Abstract
Expression of the structural genes for the anthrax toxin proteins is coordinately controlled by host-related signals, such as elevated CO2, and the trans-acting positive regulator AtxA. In addition to these requirements, toxin gene expression is under growth phase regulation. The transition state regulator AbrB represses atxA expression to influence toxin synthesis. During the late exponential phase of growth, when AbrB levels begin to decrease, toxin synthesis increases. Here we report that toxin gene expression also requires the presence of sigH, a gene encoding the RNA polymerase sigma factor associated with development in Bacillus subtilis. In the well-studied B. subtilis system, σH is required for sporulation and other post-exponential-phase processes and is part of a feedback control pathway for abrB expression. Our data indicate that a Bacillus anthracis sigH-null mutant is asporogenous and toxin deficient. Yet the sigma factor is required for toxin gene expression in a manner that is independent of the pathway leading to post-exponential-phase gene expression. σH positively controls atxA in an AbrB-independent manner. These findings, combined with previous observations, suggest that the steady-state level of atxA expression is critical for optimal toxin gene transcription. We propose a model whereby, under toxin-inducing growth conditions, control of toxin gene expression is fine-tuned by the independent effects of σH and AbrB on the expression of atxA.
Expression of the structural genes for the anthrax toxin proteins PagA (protective antigen), Cya (edema factor), and Lef (lethal factor) is coordinately controlled by host-related signals and trans-acting regulatory genes. The toxin genes are located noncontiguously within a 30-kb region of the 182-kb Bacillus anthracis virulence plasmid pXO1. Two pXO1-encoded regulators of toxin gene expression have been reported, PagR and AtxA. The pagR gene encodes a weak repressor of the bicistronic pagAR operon (25) and negatively affects the expression of the chromosomal S-layer genes sap and eag (34). Purified PagR has been shown to bind sequences within the pagA, sap, and eag promoter regions; however, alignment of the protected sequences does not reveal a PagR consensus sequence (34).
AtxA is a positive regulator of pagAR, cya, and lef as well as a number of other plasmid- and chromosome-carried genes (6, 24, 29). AtxA controls gene expression in trans, but the molecular mechanism for this regulation is unknown. AtxA is predicted to be a 56-kDa basic protein with a weak helix-turn-helix motif located at the amino terminus. Yet specific nucleic acid binding activity has not been ascribed to this protein, and there are no obvious similarities in the promoter regions of the AtxA-controlled genes. Moreover, consensus sequences for recognition by RNA polymerase sigma factors are generally not apparent for AtxA-dependent transcription start sites.
In addition to the pXO1-encoded regulators, a chromosomal gene, abrB, has been reported to affect anthrax toxin gene expression. Extensive studies in Bacillus subtilis have revealed that AbrB is a transition state regulator that plays a critical role in the suppression of post-exponential-phase gene expression during the logarithmic phase of growth (42). Optimal toxin synthesis by B. anthracis occurs during growth at 37°C in Casamino Acids medium containing bicarbonate (11, 48). In these conditions, toxin gene expression reaches a maximum during the late exponential phase of growth, coinciding with a decrease in abrB transcription. A B. anthracis abrB deletion mutant produces higher levels of all three toxin proteins, and toxin gene expression peaks earlier during growth (46).
The abrB effect on toxin gene expression may be due in part to the repression of atxA transcription. Saile and Koehler (46) demonstrated elevated transcripts of atxA in an abrB-null mutant of B. anthracis, and Strauch et al. (54) reported recently that B. anthracis AbrB binds to specific DNA sequences in the atxA promoter region. Nevertheless, there is evidence that abrB controls toxin gene expression in an atxA-independent manner. Baillie et al. (3) reported that a B. subtilis abrB-null mutant harboring the cloned pagA gene produced elevated levels of protective antigen. Considering that B. subtilis does not appear to contain an atxA homologue, this effect is independent of atxA.
The B. subtilis AbrB is a pleiotropic regulator and binds to the promoter regions of multiple target genes (42). One AbrB target is sigH (19, 53), a gene encoding the alternative sigma factor σH, which plays an important role in post-exponential-phase gene expression. The σH RNA polymerase holoenzyme recognizes and transcribes genes associated with the transition to the stationary phase of growth, including genes for cytochrome biogenesis, generation of potential nutrient sources, transport, and cell wall metabolism (7), as well as genes important for competence and sporulation initiation (16). In B. subtilis, a complex signal transduction phosphorelay system keeps the initiation of sporulation under stringent control by regulating the phosphorylation of the master response regulator, Spo0A (23, 38-41).
σH, AbrB, and Spo0A are all part of a feedback mechanism that ultimately controls the expression of each regulator and is critical for sporulation initiation. During the logarithmic phase of growth, sigH expression is repressed by AbrB, resulting in relatively low-level expression of σH targets (19, 53). As a culture transitions into the stationary phase, phosphorylation of Spo0A increases, leading to gradual activation of Spo0A (50). Phosphorylated Spo0A represses the transcription of abrB (21, 52), resulting in increased levels of σH. The spo0A gene has two promoters, one recognized by the housekeeping sigma factor, σA, and the other recognized by σH (43). The elevated Spo0A levels in the stationary phase are attributed to enhanced spo0A transcription when σH levels are high. Therefore, abrB expression in B. subtilis is increased in a sigH-null mutant that produces low levels of Spo0A (54).
In B. anthracis, AbrB represses the toxin regulator atxA (46, 54). As predicted from the B. subtilis model in which abrB expression is subject to control by σH and Spo0A, the activity of a reporter gene driven by the atxA promoter is reduced in B. subtilis spo0A and sigH mutants (54). Moreover, the increased atxA promoter activity exhibited by a B. subtilis abrB mutant is comparable to the activities of double mutants with abrB and sigH or abrB and spo0A deleted (54).
In the work presented here, we further explored the relationship between anthrax toxin gene expression and the multicomponent system controlling growth phase-specific gene expression and development that is well studied in B. subtilis. As is true for σH function in B. subtilis, the B. anthracis sigH gene is required for sporulation, demonstrating a role for this alternative sigma factor in post-exponential-phase gene regulation. However, our data show that SigH controls toxin gene expression independently of AbrB, indicating an additional function for SigH in B. anthracis. Thus, SigH joins AbrB as another key player in the disparate processes of sporulation and toxin synthesis by B. anthracis.
MATERIALS AND METHODS
Growth conditions.
