Abstract
A newly isolated bacterium, Cohnella laevoribosii RI-39, could grow in a defined medium with l-ribose as the sole carbon source. A 21-kDa protein isomerizing l-ribose to l-ribulose, as well as d-lyxose to d-xylulose, was purified to homogeneity from this bacterium. Based on the N-terminal and internal amino acid sequences of the purified enzyme obtained by N-terminal sequencing and quantitative time of flight mass spectrometry-mass spectrometry analyses, a 549-bp gene (lyxA) encoding d-lyxose (l-ribose) isomerase was cloned and expressed in Escherichia coli. The purified endogenous enzyme and the recombinant enzyme formed homodimers that were activated by Mn2+. C. laevoribosii d-lyxose (l-ribose) isomerase (CLLI) exhibits maximal activity at pH 6.5 and 70°C in the presence of Mn2+ for d-lyxose and l-ribose, and its isoelectric point (pI) is 4.2 (calculated pI, 4.9). The enzyme is specific for d-lyxose, l-ribose, and d-mannose, with apparent Km values of 22.4 ± 1.5 mM, 121.7 ± 10.8 mM, and 34.0 ± 1.1 mM, respectively. The catalytic efficiencies (kcat/Km) of CLLI were 84.9 ± 5.8 mM−1 s−1 for d-lyxose (Vmax, 5,434.8 U mg−1), 0.2 mM−1 s−1 for l-ribose (Vmax, 75.5 ± 6.0 U mg−1), and 1.4 ± 0.1 mM−1 s−1 for d-mannose (Vmax, 131.8 ± 7.4 U mg−1). The ability of lyxA to permit E. coli cells to grow on d-lyxose and l-ribose and homology searches of other sugar-related enzymes, as well as previously described sugar isomerases, suggest that CLLI is a novel type of rare sugar isomerase.
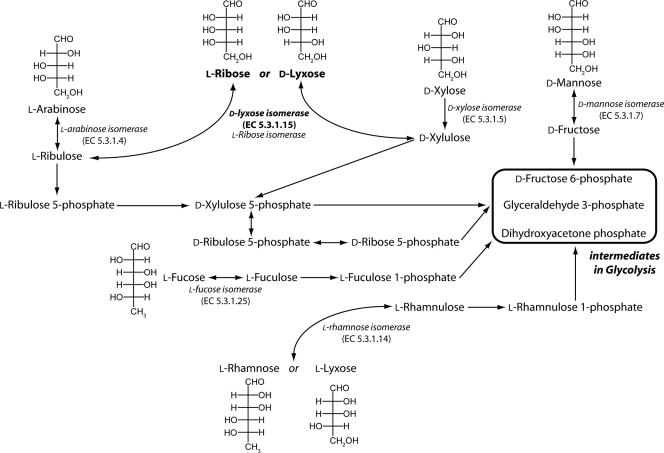
The pentose phosphate pathway that generates NADPH is a central pathway for producing metabolic energy. Glucose-6-phosphate is oxidized to ribulose-5-phosphate by the sequential action of glucose-6-phosphate dehydrogenase, lactonase, and 6-phosphogluconate dehydrogenase (15, 16, 23, 24). The ribulose-5-phosphate is epimerized to xylulose-5-phosphate and is also converted to ribose-5-phosphate, which can be transformed into glycolytic intermediates in response to physiological requirements. Prokaryotes can use a variety of sugars as carbon sources for growth, and several rare pathways of sugar metabolism leading to intermediates of the pentose phosphate have been suggested by genetic and physiological studies (Fig. 1.) (1, 16, 21, 38). However, the identities of the enzymes involved remain obscure.
FIG. 1.
Possible pathways of pentose metabolism. d-Lyxose can be isomerized to d-xylulose, an intermediate in the pentose phosphate pathway. l-Ribose can be converted to l-ribulose or reduced to ribitol (4), and l-lyxose can be used by the l-rhamnose pathway found in certain E. coli mutants (42). Glycolytic intermediates which can be connected to the pentose phosphate pathway are enclosed in a box.
d-Lyxose isomerase (LI) (E.C. 5.3.1.15) is defined as an enzyme that catalyzes the reversible isomerization of d-lyxose to d-xylulose, which is involved in the pentose phosphate pathway (3). However, d-lyxose (the 2-epimer of d-xylose) is not found in many microorganisms, and the mechanisms of transport and metabolism of this compound are not well defined. During the past few decades several enzymes have been proposed to catalyze the isomerization of d-lyxose to d-xylulose (3, 17, 18, 30). Some examples of these enzynes are the d-mannose isomerases (MI) (E.C. 5.3.1.7) from Agrobacterium radiobacter M-1 (18) and Mycobacterium smegmatis (17). Although these MIs are induced by the presence of d-lyxose as the sole carbon source, they exhibit very low levels of activity with this sugar, indicating that they are not primarily responsible for isomerization of d-lyxose to d-xylulose. In addition, Klebsiella pneumoniae PRL-R3 cannot grow on d-lyxose, but mutants of this organism that can grow on d-lyxose have been isolated (32). An enzyme with isomerization activity for d-lyxose and d-mannose was purified from one such mutant, but it seemed to be an MI rather than an LI since it had only weak isomerization activity with d-lyxose. Although an LI has been partially purified from Aerobacter aerogenes PRL-R3 (3), it has not been genetically identified. Recently, an l-ribose isomerase (RI) catalyzing the reversible conversion of l-ribulose to l-ribose, as well as the conversion of d-lyxose to d-xylulose, was isolated from Acinetobacter sp. strain DL-28 (30, 35). No enzyme of this type was previously known to exist. This enzyme was induced by d-lyxose as a carbon source, and it was 47% and 4% as active on d-lyxose and d-mannose, respectively, as it was on l-ribose indicating that it may permit growth not only on l-ribose but also on d-lyxose as a carbon source (30). However this observation needs to be confirmed since Escherichia coli mutants that are able to utilize l-ribose for growth do so not via an l-ribose and/or d-lyxose isomerase but via the ribitol pathway (42).
The nonnaturally occurring aldopentose sugars, such as l-ribose and lyxose, have attracted much attention as synthetic intermediates for the production of pharmaceutical drugs (2, 10, 34, 45). l-Ribose in particular is a good starting material for the synthesis of l-nucleosides, which are being used to develop antiviral agents that are less toxic than the corresponding d-nucleosides (14, 22). In addition, oligonucleotides composed of 2-deoxy-l-ribose are resistant to digestion by certain nucleases (10). Recently, it was reported that 2-deoxy-l-ribose and its analogs enhanced apoptosis and suppressed the growth of tumors by competitively inhibiting reactions involving 2-deoxy-d-ribose, and it was suggested that these analogs held promise as antitumor agents (5, 7, 29, 43). d-Lyxose can also be used as starting material for the production of antitumor and immunostimulatory α-galactosylceramide agents that have been found to be active against several murine tumors (31, 39). In light of this, a great deal of effort has been directed toward the chemical synthesis of l-ribose and d-lyxose (6, 10, 34, 45). However, since chemical synthesis methods have a number of drawbacks, such as the complexity of the steps involved, long reaction times, and the formation of unnecessary by-products, the possibility of producing l-ribose using microorganisms and/or enzymes has been raised by Granström et al. (13).
