C., C. Verreth, I. Hernández-Lucas, E. Martínez-Romero, and J. Vanderleyden. 2003. Identification of a third sulfate activation system in Sinorhizobium sp. strain BR816: the CysDN sulfate activation complex. Appl. Environ. Microbiol. 69:2006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorensen, M. A., and S. Pedersen. 1991. Cysteine, even in low concentrations, induces transient amino acid starvation in Escherichia coli. J. Bacteriol. 173:5244-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soutourina, J., S. Blanquet, and P. Plateau. 2001. Role of d-cysteine desulfhydrase in the adaptation of Escherichia coli to d-cysteine. J. Biol. Chem. 276:40864-40872. [DOI] [PubMed] [Google Scholar]
- 52.Speert, D. P. 2002. Advances in Burkholderia cepacia complex. Paediatr. Respir. Rev. 3:230-235. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. W. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 55.van der Ploeg, J. R., M. A. Weiss, E. Saller, H. Nashimoto, N. Saito, M. A. Kertesz, and T. Leisinger. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 178:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 57.van der Ploeg, J. R., E. Eichhorn, and T. Leisinger. 2001. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch. Microbiol. 176:1-8. [DOI] [PubMed] [Google Scholar]
- 58.van der Ploeg, J. R., R. Iwanicka-Nowicka, T. Bykowski, M. M. Hryniewicz, and T. Leisinger. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 41:29358-29365. [DOI] [PubMed] [Google Scholar]
- 59.van der Ploeg, J. R., R. Iwanicka-Nowicka, M. A. Kertesz, T. Leisinger, and M. M. Hryniewicz. 1997. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J. Bacteriol. 179:7671-7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warren, M. J., E. L. Bolt, C. A. Roessner, A. I. Scott, J. B. Spencer, and S. C. Woodcock. 1994. Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem. J. 302:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]