Escherichia coli strains JM109 and GM2163 (New England Biolabs, Beverly, MA) were grown in Luria-Bertani (LB) broth (47) and used as hosts for cloning. B. anthracis strains were grown in LB broth to obtain cells for transductions, electroporations, and DNA extraction. B. anthracis culture supernatants and cell extracts for Western hybridization and β-galactosidase assays were obtained from cells grown as follows. A sample from an overnight culture grown at 30°C with agitation in LB broth containing 0.5% glycerol and appropriate antibiotics was used to inoculate 45 ml of CACO3 medium (CA medium [56] buffered with 100 mM HEPES [pH 8.0] and 0.8% [wt/vol] sodium bicarbonate) in a 250-ml Erlenmeyer flask. Cultures were incubated at 37°C in an atmosphere containing 5% CO2. Antibiotics were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Fairlawn, NJ) and added to media (concentrations are indicated in parentheses) when appropriate: ampicillin (100 μg/ml), erythromycin (150 μg/ml for E. coli; 5 μg/ml for B. anthracis), kanamycin (20 μg/ml for E. coli; 100 μg/ml for B. anthracis), and spectinomycin (50 μg/ml for E. coli; 100 μg/ml for B. anthracis). All other chemicals were purchased from Sigma Aldrich unless indicated otherwise.
DNA isolation and manipulation.
Extraction of chromosomal DNA from B. anthracis cultures was carried out using a Mo Bio genomic isolation kit (Mo Bio Laboratories, Solana Beach, CA). Preparation of plasmid DNA from E. coli, transformation of E. coli, and recombinant DNA techniques were performed using standard procedures (2). B. anthracis was electroporated with unmethylated plasmid DNA from E. coli GM2163 as described elsewhere (29). Restriction enzymes and T4 DNA ligase were purchased from Promega (Madison, WI) and Fisher Scientific, and Taq DNA polymerase was purchased from New England Biolabs (Beverly, MA).
Strain construction.
Table 1 contains a complete list of B. anthracis strains used, including relevant characteristics. The B. anthracis sigH-null mutant UT198 was constructed by replacing the sigH gene (NC003997.3; nucleotides [nt] 102990 to 103646) with the Ω-km2 element, using a previously described protocol (29). B. anthracis strain UT301 was created by double-crossover recombination of sigH and upstream regulatory elements into the chromosomal plcR locus (NC003997.3; nt 1133897 to 1134781). To create UT301, plasmid pUTE719, containing the Ω-spc element flanked by DNA sequences found upstream and downstream of the plcR gene, was constructed. The plcR-flanking sequences were amplified with primer pairs CR123 (5′-GAGCTCGGATCCCGATTCAATTCGGCTCACTT-3′)/ES40 (5′-AACTCCAGTGTTGCGGAAACGTTAAAGA-3′) and CR124(5′-GAGCTCTTGAAAACGCAATTGCAAAC-3′)/ES43 (5′-ACGCGTCGACTCGTATCTCCTGCCCAATTC-3′) and Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). The plcR-flanking regions and the Ω-spc element were cloned into pUTE583 (12) such that a unique SacI site was located between the Ω-spc element and the downstream flanking region. Primers MH187 (5′-CCCGAGCTCGGAAGAATCAAACTGCAGATG-3′) and MH188 (5′-CCCGAGCTCGTATCTTTCATCGTAGAG-3′) were used to amplify a 1,209-bp fragment containing the sigH gene and its predicted promoter region. The PCR product was digested with SacI and cloned into pUTE719. The resulting construct, pUTB1, was electroporated into UT198, and the resulting strain was cultured to facilitate recombination as described previously (46).
TABLE 1.
B. anthracis strains used in this study
| Straina | Phenotype or genotype | Relevant characteristicsb | Source or reference |
|---|---|---|---|
| UM44 | Ind− | Weybridge strain; pXO1+ pXO2− | C. Thorne |
| UT53 | atxA | Tox− Kmr | 15 |
| UT133 | cya::lacZ | Emr Kmr; UM44 transduced with CP51 (55) propagated on RBAF 144 (48) | This work |
| UT147 | pag::lacZ | Emr Kmr; UM44 transduced with CP51 (55) propagated on RBAF 140 (48) | This work |
| UT148 | lef::lacZ | Emr Kmr; UM44 transduced with CP51 (55) propagated on RBAF 143 (48) | This work |
| UT157 | spo0A | Spo− Spcr | 46 |
| UT166 | abrB | Spcr | 46 |
| UT198 | sigH | Spo− Kmr | This work |
| UT199 | sigH pagA::lacZ | Spo− Kmr Emr; UT147 transduced with CP51 (55) propagated on UT198 | This work |
| UT200 | sigH lef::lacZ | Spo− Kmr Emr; UT148 transduced with CP51 (55) propagated on UT198 | This work |
| UT201 | sigH cya::lacZ | Spo− Kmr Emr; UT133 transduced with CP51 (55) propagated on UT198 | This work |
| UT285 | abrB spo0A | Spo− Spcr Kmr; abrB was deleted in UT157 and is replaced by Ω-km2 | This work |
| UT290 | spo0A sigH | Spo− Spcr Kmr; UT157 was transduced using CP51 phage (55) propagated on UT198 | This work |
| UT291 | abrB sigH | Spo− Spcr,Kmr; UT66 was transduced using CP51 phage (55) propagated on UT198 | This work |
| UT301 | sigH plcR::sigH | Spo+ Spcr Kmr; sigH gene introduced in the plcR locus using pUTB1 | This work |
All UT mutants listed are derivatives of Weybridge strain UM44.
Abbreviations: Ind−, indole auxotrophy; Tox−, toxin production deficiency; Emr, erythromycin resistance; Kmr, kanamycin resistance; Spcr, spectinomycin resistance; Spo−, sporulation deficiency.
Microscopy.
B. anthracis cells were visualized using a Labophot Nikon microscope, and pictures were obtained using a COOLPIX995 digital camera.
Western blot analysis.
Culture supernatants were filtered through 0.2-μm-pore-size syringe filters (Corning, Corning, NY). For detection of protective antigen (PA), lethal factor (LF), and edema factor (EF), samples were treated and visualized as described previously (46). Equal volumes of culture supernatants were used to compare toxin levels in the parent and mutant strains. Cell extracts were prepared and probed for the presence of σH as follows. Cell pellets from 1-ml samples were resuspended in 1× phosphate-buffered saline (PBS) and mechanically sheared by using a mini-bead beater 8 (MBB8) (Biospec Products, Bartlesville, OK), incubated for 5 min at 65°C, and sheared again for 1 min. Cell debris was pelleted at 16,000 × g for 10 min. Supernatant fractions were treated with 40 μl of 25× stock protease inhibitor cocktail (Roche, Indianapolis, IN). The protein concentration was determined by using a BCA assay kit (Pierce, Rockford, IL) with bovine serum albumin as the standard, following the manufacturer's instructions. Samples containing 4 μg of total protein were resolved on 15% sodium dodecyl sulfate-polyacrylamide gels, transferred, and blocked in 1× TBS-T (20 mM Tris base, 137 mM NaCl, 0.1% Tween 20 [pH 7.6]) containing 5% milk as described previously (46). The membranes were probed using rabbit antisera raised against B. subtilis σH and SigA, at dilutions of 1:500 and 1:1,000, respectively, in 1× TBS-T-5% milk (46).