In this study, we attempted to isolate a strain that is able to use l-ribose as a carbon source and to identify the gene conferring this rare ability. We cloned and characterized a gene encoding a potential d-lyxose (l-ribose) isomerase from a novel strain, Cohnella laevoribosii RI-39, isolated from hot springs in a volcanic area. This enzyme exhibited high isomerizing activity for d-lyxose, d-mannose, and l-ribose. Here we report the cloning, expression, purification, and characterization of this novel d-lyxose (l-ribose) isomerase.
MATERIALS AND METHODS
Materials.
The pBluescript II KS(+) vector for construction of genomic DNA libraries was obtained from Stratagene, the pGEM-T Easy vector for construction of an expression vector and for sequencing was obtained from Promega, and the pET22b(+) expression vector was obtained from Novagen. Ex-Taq DNA polymerase, deoxynucleoside triphosphates (dNTPs), chemicals for PCR, and restriction enzymes were obtained from Takara Biomedicals. T4 DNA ligase was obtained from Promega. Oligonucleotides were synthesized by Cosmo. Thermal cycling was performed using a ThermoHybaid PCR system. Genomic tip and plasmid miniprep kits were purchased from QIAGEN, and all columns for purification were obtained from Amersham Biosciences. Centriprep YM-10 concentrators and Millex syringe-driven filter units (0.22 μm) were purchased from Millipore. Electrophoresis reagents were obtained from Bio-Rad, and all chemicals used for enzyme assays and characterization were obtained from Sigma.
Bacterial strains and culture conditions.
To isolate microorganisms able to grow on l-ribose, hot spring water samples collected from Likupang, Indonesia, were screened on modified minimal salts agar plates supplemented with 0.5 to 1% l-ribose as the sole carbon source incubated at 50°C. In order to isolate single colonies and confirm that they utilize l-ribose, screened strains were serially diluted and subcultured onto a modified EM-1 medium (28) supplemented with (per liter) 5 g of l-ribose, 3 g of yeast extract, 2 g of NaCl, 0.1 g of CaCl2, 0.5 g of MgSO4, 0.5 g of MgCl2, 0.5 g of KH2PO4, 0.125 g of K2HPO4, 0.5 g of (NH2)2SO4, and 0.1 g of KCl. The strain with the highest l-ribose activity was isolated and designated C. laevoribosii sp. nov. strain RI-39 (E. A. Cho, J. S. Lee, K. C. Lee, H. C. Jung, J. G. Pan, and Y. R. Pyun, submitted for publication). The newly isolated organism was grown aerobically in 10 liters of EM-1 medium at 50°C for 56 h and harvested by centrifugation at 10,000 × g for 20 min, and the cell pellet was washed twice with 50 mM sodium phosphate buffer (pH 6.5). The washed cells were stored at −80°C until they were used. E. coli TOP10 was used to construct a genomic DNA library, E. coli DH5α was used to construct the expression vector, and E. coli BL21(DE3) was used for expression. Each of the E. coli strains was grown overnight in Luria-Bertani medium with ampicillin (100 μg/ml) in a rotary shaker at 37°C. Complementation experiments were performed in minimal M9 medium supplemented with each carbon source at a concentration of 0.4% and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Growth was monitored by determining the absorbance at 660 nm with an HP8452A spectrophotometer (Hewlett-Packard).
Purification of the l-ribose-isomerizing enzyme from C. laevoribosii.
All purification steps were performed at 4°C. Pelleted cells (from 10 liters) were resuspended in 200 ml of 50 mM sodium phosphate buffer (pH 6.5) and disrupted by sonication. The lysate was centrifuged at 10,000 × g for 30 min to remove cell debris, and the supernatant was loaded onto a DEAE-Sephacel ion-exchange column (20 ml) equilibrated with the same buffer. The column was washed with 20 column volumes of the same buffer, and a linear gradient of NaCl (from 0 to 0.5 M) was applied to elute the absorbed proteins. The fractions (60 ml, 150 mM NaCl) containing enzyme activity were pooled, (NH4)2SO4 was added to a concentration of 1 M, the suspension was filtered through a 0.2-μm filter, and the filtrate was loaded onto a RESOURCE PHE column (1 ml) equilibrated with 50 mM sodium phosphate (pH 6.5) buffer containing 1 M (NH4)2SO4. The absorbed proteins were eluted with a descending gradient of (NH4)2SO4 (from 1 to 0 M) at a flow rate of 0.5 ml/min. Fractions containing the l-ribose-isomerizing enzyme [6 ml, 1 to 0.5 M (NH4)2SO4] were pooled and dialyzed against 50 mM sodium phosphate (pH 6.5) buffer. The purity of the preparations was checked by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (26). Protein concentrations were determined by the bicinchoninic acid method (36) with bovine serum albumin as the standard. The purified enzyme was divided into aliquots and stored at −20°C. Under these conditions the enzyme activity remained almost constant for several months.
Cloning and expression of C. laevoribosii l-ribose-isomerizing enzyme in E. coli.
Based on the N-terminal and internal amino acid sequences of the purified l-ribose-isomerizing enzyme (Fig. 2), we designed two degenerate primers, forward primer F-RI-n-term (5′-GGATGMGIGGNACNGA-3′) and reverse primer R-RI-intern (5′-GGTCIGTRTTIADRTANGTNAC-3′), to amplify a sequence encoding a region from the N terminus (MRGTE) to the internal sequence (DTNLYTV) of the protein. Genomic DNA of C. laevoribosii extracted with a genomic tip kit (QIAGEN) was used as the template for the PCR. The PCR mixture (50 μl) contained 300 ng of genomic DNA, 10 pmol of each primer, 1× PCR buffer containing 2 mM MgCl2, each dNTP at a concentration of 0.2 mM, and 2.5 U of Ex-Taq DNA polymerase. The PCR was carried out for 30 cycles consisting of 50 s of denaturation at 94°C, 40 s of annealing at 50°C, and 20 s of extension at 72°C. The 153-bp PCR product was extracted from an agarose gel, subcloned into the pGEM-T Easy vector, and sequenced (Fig. 2).
FIG. 2.
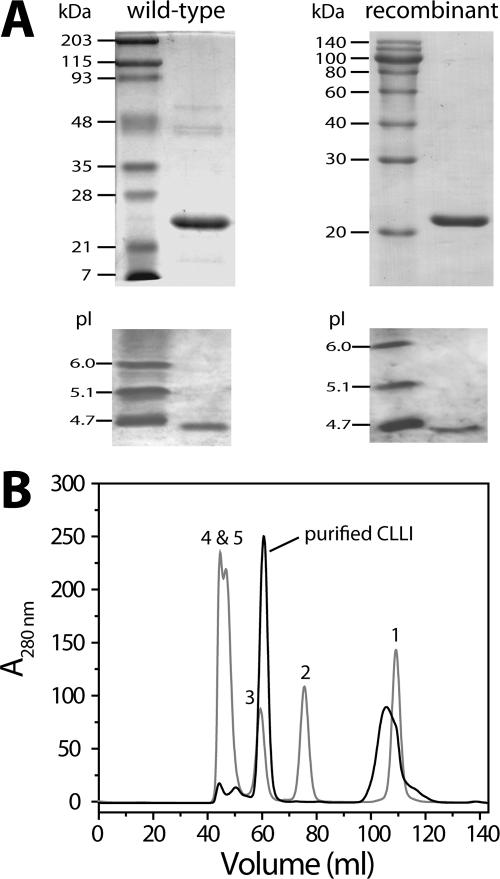
SDS-PAGE, isoelectric focusing, and gel filtration analyses of CLRI. (A) Purified wild-type and recombinant CLRIs were analyzed by SDS-12% PAGE (top), and purified proteins were also analyzed by isoelectric focusing (pH 3.0 to 10.0) (bottom). Left lane, molecular weight standards; right lane, purified CLRI. (B) Pooled fractions showing RI activity from the previous step were applied to Superdex 75 calibrated with the standard proteins (Bio-Rad). Peak 1, vitamin B12 (1.359 kDa); peak 2, myoglobin (17 kDa); peak 3, ovalbumin (44 kDa); peak 4, globulin (158 kDa); peak 5, thyroglobulin (670 kDa).
Next, in order to obtain the entire RI coding sequence, we constructed a genomic library from genomic DNA partially digested with Sau3AI. DNA fragments (2.5 to 5 kb) purified with a QIAEX II gel extraction kit were ligated into the BamHI site of the pBluescript II KS(+) vector, and the resulting genomic library was introduced into E. coli TOP10 by electroporation using a Gene-Pulser (Bio-Rad). We designed four primers, primers AF (5′-GGTGGCGGAGAT-GTTTC-3′), CF (5′-GAACTGGAAAAGGTGGAGG-TCG-3′), BR (5′-GCCAAGTTCCCCAA-TCC-3′), and CR (5′-CGATGGCGTCAGGGCGATG-3′), based on the 153-bp DNA sequence obtained. After this, nested PCR was performed using pairs of these primers together with primers containing the T3 and T7 promoter coding sequences of the pBluescript II KS(+) vector, AR-T3 (5′-AATTAACCCTCACTAAAG-3′) and BF-T7 (5′-TAATACGACTCACTATAGGG-3′), to amplify DNA fragments which we hoped would contain the complete RI coding sequence. Each reaction mixture (50 μl) contained 2 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 500 ng of DNA, 10 pmol of each primer in 1× PCR buffer, and 2.5 U of Ex-Taq DNA polymerase. The thermal cycler parameters were as follows: 30 cycles of 50 s at 94°C, 40 s at 45 to 55°C, and 90 to 300 s at 72°C. The various nested PCR products (0.3, 0.5, 0.6, 0.9, 1, 1.2, and 2.5 kb) were extracted from an agarose gel, cloned into the pGEM-T Easy vector, and sequenced by the procedure described above. In this way we generated a 549-bp open reading frame (ORF) and compared it to sequences obtained from a BLAST search. On the basis of the DNA sequence, primers NDE-RI-F (5′-CCATATGCGGGGGACGGAATGGCGG-3′) and XHO-RI-R (5′-CCTCGAGTCAATCG-TCAAACTCGATGACCG-3′), which incorporated NdeI and XhoI restriction enzyme sites (underlined) at each end of the 549-bp DNA gene, were used to amplify the complete DNA encoding RI and cloned it into pET-22b(+) digested with NdeI and XhoI to obtain pET-CLRI-549. This plasmid was transformed into E. coli BL21(DE3). For expression, a transformant was grown at 37°C in 1 liter of Luria-Bertani medium containing 100 μg of ampicillin per ml and induced in the mid-exponential phase (A600, 0.5) by adding IPTG (final concentration, 1 mM). After a further 5 h of growth, the cells were harvested by centrifugation (10,000 × g, 20 min, 4°C).
To purify the recombinant enzyme, the pellet was resuspended in 50 ml of 50 mM sodium phosphate buffer (pH 6.5) and disrupted by sonication. The lysate was centrifuged (10,000 × g, 30 min) to remove cell debris, and the supernatant was filtered though a 0.2-μm filter, applied to a DEAE-Sephacel column (20 ml) equilibrated with 50 mM sodium phosphate (pH 6.5), and eluted with a linear gradient of NaCl (0 to 500 mM). The fractions containing RI activity (50 ml, 150 mM NaCl) were pooled and concentrated with a Centriprep 10K (Millipore). The concentrate was loaded onto a Superdex 75 column (HiLoad 16/60 prep grade; Pharmacia) preequilibrated with 50 mM sodium phosphate (pH 6.5) containing 150 mM NaCl. The purified enzyme was dialyzed against 50 mM sodium phosphate buffer (pH 6.5) and stored at −20°C.
Assay of enzyme activity.
Sugar-isomerizing activity was determined by measuring the accumulation of ketose. The standard reaction mixture (125 μl) contained 50 mM sodium phosphate buffer (pH 6.5), 1 mM MnCl2 (or NiCl2), 25 μl of an enzyme preparation at a suitable dilution, and 20 mM d-lyxose (or l-ribose) and was incubated at 70°C for 10 min. The reaction was stopped by cooling on ice. The ketose (l-ribulose from l-ribose and d-xylulose from d-lyxose) was quantified by the cysteine-carbazole method (9), and the A560 was determined as previously described (27). One unit of enzyme activity was defined as the amount of enzyme that produced 1 μl of product per min under the assay conditions.
Determination of N-terminal and internal amino acid sequences.
The purified protein was analyzed by 12% SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane for N-terminal and internal sequence determination. N-terminal sequencing of the purified protein was performed at the Analytical Core Facility of Tufts University (Boston, MA). Internal amino acid sequencing was carried out by mass spectrometry (MS) as follows. Four of the peptide fragments obtained by tryptic digestion for 4 h at 37°C were sequenced by the quantitative time of flight MS-MS method. Isoelectric focusing was carried out with a protein II Ready Gel precast system (pH range, 3.0 to 10.0; Bio-Rad) in a stepped fashion, as follows: 15 min at 100 V, 15 min at 200 V, and 60 min at 450 V, all at 4°C.
Physicochemical characterization.
The effect of temperature on isomerization was determined using the standard protocol. To determine the effect of pH on enzyme activity, the sodium phosphate buffer was replaced by 50 mM sodium acetate buffer (pH 4.0 to 6.0), phosphate buffer (pH 6.0 to 7.5), HEPES buffer (pH 7.0 to 8.0), Tris-HCl buffer (pH 7.5 to 9.0), and glycine-NaOH buffer (pH 9.0 to 10.0). To assess the substrate specificity of the purified enzyme, various sugar substrates were added to reaction mixtures.
Effect of divalent metals on activity.
Enzyme samples were incubated with 10 mM EDTA at 25°C for 3 h, followed by overnight dialysis against 50 mM sodium phosphate buffer (pH 6.5) at 4°C with several changes of buffer. The effects of various metal ions were assessed by adding CoCl2 · 6H2O, MnCl2 · 4H2O, MgCl2 · 6H2O, CaCl2 · 2H2O, ZnCl2 · 6H2O, CuCl2 · 2H2O, FeCl2 · 6H2O, or NiCl2 · 6H2O at concentrations of 1 and 10 mM to the dialyzed enzyme and assaying the isomerizing activity under standard conditions without 1 mM MnCl2.