β-Galactosidase assays.
One-milliliter culture samples were collected hourly from the early logarithmic (3 h) to the 3-h post-stationary phase of growth (10 h). β-Galactosidase assays were performed according to Miller (36). Cell extracts were prepared as described for Western blot analysis (above). At least three independent cultures were assayed for enzyme activity. The figures show data from representative experiments.
Bioinformatics analyses.
The predicted amino acid sequences of σH from B. anthracis and B. subtilis were aligned using ClustalW. The NCBI BlastP program was used to identify B. anthracis homologs of B. subtilis genes known to be transcribed by σH RNA polymerase. The predicted amino acid sequences for the B. subtilis Spo0A, Spo0F, Spo0M, CitG, KinA, PhrC, PhrI, Spo0M, SpoVG, SpoVS, and SigA proteins were compared by BLAST analysis against the sequence of the B. anthracis Ames genome (NC003997). Genes encoding the proteins with the highest percent identity were considered for analysis. Upon identification of a candidate gene, the sequence of the 500 nucleotides upstream of the predicted translational start was examined for the presence of a σH consensus. Wherever possible, the promoter regions of the B. subtilis genes were aligned with the B. anthracis sequences using ClustalW.
Purification of B. anthracis σH.
Sequences corresponding to the B. anthracis sigH locus were amplified from genomic DNA using PCR with primers YC85 (5′-CACCATGGAAGCAGGCTTCGTAAGTG-3′) and YC86 (5′-ATTTGAAGTGGTACTCTCTCTC-3′). The resulting product was cloned into the protein expression vector pET101/D-TOPO (Invitrogen, Carlsbad, CA) to give plasmid pUTE493. The plasmid was constructed such that C-terminally His-tagged σH was expressed from a T7 promoter. pUTE493 was introduced into E. coli BL21(DE3), and the recombinant B. anthracis protein was expressed and purified from the strain under denaturing conditions according to the manufacturer's instructions (QIAexpress; QIAGEN, Valencia, CA). Recombinant σH was dialyzed overnight in refolding buffer following the protocol described by Haldenwang et al. (22).
In vitro runoff transcription assays.
To create templates for in vitro runoff transcription reactions, blunt-ended DNA fragments corresponding to the pagA, lef, and cya promoter regions from positions −90 to +88, −70 to +132 and −122 to +63, respectively, relative to the main transcriptional start sites, and to the spoVG and atxA promoter regions from positions −243 to +1 and −870 to +1, respectively, relative to the translational start, were amplified by PCR using Deep Vent DNA polymerase. Each PCR product was purified using Zymoclean reagents (Orange, CA), concentrated 4 times and quantified by measuring absorbance at an optical density of 260 nm.
Recombinant B. anthracis σH was used in combination with E. coli core RNA polymerase purchased from Epicenter (Madison, WI) for in vitro transcription reactions. For each reaction, 0.1 pmol of template DNA was added to 1× in vitro transcription buffer (Promega) supplemented with 200 mM KCl and containing 77.25 nM E. coli core RNA polymerase and 480 nM σH in a total volume of 10 μl. Following incubation at 37°C for 30 min, ribonucleotides and [α-32P]UTP were added to a final concentration of 0.5 mM in a 20-μl volume. After incubation for an additional 15 min at 37°C, reactions were terminated by purification through G-30 columns (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. Eluates were mixed with equal volumes of RNA loading buffer (0.05% bromophenol blue, 20 mM EDTA in deionized formamide [Sigma-Aldrich]) and heated at 80°C for 10 min prior to electrophoresis. Samples (7 μl) were electrophoresed at 1,400 V for 2.5 h in 8% acrylamide-7 M urea denaturing gels. Gels were dried for 4 h using a model 583 Bio-Rad gel dryer prior to exposure to photographic film at −80°C overnight. Molecular size markers were generated by in vitro transcription of an RNA century marker template set (Ambion, Austin, TX), following the manufacturer's instructions.
RESULTS
Creation of a B. anthracis sigH-null mutant.
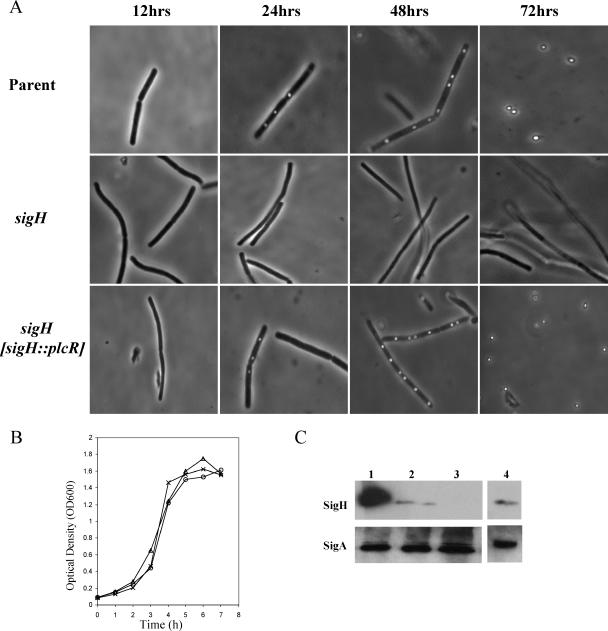
We identified the sigH gene of B. anthracis (NC003997.3; nt 102990 to 103646) on the basis of sequence homology to B. subtilis sigH (NC000964.2; nt 116597 to 117253). A protein BLAST search (BlastP) revealed that BA0093 is predicted to encode a protein (NP_842661.1) that bears an 81% identity to the B. subtilis σH (NP_387979). We constructed a sigH-null mutant, UT198, in which the sigH gene was replaced by a kanamycin cassette using methods described previously (46). As expected, the sigH mutant, UT198, was unable to sporulate (Fig. 1A). The sigH-null mutation did not affect the growth rate when cells were cultured in CACO3 medium at 37°C in an atmosphere containing 5% CO2, conditions that are optimal for toxin gene expression (11, 48) (Fig. 1B). The mutation in UT198 was complemented by placement of the sigH gene in another chromosomal locus, plcR (NC003997.3; nt 1133897 to 1134781). The B. anthracis plcR gene is predicted to encode a truncated, nonfunctional protein (1, 35, 49). Recombinant strain UT301 was derived from UT198 and harbors sigH under the control of its native promoter in the plcR locus. Sporulation and σH synthesis by UT301 are comparable to those of the parent strain, UM44 (Fig. 1A and C).