Determination of kinetic parameters.
Reactions were performed at 60°C with l-ribose, d-lyxose, and d-mannose as substrates as described above, except that isomerization activity was assayed after incubation for 3 min to obtain the initial reaction rates. The substrate concentrations ranged from 1 to 300 mM. To determine Vmax and Km values, the Levenberg-Marquardt algorithm (KaleidaGraph program; Synergy Software) was used to fit the kinetic data to the Michaelis-Menten equation using nonlinear-squares regression.
Analysis of isomerization products.
The isomerization mixtures (1 ml) contained 10 mM d-lyxose (or l-ribose), 1 mM MnCl2, and 0.5 mg of purified enzyme in 50 mM sodium phosphate buffer (pH 6.5). After various periods (0 to 6 h) of incubation at 60°C, they were freeze-dried under a vacuum. For gas chromatography (GC)-MS analysis, all products were converted to trimethylsilyl ethers by adding Sylon BTZ [N,O-bis(trimethylsilyl)acetamide—chlorotrimethylsilane-N-trimethylsilylimidazole, 3:2:3; Supelco, Bellefonte, PA] silylating reagent, using the procedure supplied with the reagent (41). Pyridine (200 μl) and Sylon BTZ (100 μl) were added to the freeze-dried residue, which was allowed to react for 5 min at room temperature. GC was performed using a Hewlett-Packard series HP6890 gas chromatograph equipped with a split injector and mass spectrometer detection system (HP5973). The column was an HP-5MS column (60 m by 0.25 mm [inside diameter]; film thickness, 0.25 μm), the injection temperature was 290°C, and the sample injection volume was 1 μl with a split ratio of 50:1. The carrier gas was helium at a flow rate of 0.8 ml/min. The following temperature gradient was used for eluting the products: 100°C for 5 min, an increase to 150°C at a rate of 10°C/min, maintenance at 150°C for 15 min, an increase to 250°C at a rate of 10°C/min, maintenance at 250°C for 15 min, and finally an increase to 300°C at a rate of 10°C/min and maintenance at 300°C for 60 min (total run time, 115 min). Mass spectra were collected by using the scan mode, and the solvent delay time was 15 min. For qualitative analysis, the samples and authentic standard substances (l-ribose, d-ribose, d-lyxose, d-ribulose, l-arabinose, d-arabinose, and d-xylulose) were verified by comparing the GC retention times and the total ion chromatogram. Using overlapped peaks, all of the mass spectra were compared with the spectra in a mass spectrum library (Wiley 275) or with the spectra of the trimethylsilyl derivatives of authentic standard compounds.
Nucleotide sequence accession number.
The GenBank/EMBL accession number for the C. laevoribosii lyxA gene is DQ978225.
RESULTS
Isolation of l-ribose-utilizing C. laevoribosii and purification of an l-ribose-isomerizing enzyme.
We examined 50 l-ribose-utilizing strains isolated from Indonesian hot spring water samples after 7 to 14 days of incubation on minimal EM-1 medium containing 1% l-ribose at 45°C and pH 6.5. Isolate RI-39 grew to the maximum cell density on l-ribose as the sole carbon source in only 2 to 3 days in several subcultures on minimal EM-1 medium containing 0.5% l-ribose. The newly isolated l-ribose-utilizing strain was identified as a new species of the genus Cohnella on the basis of morphological, physicochemical, chemotaxonomic, and molecular criteria, and it was designated C. laevoribosii RI-39 (Cho et al., submitted). Its l-ribose-isomerizing activity was induced by l-ribose and d-lyxose but not by glucose and other aldopentoses tested.
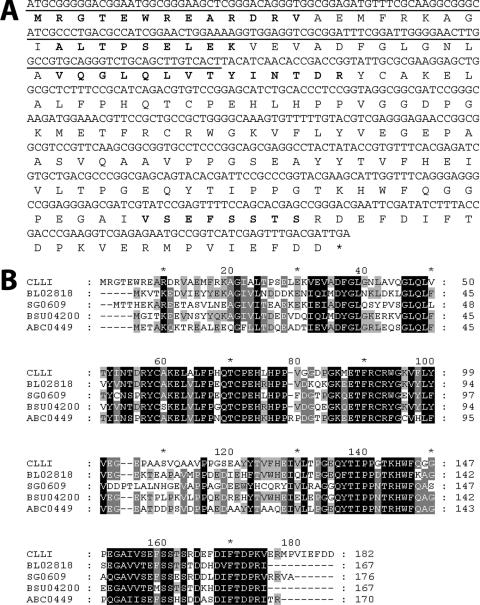
Table 1 summarizes the results of a typical purification from a 10-liter culture of C. laevoribosii RI-39. C. laevoribosii l-ribose isomerase (CLRI) was purified to homogeneity by DEAE-Sephacel ion-exchange chromatography, followed by RESOURCE PHE hydrophobic chromatography. This procedure yielded purified CLRI with a specific activity of 2 U/mg. As shown in Fig. 2A, the purified enzyme had a molecular mass of approximately 21 kDa based on SDS-PAGE and a pI of about 4.2 based on isoelectric focusing. The presence of a single protein was confirmed by N-terminal amino acid sequencing. The sequence (MRGTEWREARDRVAEMFRKA) exhibited no homology with the sequences of other sugar isomerases that have been described. To determine internal amino acid sequences, the protein band isolated by SDS-PAGE was proteolytically digested with trypsin, and the resulting peptides were separated by reversed-phase high-performance liquid chromatography. Four internal peptide sequences were determined by automated Edman degradation using quantitative time of flight MS-MS; these sequences were LVTYLNTDR, VSEFSSTS, ALTPSELEK, and VQGLQLVTYINTDR (Fig. 3).
TABLE 1.
Purification of RI from C. laevoribosii RI-39 and recombinant CLRI expressed in E. colia
| Enzyme | Purification step | Protein (mg) | Activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Native CLRI | Cell extract | 880 | 52.7 | 0.06 | 100 | 1 |
| DEAE-Sephacel | 22 | 23.2 | 1.05 | 44 | 18 | |
| RESOURCE PHE | 4.2 | 8.4 | 2.0 | 16 | 33 | |
| Recombinant CLRI | Cell extract | 200 | 120 | 0.6 | 100 | 1 |
| DEAE-Sephacel | 22 | 63.7 | 2.9 | 46 | 5 | |
| Superdex 75 | 12 | 50.2 | 4.2 | 41 | 7 |
Enzyme activity was measured at 70°C using l-ribose as the substrate (see Materials and Methods).
FIG. 3.
(A) Nucleotide sequence of the CLRI gene and deduced amino acid sequence of CLRI. N-terminal and internal amino acid sequences obtained from the native enzyme are in bold type. An asterisk indicates the translation stop codon. (B) Alignment of the amino acid sequences of CLRI and microbial homologues. Conserved residues are highlighted with a black background; partially conserved residues are highlighted with a gray background. The alignment was generated using CLUSTAL X (40). Database searching was performed using the BLAST program at the NCBI website (www.ncbi.nlm.nih.gov/BLAST). CLRI, C. laevoribosii RI (lyxA; NCBI accession no. DQ978225); BL02818, B. licheniformis hypothetical protein (ydaE; AAU22106); SG0609, S. glossinidius conserved hypothetical protein (BAE73884); BSU04200, B. subtilis hypothetical protein (ydaE; CAB12227); ABC0449, B. clausii conserved hypothetical protein (ydaE; BAD62991).
Cloning, expression, and purification of CLRI.
Using degenerate primers based on the N-terminal and internal sequences of CLRI, a 153-bp PCR product was obtained from C. laevoribosii genomic DNA (Fig. 3A). The sequence of this 153-bp product was used to design two pairs of forward and reverse primers together with a pair of primers based on the T7 and T3 promoter coding sequences of the pBluescript II KS(+) vector. Pairs of these primers were used in nested PCR to obtain a full-length product encoding CLRI from a genomic library constructed in the pBluescript II KS(+) vector. In the process, DNA fragments that were about 0.3 to 1.2 kb long were cloned into the pGEM-T Easy vector, sequenced, and shown to contain the putative ORF of CLRI (549 bp), which also contained the N-terminal and internal sequences obtained from purified CLRI (Fig. 3A). In order to amplify the intact 549-bp CLRI coding gene, PCR was performed using primers NDE-RI-F and XHO-RI-R (see Materials and Methods). The PCR product was cloned into the pET-22b(+) vector to generate pET-CLRI-549 and sequenced. Analysis of the sequence demonstrated that the 549-bp RI gene of C. laevoribosii encoded an 182-amino-acid polypeptide with a calculated molecular mass of 20,351 Da and a calculated pI of 4.9.
The gene for C. laevoribosii CLRI in pET-CLRI-549 was expressed upon induction with IPTG. As shown in Fig. 2, the recombinant CLRI was purified sevenfold by DEAE-Sephacel chromatography followed by Superdex 75 chromatography, and the yield was 41% (Table 1). Using these purification steps, we obtained approximately 10 mg of purified recombinant CLRI from 1 liter of E. coli BL21 culture broth. The apparent molecular mass of the protein was estimated to be 21 kDa by SDS-PAGE, which is consistent with the molecular mass calculated from the presumptive amino acid sequence. In addition, gel filtration data indicated that the native recombinant enzyme had a molecular mass of about 42 kDa and therefore was a homodimer. We performed isoelectric focusing with the wild-type and recombinant enzymes and obtained pI values of approximately 4.2, which were slightly higher than the calculated pI (Fig. 2A).
Physicochemical and functional characterization of CLRI.
The temperature dependence of the wild-type and recombinant CLRI was determined in the presence of 1 mM Mn2+ after 10 min of incubation at various temperatures. The apparent optimum temperature for both the wild-type and recombinant enzymes was 70°C. The pH dependence of the wild-type and recombinant enzymes was determined after 10 min of incubation at various pHs. The pH optimum for both enzyme preparations was 6.5. To see whether these enzymes require divalent metals as cofactors for activity, as is generally the case for sugar isomerases, the purified native and recombinant enzymes (the activities of as-isolated enzymes were defined as 100%) were treated with 10 mM EDTA for 3 h at 25°C and dialyzed against 50 mM sodium phosphate buffer (pH 6.5). Various divalent cations were then added to the apo proteins (21.5% and 14.2% for native and recombinant C. laevoribosii d-lyxose [l-ribose] isomerases [CLLIs], respectively), and the standard assay was performed with d-lyxose as a substrate. CLRI exhibited a dependence on divalent metal ions such as 1 mM Mn2+ (470% and 368%), 1 mM Ni2+ (77% and 70%), and 1 mM Co2+ (27% and 45%). The observations described above show that the physicochemical properties of the recombinant enzyme were identical to those of the native RI from C. laevoribosii.
To investigate the substrate specificity of CLRI, various aldopentoses and aldohexoses were tested as substrates. Interestingly, of the aldoses tested, the enzyme exhibited the highest isomerization activity with d-lyxose (13.4 U/mg) in the presence of 1 mM Mn2+. The enzyme was also active with l-ribose (0.1 U/mg in the presence of 1 mM Ni2+) and d-mannose (0.8 U/mg in the presence of 1 mM Mn2+), but not with the aldopentoses and hexoses tested (l-lyxose, d-xylose, l-xylose, d-arabinose, l-arabinose, d-ribose, l-fucose, d-glucose, d-galactose, d-erythrose, l-rhamnose, 2-deoxy-d-ribose, and ribose-5-phosphate) (data not shown). Although CLRI exhibited some activity with l-ribose and d-mannose, it appears to be an LI rather than an RI. Since our kinetic analyses confirmed this view (see below), we changed the designation of the enzyme from CLRI to CLLI.
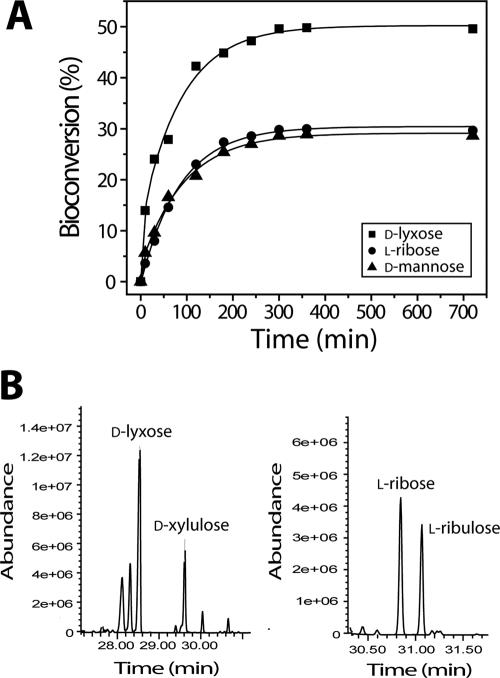
In order to investigate the ratios of conversion of d-lyxose to d-xylulose and of l-ribose and l-ribulose, aliquots of a reaction mixture were withdrawn periodically and analyzed by spectroscopic and GC-MS methods. As shown in Fig. 4A, maximum levels of conversion of d-lyxose to d-xylulose, l-ribose to l-ribulose, and d-mannose to d-fructose of 49, 28, and 27%, respectively, were obtained after 6 h of incubation at 60°C and pH 6.5. Each of the ketoses generated from the corresponding aldose was confirmed by GC-MS analysis with authentic standards (Fig. 4B). Each of the mass spectra was analyzed as displayed in the Wiley libraries.
FIG. 4.
(A) Time course of isomerization of aldopentoses (d-lyxose, l-ribose, and d-mannose) to aldoketoses (d-xylulose, l-ribulose, and d-fructose) by CLLI. The reaction mixtures (1 ml) contained 50 mM sodium phosphate buffer (pH 6.5), 10 mM d-lyxose, l-ribose, or d-mannose, 1 mM Mn2+, and 0.5 mg of CLLI. Aliquots were withdrawn after various times of incubation at 60°C. (B) GC-MS chromatograms of isomerization products from the reaction mixtures with d-lyxose and l-ribose as substrates.
Kinetic analysis of CLLI.