FIG. 1.
Creation of a B. anthracis sigH-null mutant. (A) Sporulation of the parent, mutant, and complemented strains grown in PA broth (20) at 30°C. Samples were taken at the times indicated and assessed for the presence of spores using light microscopy. (B) Growth of UM44 (parent), UT198 (ΔsigH), and UT301 (ΔsigH sigH::plcR) in CACO3 broth in 5% CO2 at 37°C. OD600, optical density at 600 nm. (C) Synthesis of σH by the parent and mutant strains. The culture samples were obtained during the late exponential phase of growth (5 h). Solubilized cellular protein (4 μg) was subjected to Western blot analysis using rabbit anti-σH antibody raised against B. subtilis σH or rabbit anti-σA antibody raised against B. subtilis σA (as a loading control). Lane 1, recombinant protein; lane 2, UM44; lane 3, UT198; lane 4, UT301.
Deletion of sigH results in a toxin-deficient phenotype.
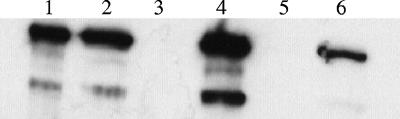
Considering that abrB and spo0A affect toxin gene expression in B. anthracis (46), and considering the relationships between sigH and these regulators in B. subtilis (50), we assessed toxin synthesis by the B. anthracis parent and the sigH-null mutant. Equal volumes of culture supernatant fractions of strains grown in conditions that promote toxin gene expression were probed for the relative amounts of each toxin protein. As shown in Fig. 2A, at the late exponential growth phase the levels of PA, LF, and EF in the supernatant of the UT198 (sigH) strain were significantly lower than those in the supernatant of the UM44 (parent) culture. The toxin deficiency of UT198 was complemented by the addition of sigH in trans (UT301). UT301 produced greater amounts of PA and LF than UT198, but less PA and LF than UM44 (Fig. 2A, lane 4). This result was surprising, given that σH synthesis by UT301 was comparable to that of the parent strain (Fig. 1B).
FIG. 2.
Reduced toxin gene expression by a B. anthracis sigH-null mutant. (A) Synthesis of the toxin proteins. Supernatant samples for Western hybridization analysis were obtained during the late exponential phase of growth (5 h). Equal volumes of supernatant were probed using rabbit antisera raised against the toxin proteins as indicated. Lane 1, recombinant protein; lane 2, UM44 (parent); lane 3, UT198 (sigH); lane 4, UT301 (sigH plcR::sigH). (B) β-Galactosidase activities of promoter-lacZ fusions in parent (circle) and sigH (square) backgrounds. pagA::lacZ, UT147 (parent) and UT199 (sigH); lef::lacZ, UT148 (parent) and UT200 (sigH); cya::lacZ, UT133 (parent) and UT201 (sigH). Specific β-galactosidase activity (nmol o-nitrophenyl-β-d-galactopyranoside [ONPG]/min/mg protein) is shown.
Transcriptional analysis also revealed significantly reduced expression of the toxin genes in the sigH-null mutant compared to that in the parent strain. UM44 (parent)- and UT198 (sigH)-derived strains harboring the individual transcriptional fusions pagA::lacZ, lef::lacZ, and cya::lacZ at the toxin gene loci were created, and toxin gene promoter activity was assessed as the β-galactosidase activity of cells grown in conditions optimal for toxin synthesis. Throughout growth, the enzyme activities associated with the pag-lacZ, cya-lacZ, and lef-lacZ reporter genes in sigH-null strains were 4% or less of those observed for the parent strains (Fig. 2B). These results demonstrate that sigH affects toxin gene expression at the transcriptional level.
σH affects toxin synthesis independently of AbrB.
The B. anthracis abrB gene exerts a negative effect on toxin gene expression. An abrB-null mutant expresses cya, lef, and pagA earlier than the parent strain during growth in batch culture, indicative of growth phase-dependent control (46). This phenotype has been attributed to specific binding of the AbrB protein to the promoter of the toxin gene regulator atxA (54). AtxA is a strong positive regulator of the toxin genes; the toxin-deficient phenotype of an atxA-null mutant is comparable to that of the sigH mutant (15). In B. subtilis, steady-state levels of AbrB are modulated by a feedback mechanism whereby AbrB represses sigH, σH RNA polymerase transcribes spo0A, and phosphorylated Spo0A represses abrB. In B. subtilis, deletion of sigH results in elevated levels of AbrB during the stationary phase of growth (19, 43, 50).
We reasoned that if the toxin-deficient phenotype of the B. anthracis sigH-null mutant was due solely to elevated AbrB levels, then the toxin phenotype of a sigH abrB double mutant should match that of an abrB mutant. We assessed toxin protein levels in culture supernatants of sigH, abrB, and abrB sigH mutants. Figure 3 shows the results of Western hybridizations comparing LF production by the parent and mutant strains. As reported previously (46), LF production by the abrB-null mutant was elevated relative to that of the parent strain. However, the sigH abrB double mutant exhibited an LF-deficient phenotype comparable to that of the sigH-null mutant. Similar results were obtained when supernatants were probed for the other toxin components, EF and PA (data not shown). These data indicate that the SigH effect on toxin synthesis cannot be attributed exclusively to SigH control of abrB.
FIG. 3.
Synthesis of LF by parent and mutant strains. Supernatant samples for toxin analysis were taken during the late exponential phase of growth (5 h). Lane 1, recombinant protein; lane 2, UM44 (parent); lane 3, UT198 (sigH); lane 4, UT166 (abrB); lane 5, UT291 (abrB sigH); lane 6, UT301 (sigH sigH::plcR).
σH positively regulates atxA.
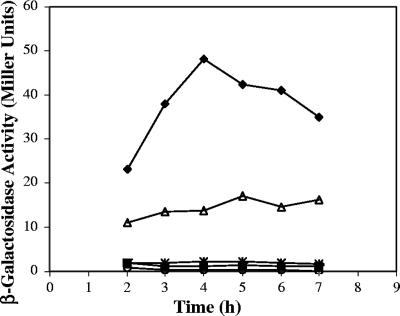
Other than AbrB, no trans-acting regulators have been associated with expression of the atxA gene. To assess the effect of SigH on atxA expression, we examined atxA promoter activity in the parent and mutant strains. As shown in Fig. 4, atxA promoter activity was highest during the mid-exponential growth phase and elevated in the abrB-null mutant, consistent with repression of atxA by AbrB (46, 54). Expression of atxA was barely detectable in the sigH mutant, in agreement with the low level of toxin synthesis by this mutant. Nevertheless, the atxA promoter activity of the abrB sigH double mutant was comparable to that of the sigH mutant, indicating that SigH positively controls atxA independently of AbrB.