The Michaelis-Menten kinetic parameters (Km and Vmax) of CLLI for d-lyxose, l-ribose, and d-mannose were determined from linear Lineweaver-Burk plots. As shown in Table 2, the apparent Km and Vmax for l-ribose were 121.7 ± 10.8 mM and 75.5 ± 6.0 U/mg, respectively, whereas the apparent Km and Vmax for d-lyxose were 22.4 ± 1.5 mM and 5,434.8 U/mg, respectively. Consequently, the catalytic efficiency (kcat/Km) of CLLI for d-lyxose was 390-fold higher than that for l-ribose. In addition, the activity for d-mannose was approximately 63-fold lower than that for d-lyxose. Thus, the results indicate that this aldol-keto enzyme is essentially a d-lyxose isomerase.
TABLE 2.
Kinetic parameters of recombinant CLLIa
| Substrate | Km (mM) | Vmax (U/mg) | kcat (s−1) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|---|
| d-Lyxose | 22.4 ± 1.5 | 5,434.8 | 1,902 ± 0 | 84.9 ± 5.8 |
| l-Ribose | 121.7 ± 10.8 | 75.5 ± 6.0 | 26.4 ± 2.1 | 0.2 ± 0 |
| d-Mannose | 34.0 ± 1.1 | 131.8 ± 7.4 | 46.1 ± 2.6 | 1.4 ± 0.1 |
The data are means ± standard deviations.
Identification of the lyxA gene encoding CLLI.
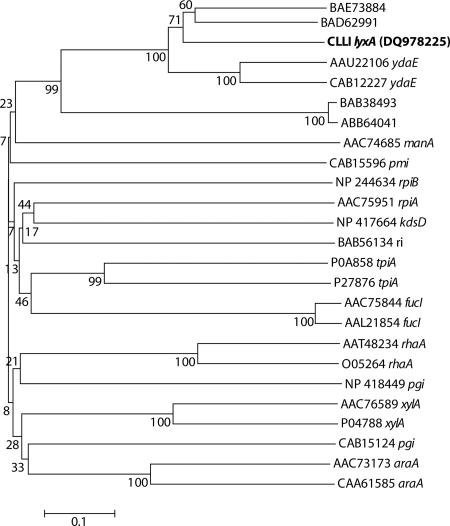
Based on the functional characterization described above, the 549-bp gene encoding LI from C. laevoribosii was designated lyxA. The deduced amino acid sequence of CLLI exhibited no significant homology to any other sugar isomerase, including the RI from Acinetobacter sp. (30). However, a BLAST search with lyxA revealed that CLLI exhibited significant levels of sequence homology (≥60%) with proteins from Bacillus species (Fig. 3B), such as BL02818 (ydaE) from Bacillus licheniformis (ATCC 14580), BSU04200 (ydaE) from Bacillus subtilis, and ABC0449 (a conserved hypothetical protein) from Bacillus clausii KSM-K16, and with SG0609 (a conserved hypothetical protein) from Sodalis glossinidius. These homologues have not been functionally characterized yet. Moreover, in order to analyze the phylogenetic relationships of CLLI in phosphate and unphosphorylated sugar isomerase families, a phylogenetic analysis was performed by the neighbor-Joining (NJ) method (33). A phylogram of bacterial sugar isomerases, including the hypothetical proteins homologous to CLLI mentioned above, based on an NJ tree is shown in Fig. 5. The topology obtained by the NJ method is not supported by very good bootstrap values due to very low levels of sequence identity among different sugar isomerase families. Nevertheless, CLLI and its homologues form a separate cluster, which does not include any other known sugar isomerases. Thus, together with the sequence analysis, this phylogenetic analysis indicated that CLLI is a novel type of d-lyxose isomerase.
FIG. 5.
Phylogenetic tree of CLLI and other sugar isomerases. Bootstrap values are indicated at the branch points. The bar indicates a branch length equivalent to 0.1 change per amino acid. All sequences were obtained from GenBank. BAE73884, S. glossinidius conserved hypothetical protein; BAD62991, B. clausii conserved hypothetical protein; CLLI, C. laevoribosii d-lyxose isomerase (NCBI protein database accession no. DQ978225); AAU22106 ydaE, B. licheniformis ydaE-encoded protein; CAB12227 ydaE, B. subtilis ydaE-encoded protein; BAB38493, E. coli hypothetical protein; ABB64041, Shigella dysenteriae conserved hypothetical protein; AAC74685 manA, E. coli mannose-6-phosphate isomerase; CAB15596 pmi, B. subtilis phosphomannose isomerase; NP 244634 rpiB, Bacillus halodurans ribose-5-phosphate isomerase; AAC75951 rpiA, E. coli ribose-5-phosphate isomerase; NP 417664 kdsD, E. coli putative isomerase; BAB56134 ri, Acinetobacter sp. strain DL-28 l-ribose isomerase; P0A858 tpiA, E. coli triose phosphate isomerase; P27876 tpiA, B. subtilis triose phosphate isomerase; AAC75844 fucI, E. coli fucose isomerase; AAL21854 fucI, Salmonella enterica serovar Typhimurium LT2 fucose isomerase; AAT48234 rhaA, E. coli l-rhamnose isomerase; O05264 rhaA, B. subtilis l-rhamnose isomerase; NP 418449 pgi, E. coli glucose-6-phosphate isomerase; AAC76589 xylA, E. coli xylose isomerase; P04788 xylA, B. subtilis xylose isomerase; CAB15124 pgi, B. subtilis glucose-6-phosphate isomerase; AAC73173 araA, E. coli l-arabinose isomerase; CAA61585 araA, B. subtilis l-arabinose isomerase. Amino acid sequences were aligned with the Vector NTI AlignX software (Suite 9.0.0; Invitrogen, Carlsbad, CA). Phylogenetic trees were constructed by the neighbor-joining method (33), using the MEGA software, version 3.0 (25). The p-distance correction substitution model was used in a tree-building analysis. Bootstrap values were calculated based on 1,000 replicates of the data (11).
DISCUSSION
To date, the genomes of more than 380 microorganisms have been sequenced, and their genes and enzymes used in carbohydrate metabolism have been analyzed (8, 12, 19, 37). The enormous amount of information not only has confirmed previous conclusions but also has revealed novel genes controlling metabolic pathways. Nonetheless, a number of mysterious ORFs remain (44). This is especially true of a number of ORFs encoding stress-regulated hypothetical proteins. If a nonnaturally occurring sugar is available as a sole carbon source, microorganisms have to turn on specific metabolic pathways or modify some normal metabolic pathway to utilize it. An example of the latter situation is the observation that wild-type E. coli cannot grow on the rare sugar l-lyxose but mutants that are able to utilize this sugar via the rhamnose metabolic pathway as a result of mutations in the rhamnose kinase can be isolated; these mutants can phosphorylate the l-xylulose produced from l-lyxose (4). We cannot exclude the possibility that there are heretofore unknown metabolic pathways for very specific substrates since numerous hypothetical proteins produced under stress conditions have not been functionally characterized yet. This possibility is exemplified by several cases of the metabolism of rare sugars (13, 32). In some cases, a new enzymatic activity can be shown to be derived from a previously known enzyme with a different substrate (44).
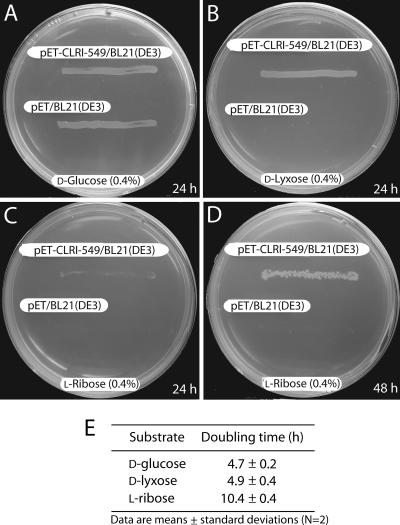
In this report we describe the cloning, purification, and characterization of an LI encoded by a 549-bp gene designated lyxA in a newly isolated species, C. laevoribosii, which can grow on l-ribose. This enzyme is the first genetically identified isomerase that can catalyze the interconversion of d-lyxose to d-xylulose with a high specific activity. Although CLLI also converted d-mannose and l-ribose to d-fructose and l-ribulose, respectively, the amino acid sequence of the enzyme does not exhibit significant homology to MIs or RIs that can also isomerize d-lyxose (18, 30). In addition, the substrate specificity of CLLI is different from the substrate specificities of MIs and RIs, suggesting that the enzyme is a novel LI. Moreover, the fact that the lyxA gene of C. laevoribosii (in pET-CLRI-549) can confer on E. coli the ability to use d-lyxose and l-ribose as carbon sources (Fig. 6) strongly suggests that CLLI converts d-lyxose to d-xylulose, thus generating a precursor of the pentose phosphate pathway (Fig. 1). It has also been observed that BL21/pET-CLRI-549 grows much more rapidly on d-lyxose than on l-ribose, as expected from the characterization of CLLI described above.
FIG. 6.
lyxA gene of C. laevoribosii enables E. coli BL21(DE3) to grow on d-lyxose or l-ribose as a sole carbon source. (A to D) Cells were streaked on minimal M9 agar plates supplemented with d-glucose, d-lyxose, or l-ribose as the sole carbon source and incubated at 37°C (see Materials and Methods). (E) To obtain growth curves of E. coli BL21(DE3)/pET-CLRI-549 on minimal M9 media with various sugars as carbon sources, inocula (0.14 × 108 cells) were grown on minimal M9 medium containing 0.4% glucose, washed, and transferred into minimal M9 medium containing 0.4% l-ribose, d-lyxose, or d-glucose, and then the absorbance at 600 nm of the cultures was monitored.
Interestingly, an LI from A. aerogenes PRL-R3 has been reported to be induced by d-mannose as well as d-lyxose (3). In addition, an MI from M. smegmatis (17) and an RI from Acinetobacter sp. strain DL-28 (30) were also induced by d-lyxose. It is noteworthy that the enzymes mentioned above, like CLLI from C. laevoribosii, which are induced not only by l-ribose (0.5 U/unit of optical density at 600 nm [OD600]) and d-lyxose (2.6 U/unit of OD600) but also by d-mannose (1.3 U/unit of OD600), have similar affinities for the three sugars (d-lyxose, d-mannose, and l-ribose), although they have different substrate preferences. From these results, it can be speculated that these three sugars have a common configuration able to induce LIs, RIs, and MIs. This intriguing metabolic phenomenon can be explained by a proposal of Izumori and Yamanaka (20). Based on induction experiments, these authors proposed that the equivalent configurations of the C-1 to C-4 OH groups of d-mannose and d-lyxose are capable of inducing the corresponding isomerases, and this idea seems also to apply to l-ribose. If this is true, this property could be valuable for engineering the substrate specificity of a sugar isomerase, as well as for understanding the pathways of metabolism of these rare sugars. This speculation tempts us to suggest that the three different sugar isomerases may have a common evolutionary origin even though they exhibit no significant homology and that they may have diverged to act on different substrates. In order to investigate the substrate specificity of CLLI in more detail, we are currently performing a three-dimensional structural analysis using nuclear magnetic resonance spectroscopy.
Supplementary Material
Acknowledgments
This work was supported by grant 2001-2-0109 from the Korea Science and Engineering Foundation and by grant AIC-08-02 from the Ministry of Commerce, Industry, and Energy, Korea.
We gratefully acknowledge Julian Gross and Han-Seung Lee for helpful discussions and for editing the manuscript.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Ahmed, Z. 2001. Production of natural and rare pentoses using microorganisms and their enzymes. Electron. J. Biotechnol. 4:103-111. [Google Scholar]
- 2.Ahmed, Z., H. Sasahara, S. H. Bhuiyan, T. Saiki, T. Shimonishi, G. Takada, and K. Izumori. 1999. Production of d-lyxose from d-glucose by microbial and enzymatic reactions. J. Biosci. Bioeng. 88:676-678. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, R. L., and D. P. Allison. 1965. Purification and characterization of d-lyxose isomerase. J. Biol. Chem. 240:2367-2372. [PubMed] [Google Scholar]
- 4.Badia, J., R. Gimenez, L. Baldomá, E. Barnes, W. D. Fessner, and J. Aguilar. 1991. l-Lyxose metabolism employs the l-rhamnose pathway in mutant cells of Escherichia coli adapted to grow on l-lyxose. J. Bacteriol. 173:5144-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Y. C. 2001. Potential use of antiviral l-nucleoside analogues for the prevention or treatment of viral associated cancers. Cancer Lett. 162:S33-S37. [DOI] [PubMed] [Google Scholar]
- 6.Cho, B. H., J. H. Kim, H. B. Jeon, and K. S. Kim. 2005. A new efficient and practical synthesis of 2-deoxy-l-ribose. Tetrahedron 61:4341-4346. [Google Scholar]
- 7.Chu, C. K., T. Ma, K. Shanmuganathan, C. Wang, Y. Xiang, S. B. Pai, G. Q. Yao, J. P. Sommadossi, and Y. C. Cheng. 1995. Use of 2′-fluoro-5-methyl-beta-l-arabinofuranosyluracil as a novel antiviral agent for hepatitis B virus and Epstein-Barr virus. Antimicrob. Agents Chemother. 39:979-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordwell, S. J. 1999. Microbial genomes and missing enzymes: redefining biochemical pathways. Arch. Microbiol. 172:269-279. [DOI] [PubMed] [Google Scholar]
- 9.Dische, Z., and E. Borefreund. 1951. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J. Biol. Chem. 192:583-587. [PubMed] [Google Scholar]
- 10.Fazio, F., and M. P. Schneider. 2000. A novel synthesis of 2-deoxy-l-ribose. Tetrahedron Asymmetry 11:1869-1876. [Google Scholar]
- 11.Felsenstein, J. 1996. Inferring phylogenies from protein sequences by parsimony, distance and likelihood methods. Methods Enzymol. 266:418-427. [DOI] [PubMed] [Google Scholar]
- 12.Flores, C. L., C. Rodriguez, T. Petit, and C. Gancedo. 2000. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol. Rev. 24:507-529. [DOI] [PubMed] [Google Scholar]
- 13.Granström, T. B., G. Takata, M. Tokuda, and K. Izumori. 2004. Izumoring: a novel and complete strategy for bioproduction of rare sugars. J. Biosci. Bioeng. 97:89-94. [DOI] [PubMed] [Google Scholar]