FIG. 4.
β-Galactosidase activities of atxA-lacZ fusions in parent and mutant strains during growth in toxin-inducing conditions. The low-copy-number plasmid pUTE411 containing a PatxA-lacZ transcriptional fusion (46) was introduced into UM44 (parent; triangle), UT198 (sigH; square), UT166 (abrB; diamond), UT291 (abrB sigH; asterisk), and UM44(pHT304-18Z) (empty vector control; circle).
The σH consensus sequence is absent from the atxA and toxin gene promoters.
In B. subtilis, genes that are recognized by σH RNA polymerase are preceded by a consensus sequence (4, 7, 31, 33, 44). Dai et al. (15) previously reported a single transcriptional start site for the atxA gene that is preceded by a consensus sequence for the housekeeping sigma factor, σA. We examined DNA sequences up to 1,000 nucleotides upstream of the predicted translational start site of atxA and detected no apparent σH consensus sequences. Sequences corresponding to the σH consensus are also not apparent in the toxin gene promoter regions. In fact, no consensus sequences for recognition by any known sigma factor are apparent in the cya and lef gene promoters or upstream of the major atxA-regulated transcription start site, P1, of the pagA gene. Weak, constitutively expressed apparent start sites for pagA, P2, and other RNAs with 5′ ends mapping downstream of P2 have been described (15, 29). A consensus sequence for σA is located upstream of P2 (15).
We questioned whether the promoter regions of B. anthracis genes transcribed by σH RNA polymerase contained sequences resembling the B. subtilis σH consensus. We identified B. anthracis homologues of B. subtilis genes known to be transcribed by σH RNA polymerase. Analysis of sequences upstream of such genes revealed the presence of sequences that closely resembled the σH consensus sequence established in B. subtilis (Table 2). Each promoter was aligned with its B. subtilis counterpart using ClustalW (13), to compare sequence similarities and positioning of the σH consensus. In the cases of spoVS, sigA, spoVG, and phrC, the distances between the consensus sequences and the predicted translational start sites for the genes were very similar in the two species. The B. subtilis consensus sequence for the −10 region (RxxGAATww; R indicates A or G; w indicates A or T [7]) was present in all of the B. anthracis promoters investigated. On the other hand, the −35 regions of the B. anthracis promoters deviated from the B. subtilis σH consensus in the third and last positions (Table 2). The −10 and −35 regions for σH recognition in B. subtilis are typically separated by an 11- to 12-nucleotide spacer. The B. anthracis promoter regions investigated here revealed spacers that ranged from 10 to 14 nucleotides.
TABLE 2.
B. anthracis homologues of B. subtilis σH RNA polymerase-transcribed genesa
| B. anthracis gene | B. subtilis homologb | Predicted −35 region | Spacing (nt) | Predicted −10 region |
|---|---|---|---|---|
| BA1767 | citG | AGAGGAATT | 13 | ATAGAATAT |
| BA4223 | kinA | NDc | AAAGAATTA | |
| BA1018 | phrC | TCAGGAGTA | 10 | TTAGCAGTG |
| BA3285 | phrI | ATGGGAATA | 13 | GCCGAATTA |
| BA4394 | spo0A | ATCGGAAAG | 14 | AGGGAATTT |
| BA5581 | spo0F | ATAGGAAAC | 11 | AAAGAATAG |
| BA2308 | spo0M | TTTGGAAAA | 20 | TGCGAATGT |
| ATGGGAGAA | 16 | ATGGAATAT | ||
| BA0047 | spoVG | AAGGGAAAA | 10 | GTGGAATTT |
| BA2154 | spoVS | GCAGGAAGA | 11 | ATCGAATGA |
| BA4515 | sigA | GAAGGATTC | 11 | GTAGAATAC |
| B. anthracis consensus | RxxGGAwWw | 10 to 14 | RxxGAATww |
Boldface indicates a conserved nucleotide. R is an A or a G, and W is an A or a T. A lowercase letter denotes a position in the consensus that is not always conserved.
The B. subtilis consensus is as follows: RxAGGAwWW (−35 region), 11 to 12 spaces, RxxGAATww (−10 region).
ND, not determined.
In vitro transcription of B. anthracis promoters by σH RNA polymerase.
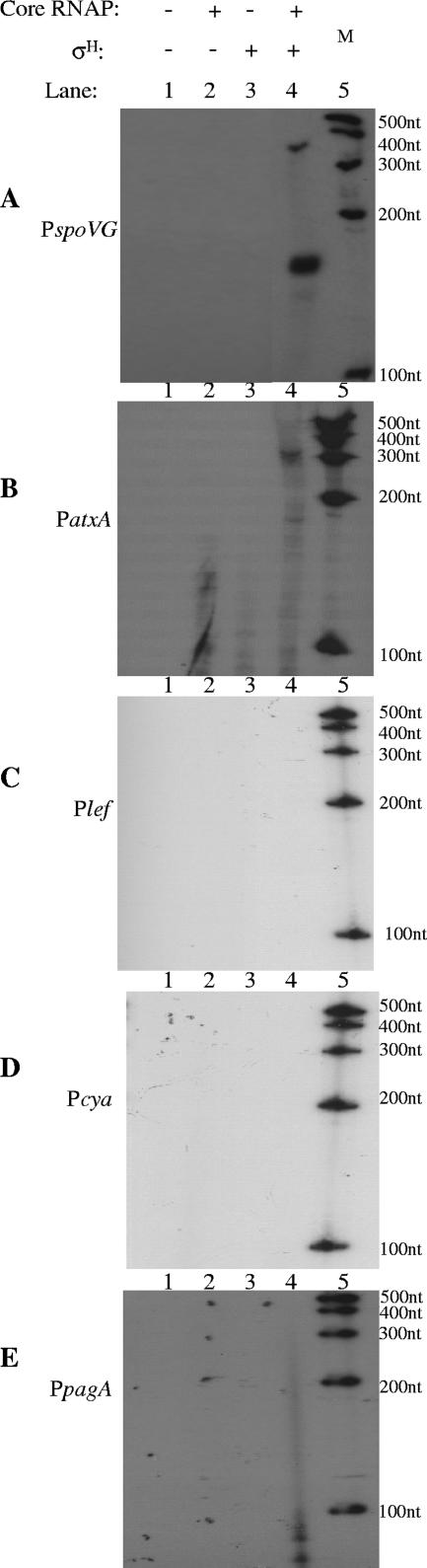
To determine if atxA or the toxin genes are transcribed directly by σH RNA polymerase, we performed in vitro transcription experiments using recombinant B. anthracis σH, E. coli core RNA polymerase, and B. anthracis promoter templates (Fig. 5). B. subtilis σ factors have been reported to function in vitro with the E. coli core enzyme (9, 17). The B. anthracis spoVG promoter, which bears the B. subtilis σH consensus and is strongly SigH dependent in B. subtilis (10, 16), was tested as a positive control. Reactions with the spoVG DNA template yielded an abundant RNA product of the predicted size for a runoff transcript initiated at the spoVG promoter (Fig. 5A). Thus, the purified recombinant B. anthracis σH protein can function with core RNA polymerase to direct transcription from a B. anthracis gene promoter harboring a σH consensus sequence.
FIG. 5.
In vitro transcription of B. anthracis promoters using recombinant σH RNA polymerase holoenzyme. C-terminally His-tagged σH purified from B. anthracis was used with core RNA polymerase (RNAP) from E. coli as described in Materials and Methods. The compositions of the reaction mixtures and DNA templates are as indicated. +, present; −, absent. Molecular size markers (M) shown in lane 5 are as indicated on the right.
Given the apparent lack of σH recognition sequences in the atxA, pagA, lef, and cya gene promoters, we predicted that transcripts would not be generated in reactions using these templates. As expected, the toxin promoter templates did not yield transcripts (Fig. 5C, D, and E), indicating that σH affects pagA, lef, and cya gene expression indirectly. In some reactions, faint bands of a size indicating nonspecific end-to-end transcription of the templates were observed (data not shown). The small species detected in some reactions containing the pagA template (Fig. 5E, lane 4) are most likely stable degradation products of end-to-end transcripts.
As shown in Fig. 5B, lane 4, an RNA transcript was detected in reactions with the atxA promoter template. The size of the in vitro-generated RNA product indicated a transcript initiating approximately 250 nt upstream of the previously reported transcriptional start site for atxA (14). This result was surprising because, as discussed above, sequences in this region do not resemble the σH consensus established for B. subtilis. Nevertheless, data from reverse transcription-PCRs employing RNA from cultured cells confirmed the presence of an RNA transcript in this region (data not shown).
DISCUSSION
The alternative sigma factors of bacteria provide mechanisms for shifting gene expression patterns in response to a changing environment or phase of growth (27). Among the large number of genes with promoters recognized by alternative sigma factors are genes encoding virulence or virulence-associated proteins. Examples include the inlA gene of Listeria monocytogenes, the ica operon in Staphylococcus aureus (27), and the exotoxin genes of various clostridial species (17). In some cases, sigma factor homologues possess species-specific functions. For example, SigN RNA polymerase transcribes genes associated with virulence in Pseudomonas aeruginosa and Pseudomonas syringae, but SigN is not required for the transcription of virulence genes in Xanthomonas campestris (26, 27). Similarly, lipase gene expression is differentially regulated by the alternative sigma factor SigB in the closely related species Staphylococcus aureus and Staphylococcus epidermidis (27). While lipase secretion is elevated in an S. aureus sigB-null mutant (30), secretion of processed lipase in an S. epidermidis sigB-null mutant is reduced significantly (28).
In the nonpathogenic Bacillus species B. subtilis, the alternative sigma factor σH is required for transcription of a large number of genes that are essential for sporulation (7, 50, 51). As expected, deletion of the sigH homolog in B. anthracis abolished the ability of the mutant to sporulate, but surprisingly, this sigma factor gene was also required for anthrax toxin gene transcription. Thus, the sigH gene, well studied in B. subtilis for its role in development, has an additional role in B. anthracis as a key player in virulence gene expression.
In B. subtilis, the promoters of genes transcribed by σH RNA polymerase contain a conserved DNA sequence that is recognized by σH. A sequence closely resembling the B. subtilis σH consensus is present in the promoter regions of B. anthracis homologues of B. subtilis genes known to be transcribed by σH RNA polymerase. For the B. anthracis homologues, the sequence in the −35 region differs slightly from the consensus established for the B. subtilis genes. RNA transcripts were obtained from in vitro reactions using the promoter of one representative σH RNA polymerase-transcribed homologue, the B. anthracis spoVG gene, as the template. The promoter region of atxA, the major regulator of anthrax toxin gene transcription, does not bear sequence similarity to the σH consensus sequence established for B subtilis. Nevertheless, in in vitro reactions, σH RNA polymerase generated transcripts from the atxA promoter template. No in vitro transcripts were detected in comparable reactions employing toxin gene templates, indicating that σH RNA polymerase-mediated transcription from the atxA promoter was specific. Taken together, these data indicate that σH controls toxin gene expression indirectly, via its direct control of atxA gene transcription. Moreover, the B. anthracis sigma factor appears to recognize promoters lacking the σH consensus sequence established for B. subtilis σH.
The biochemical activity of the B. anthracis σH protein may not be identical to that of its B. subtilis counterpart. The predicted amino acid sequences of the B. anthracis and B. subtilis σH homologues differ by two residues within region 4.2, the domain responsible for recognition and binding of the −35 region (Fig. 6). The B. anthracis σH contains serine residues in positions 160 and 172 that are occupied by glycine and valine residues, respectively, in the B. subtilis σH protein. In addition to these differences, the 13 amino-terminal residues of the two proteins exhibit relatively low amino acid sequence similarity. Comparable differences among other sets of sigma factor homologues can result in altered target specificity and binding affinity (45).
FIG. 6.
Amino acid sequence comparison of the B. anthracis (B.a) and B. subtilis (B.s) homologs. Alignment was performed using the ClustalW web-based alignment program. The underlined sequence marks the position of region 4. Region 4 makes contact with the −35 box on the target promoter. Two significant amino acid differences are boxed.
Our investigation reveals newly discovered regulatory relationships between sigH, atxA, and another anthrax toxin gene regulator, abrB, when B. anthracis is grown in conditions that favor toxin gene expression. AbrB is a transition state regulator, best characterized in B. subtilis, that affects the transcription of multiple genes in a growth phase-dependent manner (42, 50). Steady-state levels of anthrax toxin gene transcripts are increased in a B. anthracis abrB-null mutant (46), and in vitro footprint analyses have revealed AbrB binding sites within the atxA promoter region (54). Thus, AbrB appears to affect toxin gene expression by controlling the transcription of atxA.