- 14.Gumina, G., G. Y. Song, and C. K. Chu. 2001. l-Nucleosides as chemotherapeutic agents. FEMS Microbiol. Lett. 202:9-15. [DOI] [PubMed] [Google Scholar]
- 15.Gunsalus, I. C., B. L. Horecker, and W. A. Wood. 1955. Pathways of carbohydrate metabolism in microorganisms. Bacteriol. Rev. 19:79-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heath, E. C., J. Hurwitz, B. L. Horecker, and A. Ginsburg. 1958. Pentose fermentation by Lactobacillus plantarum. J. Biol. Chem. 231:1009-1029. [PubMed] [Google Scholar]
- 17.Hey-Ferguson, A., and A. D. Elbein. 1970. Purification of a d-mannose isomerase from Mycobacterium smegmatis. J. Bacteriol. 101:777-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirose, J., K. Maeda, H. Yokoi, and Y. Takasaki. 2001. Purification and characterization of mannose isomerase from Agrobacterium radiobacter M-1. Biosci. Biotechnol. Biochem. 65:658-661. [DOI] [PubMed] [Google Scholar]
- 19.Hou, S., J. H. Saw, K. S. Lee, T. A. Freitas, C. Belisle, Y. Kawarabayasi, S. P. Donachie, A. Pikina, M. Y. Galperin, E. V. Koonin, K. S. Makarova, M. V. Omelchenko, A. Sorokin, Y. I. Wolf, Q. X. Li, Y. S. Keum, S. Campbell, J. Denery, S. I. Aizawa, S. Shibata, A. Malahoff, and M. Alam. 2004. Genome sequence of the deep-sea γ-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc. Natl. Acad. Sci. USA 101:18036-18041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumori, K., and K. Yamanaka. 1977. Speculative studies on an anomeric specificity of inducers of d-lyxose isomerase. FEBS Lett. 77:133-135. [DOI] [PubMed] [Google Scholar]
- 21.Izumori, K., K. Yamanaka, and A. D. Elbein. 1976. Pentose metabolism in Mycobacterium smegmatis: specificity of induction of pentose isomerases. J. Bacteriol. 128:587-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung, M. E., and Y. Xu. 1997. Efficient synthesis of l-ribose and 2-deoxy l-ribose from d-ribose and l-arabinose. Tetrahedron Lett. 38:4199-4202. [Google Scholar]
- 23.Kletzien, R. F., P. K. W. Harris, and L. A. Foellmi. 1994. Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB J. 8:174-181. [DOI] [PubMed] [Google Scholar]
- 24.Kruger, N. J., and A. von Schaewen. 2003. The oxidative pentose phosphate pathway: structure and organization. Curr. Opin. Plant Biol. 6:236-246. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings Bioinformatics 5:150-163. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lee, D. W., H. J. Jang, E. A. Choe, B. C. Kim, S. J. Lee, S. B. Kim, Y. H. Hong, and Y. R. Pyun. 2004. Characterization of a thermostable l-arabinose isomerase (d-galactose) isomerase from the hyperthermophilic eubacterium Thermotoga maritima. Appl. Environ. Microbiol. 70:1397-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, D. W., Y. S. Koh, K. J. Kim, B. C. Kim, H. J. Choi, D. S. Kim, M. T. Suhartono, and Y. R. Pyun. 1999. Purification and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 179:393-400. [DOI] [PubMed] [Google Scholar]
- 29.Ma, T., S. B. Pai, Y. L. Zhu, J. S. Lin, K. Shanmuganathan, J. Du, C. Wang, H. Kim, M. G. Newton, Y. C. Cheng, and C. K. Chu. 1996. Structure-activity relationships of 1-(2-deoxy-2-fluoro-beta-l-arabinofuranosyl) pyrimidine nucleosides as anti-hepatitis B virus agents. J. Med. Chem. 39:2835-2843. [DOI] [PubMed] [Google Scholar]
- 30.Mizanur, R. M., G. Takata, and K. Izumori. 2001. Cloning and characterization of a novel gene encoding l-ribose isomerase from Acinetobacter sp. strain DL-28 in Escherichia coli. Biochim. Biophys. Acta 1521:141-145. [DOI] [PubMed] [Google Scholar]
- 31.Morita, M., E. Sawa, K. Yamaji, T. Sakai, T. Natori, Y. Koezuka, H. Fukushima, and K. Akimoto. 1996. Practical total synthesis of (2S,3S,4R)-1-O-(α-d-galactopyranosyl)-N-hexacosanoyl-2-amino-1,3,4-octadecanetriol, the antitumorial and immunostimulatory α-galactosylceramide, KRN7000. Biosci. Biotechnol. Biochem. 60:288-292. [DOI] [PubMed] [Google Scholar]
- 32.Mortlock, R. P. 1982. Metabolic acquisitions through laboratory selection. Annu. Rev. Microbiol. 36:259-284. [DOI] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic tress. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 34.Shi, Z. D., B. H. Yang, and Y. L. Wu. 2002. A stereospecific synthesis of l-deoxyribose, l-ribose and l-ribosides. Tetrahedron 58:3287-3296. [Google Scholar]
- 35.Shimonishi, T., and K. Izumori. 1996. A new enzyme, l-ribose isomerase from Acinetobacter sp. strain DL-28. J. Ferment. Bioeng. 81:493-497. [Google Scholar]
- 36.Smith, P. K., G. T. Krohn, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 37.Sprenger, G. A. 1995. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch. Microbiol. 164:324-330. [DOI] [PubMed] [Google Scholar]
- 38.Stevens, F. J., and T. T. Wu. 1976. Growth on d-lyxose of a mutant strain of Escherichia coli K12 using a novel isomerase and enzymes related to d-xylase metabolism. J. Gen. Microbiol. 97:257-265. [DOI] [PubMed] [Google Scholar]
- 39.Takagi, Y., K. Nakai, T. Tsuchiya, and T. Takeuchi. 1996. A 5′-(trifluoromethyl) anthracycline glycoside: synthesis of antitumor-active 7-O-(2,6-dideoxy-6,6,6-trifluoro-beta-l-lyxo-hexopyranosyl)adriamycinone. J. Med. Chem. 39:1582-1588. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tisza, S., P. Sass, and I. Molnár-Perl. 1994. Optimization of the simultaneous determination of acids and sugars as their trimethylsilyl (oxime) derivatives by gas chromatography-mass spectrometry and determination of the composition of six apple varieties. J. Chromatogr. A 676:461-468. [Google Scholar]
- 42.Trimbur, D. E., and R. P. Mortlock. 1991. Isolation and characterization of Escherichia coli mutants able to utilize the novel pentose l-ribose. J. Bacteriol. 173:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Rompay, A. R., M. Johansson, and A. Karlsson. 2003. Substrate specificity and phosphorylation of antiviral and anticancer nucleoside analogues by human deoxyribonucleoside kinases and ribonucleoside kinases. Pharmacol. Ther. 100:119-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White, R. H. 2006. The difficult road from sequence to function. J. Bacteriol. 188:3431-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yun, M. K., H. R. Moon, H. O. Kim, W. J. Choi, Y. C. Kim, C. S. Park, and L. S. Jeong. 2005. A highly efficient synthesis of unnatural l-sugars from d-ribose. Tetrahedron Lett. 46:5903-5905. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.