In experiments designed to model AbrB control of atxA in B. subtilis, Strauch et al. (54) showed that the activity of an atxA promoter-lacZ transcriptional fusion is increased in a B. subtilis abrB-null mutant. In B. subtilis, AbrB is one of the many factors that affect σH synthesis (54, 57). The sigH gene is subject to stringent transcriptional, posttranscriptional, and posttranslational control (32, 57). During the logarithmic phase of growth, when conditions are not conducive to sporulation, levels of σH are relatively low. Upon entry into the stationary growth phase, σH levels rise as a result of abrB repression by phosphorylated Spo0A, the master regulator of sporulation initiation. The spo0A gene has two transcriptional start sites, one of which is σH dependent. Thus, as a culture enters the stationary phase, the levels of Spo0A and σH, as members of the same positive feedback loop, rise (50). Strauch et al. (54) demonstrated that, when B. subtilis was cultured in conditions conducive to sporulation, the activity of an atxA promoter-lacZ transcriptional fusion was reduced in a sigH single mutant and increased in an abrB sigH double mutant. The change in atxA expression by the B. subtilis sigH mutant was attributed to σH control of abrB via Spo0A and AbrB control of atxA transcription. In contrast, our experiments investigating gene expression in B. anthracis revealed that the sigH effect on atxA expression is not dependent upon abrB. We note that the atxA promoter-lacZ transcriptional fusion used by Strauch et al. (54) contained only 200 nt that were upstream of the previously reported transcriptional start site for atxA and was thus devoid of the upstream region we have associated with σH RNA polymerase-mediated transcription. Moreover, there could be species-specific or growth condition-dependent differences in σH function. For our investigations, we grew B. anthracis in conditions favorable for toxin synthesis, CACO3 medium in 5% CO2. When cultured in this manner, B. anthracis cells produce high levels of the anthrax toxin proteins yet sporulate poorly even after prolonged growth (M. Hadjifrangiskou and T. M. Koehler, unpublished data) (37).
The implication of genes associated with the complex network of developmental control in B. anthracis toxin gene expression is one of a few intriguing insights into relationships between B. anthracis sporulation and virulence. Perego and coworkers (5, 8) recently reported genetic and biochemical analyses of components of the phosphorelay signal transduction system controlling development in Bacillus species. Nine sporulation kinase genes were identified in B. anthracis. Two of these contained frame shifts in all B. anthracis strains investigated, and one of them was also inactivated in a pathogenic strain of B. cereus harboring the B. anthracis toxin plasmid pXO1 (8). Their results suggest that acquisition of pXO1 and, possibly, virulence genes is associated with loss of sporulation sensor histidine kinase activities. Interestingly, in B. subtilis the sensor kinase genes kinA and kinE have σH-regulated promoters. We examined the promoter regions of the B. anthracis kinase genes for the σH consensus sequence. With the exception of BA4223, which has an apparent −10 sequence matching that of the σH consensus, none of the B. anthracis kinase genes are predicted on the basis of sequence data to be transcribed by σH RNA polymerase. Despite the lack of a σH consensus sequence in the promoter regions of these genes, the kinases may be transcribed directly by σH RNA polymerase in B. anthracis, as we have found for atxA. Alternatively, if the kinase genes are not recognized by σH RNA polymerase, this may be another indication that the pathway for B. anthracis development deviates from the pathway established for B. subtilis.
Additional evidence implicating pXO1 in B. anthracis development is the apparent incompatibility between atxA and plcR, a pleiotropic regulator of virulence genes in B. cereus and B. thuringiensis (1). The atxA gene, located on pXO1, is not found in most B. cereus and B. thuringiensis strains. The plcR gene is found in all three species but contains a nonsense mutation in all B. anthracis strains examined (49). Mignot et al. (35) reported that coexpression of plcR and atxA in a pXO1+ background prevented B. anthracis from sporulating efficiently. The sporulation defect was rescued in an atxA-null mutant, suggesting that the plcR and atxA regulons cannot successfully coexist in B. anthracis.
The results reported here, combined with previously published data, point to multiple means of control of atxA expression. Stringent control of atxA expression is in agreement with the results of a previous study indicating that steady-state levels of AtxA in B. anthracis are critical for optimal toxin synthesis. A recombinant strain harboring multiple copies of atxA and producing elevated levels of AtxA exhibited reduced pagA expression (14). We have found that a recombinant strain that overexpresses sigH produces reduced levels of the toxin proteins. Furthermore, as is true for Spo0A in B. subtilis (18), overexpression of sigH results in a delay in the onset of sporulation when B. anthracis is cultured in conditions that favor sporulation (Hadjifrangiskou and Koehler, unpublished data). The regulatory relationships between sigH and other genes controlling development may be species specific and/or vary with respect to growth conditions. Altered expression and/or function of these developmental regulators in B. anthracis may enable the bacterium to maximize toxin production during growth within a host where sporulation does not occur, while maintaining the ability to initiate sporulation upon a change in environment.
Acknowledgments
This work was sponsored by Public Health Service grant AI33537 from the National Institutes of Health.
We thank Masaya Fujita for providing anti-σH and anti-σA antisera, technical advice regarding in vitro transcription reactions, and critical reading of the manuscript. We also thank Jesus Eraso and the laboratory of Samuel L. Kaplan for helpful discussions and provision of reagents.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Okstad, A. B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M. 1993. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, NY.
- 3.Baillie, L., A. Moir, and R. Manchee. 1998. The expression of the protective antigen of Bacillus anthracis in Bacillus subtilis. J. Appl. Microbiol. 84:741-746. [DOI] [PubMed] [Google Scholar]
- 4.Banner, C. D. B., C. P. Moran, Jr., and R. Losick. 1983. Deletion analysis of a complex promoter for a developmentally regulated gene from B. subtilis. J. Mol. Biol. 168:351-365. [DOI] [PubMed] [Google Scholar]
- 5.Bongiorni, C., R. Stoessel, D. Shoemaker, and M. Perego. 2006. Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J. Bacteriol. 188:487-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourgogne, A., M. Drysdale, S. G. Hilsenbeck, S. N. Peterson, and T. M. Koehler. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 71:2736-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunsing, R. L., C. La Clair, S. Tang, C. Chiang, L. E. Hancock, M. Perego, and J. A. Hoch. 2005. Characterization of sporulation histidine kinases of Bacillus anthracis. J. Bacteriol. 187:6972-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brutsche, S., and V. Braun. 1997. SigX of Bacillus subtilis replaces the ECF sigma factor fecI of Escherichia coli and is inhibited by RsiX. Mol. Gen. Genet. 256:416-425. [DOI] [PubMed] [Google Scholar]
- 10.Carter, H. L., III, and C. P. Moran, Jr. 1986. New RNA polymerase sigma factor under spo0 control in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 83:9438-9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cataldi, A., A. Fouet, and M. Mock. 1992. Regulation of pag gene expression in Bacillus anthracis: use of a pag-lacZ transcriptional fusion. FEMS Microbiol. Lett. 77:89-93. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y., F. C. Tenover, and T. M. Koehler. 2004. β-Lactamase gene expression in a penicillin-resistant Bacillus anthracis strain. Antimicrob. Agents Chemother. 48:4873-4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai, Z., and T. M. Koehler. 1997. Regulation of anthrax toxin activator gene (atxA) expression in Bacillus anthracis: temperature, not CO2/bicarbonate, affects AtxA synthesis. Infect. Immun. 65:2576-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai, Z., J. C. Sirard, M. Mock, and T. M. Koehler. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 16:1171-1181. [DOI] [PubMed] [Google Scholar]
- 16.Dubnau, E., J. Weir, G. Nair, L. Carter III, C. Moran, Jr., and I. Smith. 1988. Bacillus sporulation gene spo0H codes for σ30 (σH). J. Bacteriol. 170:1054-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupuy, B., S. Raffestin, S. Matamouros, N. Mani, M. R. Popoff, and A. L. Sonenshein. 2006. Regulation of toxin and bacteriocin gene expression in Clostridium by interchangeable RNA polymerase sigma factors. Mol. Microbiol. 60:1044-1057. [DOI] [PubMed] [Google Scholar]
- 18.Fujita, M., and R. Losick. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita, M., and Y. Sadaie. 1998. Feedback loops involving Spo0A and AbrB in in vitro transcription of the genes involved in the initiation of sporulation in Bacillus subtilis. J. Biochem. (Tokyo) 124:98-104. [DOI] [PubMed] [Google Scholar]
- 20.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene, E. A., and G. B. Spiegelman. 1996. The Spo0A protein of Bacillus subtilis inhibits transcription of the abrB gene without preventing binding of the polymerase to the promoter. J. Biol. Chem. 271:11455-11461. [DOI] [PubMed] [Google Scholar]
- 22.Haldenwang, W. G., N. Lang, and R. Losick. 1981. A sporulation-induced sigma-like regulatory protein from B. subtilis. Cell 23:615-624. [DOI] [PubMed] [Google Scholar]
- 23.Hoch, J. A. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47:441-465. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmaster, A. R., and T. M. Koehler. 1997. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect. Immun. 65:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmaster, A. R., and T. M. Koehler. 1999. Control of virulence gene expression in Bacillus anthracis. J. Appl. Microbiol. 87:279-281. [DOI] [PubMed] [Google Scholar]
- 26.Horns, T., and U. Bonas. 1996. The rpoN gene of Xanthomonas campestris pv. vesicatoria is not required for pathogenicity. Mol. Plant-Microbe Interact. 9:856-859. [DOI] [PubMed] [Google Scholar]
- 27.Kazmierczak, M. J., M. Wiedmann, and K. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kies, S., M. Otto, C. Vuong, and F. Gotz. 2001. Identification of the sigB operon in Staphylococcus epidermidis: construction and characterization of a sigB deletion mutant. Infect. Immun. 69:7933-7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its functions as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewandoski, M., E. Dubnau, and I. Smith. 1986. Transcriptional regulation of the spo0F gene of Bacillus subtilis. J. Bacteriol. 168:870-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, J., W. M. Cosby, and P. Zuber. 1999. Role of lon and clpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol. Microbiol. 33:415-428. [DOI] [PubMed] [Google Scholar]
- 33.McQuade, R. S., N. Comella, and A. D. Grossman. 2001. Control of a family of phosphatase regulatory genes (phr) by the alternate sigma factor sigma-H of Bacillus subtilis. J. Bacteriol. 183:4905-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mignot, T., M. Mock, and A. Fouet. 2003. A plasmid-encoded regulator couples the synthesis of toxins and surface structures in Bacillus anthracis. Mol. Microbiol. 47:917-927. [DOI] [PubMed] [Google Scholar]
- 35.Mignot, T., M. Mock, D. Robichon, A. Landier, D. Lereclus, and A. Fouet. 2001. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol. Microbiol. 42:1189-1198. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, New York, NY.
- 37.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 38.Perego, M. 1998. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 6:366-370. [DOI] [PubMed] [Google Scholar]
- 39.Perego, M., P. Glaser, and J. A. Hoch. 1996. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol. Microbiol. 19:1151-1157. [DOI] [PubMed] [Google Scholar]
- 40.Perego, M., and J. A. Hoch. 1996. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:1549-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perego, M., and J. A. Hoch. 1996. Protein aspartate phosphatases control the output of two-component signal transduction systems. Trends Genet. 12:97-101. [DOI] [PubMed] [Google Scholar]
- 42.Phillips, Z. E., and M. A. Strauch. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 59:392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Predich, M., G. Nair, and I. Smith. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing σH. J. Bacteriol. 174:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi, F. X., and R. H. Doi. 1990. Localization of a second SigH promoter in the Bacillus subtilis sigA operon and regulation of dnaE expression by the promoter. J. Bacteriol. 172:5631-5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez-Romero, M. A., I. Masulis, M. A. Cevallos, V. Gonzalez, and G. Davila. 2006. The Rhizobium etli σ70 (SigA) factor recognizes a lax consensus promoter. Nucleic Acids Res. 34:1470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Sirard, J. C., M. Mock, and A. Fouet. 1994. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J. Bacteriol. 176:5188-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slamti, L., S. Perchat, M. Gominet, G. Vilas-Boas, A. Fouet, M. Mock, V. Sanchis, J. Chaufaux, M. Gohar, and D. Lereclus. 2004. Distinct mutations in PlcR explain why some strains of the Bacillus cereus group are nonhemolytic. J. Bacteriol. 186:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonenshein, A. L. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561-566. [DOI] [PubMed] [Google Scholar]
- 51.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 52.Strauch, M., V. Webb, G. Spiegelman, and J. A. Hoch. 1990. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. USA 87:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strauch, M. A. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J. Bacteriol. 177:6999-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strauch, M. A., P. Ballar, A. J. Rowshan, and K. L. Zoller. 2005. The DNA-binding specificity of the Bacillus anthracis AbrB protein. Microbiology 151:1751-1759. [DOI] [PubMed] [Google Scholar]
- 55.Thorne, C. B. 1968. Transduction in Bacillus cereus and Bacillus anthracis. Bacteriol. Rev. 32:358-361. [PMC free article] [PubMed] [Google Scholar]
- 56.Thorne, C. B., and F. C. Belton. 1957. An agar-diffusion method for titrating Bacillus anthracis immunizing antigen and its application to a study of antigen production. J. Gen. Microbiol. 17:505-516. [DOI] [PubMed] [Google Scholar]
- 57.Weir, J., M. Predich, E. Dubnau, G. Nair, and I. Smith. 1991. Regulation of spo0H, a gene coding for the Bacillus subtilis sigma H factor. J. Bacteriol. 173:521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]