Abstract
In Chlamydiaceae, the nucleotide sequence between the 5S rRNA gene and the gene for subunit F of the Na+-translocating NADH-quinone reductase (nqrF or dmpP) has varied lengths and gene contents. We analyzed this site in 45 Chlamydiaceae strains having diverse geographical and pathological origins and including members of all nine species.
The complete genomes of six Chlamydiaceae species (1, 3, 12, 19-21, 23, 24) have revealed a high level of synteny, with large-scale rearrangements and niche-specific genes restricted to a “plasticity zone” located at the replication termination region. Analysis of the rrn-nqrF intergenic segments in these sequences reveals significant variation in length and gene content that is inconsistent with host range, tissue tropism, DNA-based phylogenies, and disease spectrum in humans and animals. Therefore, this segment may represent a new genomic plasticity zone in these bacteria. We were especially interested in a frameshifted invasin/intimin-like protein gene (ilp) at this location in Chlamydia caviae (20). Fragments of ilp in several Chlamydia suis strains were also identified by Dugan et al. (5). These observations led us to a comprehensive analysis of the rrn-nqrF intergenic sequences across the Chlamydiaceae. Strains are listed in Table 1. Data on PCR and sequencing primers are available upon request. Our findings are provided in a graphic summary (Fig. 1).
TABLE 1.
Strains included in this study
| Group, organism, and strain(s) or serovar(s) | Source | Accession no. | rrn-nqrF intergenic segment length (bp) | Reference |
|---|---|---|---|---|
| Group 1 | ||||
| C. pneumoniae | ||||
| AR39 | Human | AE002161 | 2,047 | 19 |
| J138 | Human | NC002491 | 2,047 | 21 |
| CWL029 | Human | NC000922 | 2,047 | 12 |
| TW183 | Human | NC005043 | 2,047 | This study |
| C. psittaci | ||||
| 6BC | Parrot | DQ076132 | 2,334 | This study |
| Parakeet | Parrot | DQ076133 | 2,218 | This study |
| I-10 | Pigeon | DQ076144 | 2,383 | This study |
| I-6 and I-7 | Parrots | 2,200d | This study | |
| I-9 and I-11 | Pigeons | 2,200d | This study | |
| C. abortus | ||||
| S26/3 | Sheep | CR848038 | 2,320 | 24 |
| B577 | Sheep | DQ076146 | 2,343 | This study |
| LW508 | Cow | DQ076147 | 2,346 | This study |
| SV139 and I-17 | Cattle | 2,200d | This study | |
| I-1 | Sheep | 2,300d | This study | |
| I-5 | Goat | 2,300d | This study | |
| C. felis | ||||
| Fe/C-56 | Cat | AP006861 | 2,229 | 1 |
| FEPN sch562F4 | Cat | 2,200d | This study | |
| FEPN Baker | Cat | DQ076145 | 2,347 | This study |
| Group 2 | ||||
| C. caviae | ||||
| Yolk sac | Guinea pig | NC003361 | 4,463 | 20 |
| Yolk sac 62H454 and 73H301 | Guinea piga | 4,463 | This study | |
| C. muridarum | ||||
| MoPn | Mouse | AE002160 | 457 | 19 |
| C. suis | ||||
| S45 | Pig | DQ076148 | 4,184 | This study |
| R19 | Pig | AY428550 | 16,000d | 5 |
| R27 | Pig | AY428551 | 9,000d | 5 |
| Group 3 | ||||
| C. trachomatis | ||||
| A/HAR-13 | Human | NC007429 | 519 | 3 |
| D/UW-3/CX | Human | NC000117 | 518 | 23 |
| E | Human | DQ851868 | 525 | This study |
| G | Humanb | DQ851869 | 518 | This study |
| K | Humanb | DQ851870 | 518 | This study |
| D, H, and I | Humansb | 518 | This study | |
| L2/LGV434c | Human | 519 | ||
| Group 4 | ||||
| C. pecorum | ||||
| I-15 | Cow | DQ076134 | 518 | This study |
| I-12 | Cow | DQ076135 | 469 | This study |
| 1710S | Pig | DQ076136 | 445 | This study |
| I-19 | Water buffalo | DQ076137 | 427 | This study |
| LW679 | Cow | DQ076138 | 419 | This study |
| I-4 | Goat | DQ076139 | 404 | This study |
| L-17 | Pig | DQ076140 | 388 | This study |
| 1708 | Pig | DQ076141 | 380 | This study |
| JP-1-751 | Cow | DQ076142 | 356 | This study |
| I-13 | Cow | DQ076143 | 252 | This study |
The original isolate was from a guinea pig; no other C. caviae strains were available for examination.
Clinical isolate(s) from the genital tract.
The sequence for this serovar was retrieved from the Sanger Institute website at http://www.sanger.ac.uk.
The length of the rrn-nqrF segment is estimated by PCR and partial sequencing.
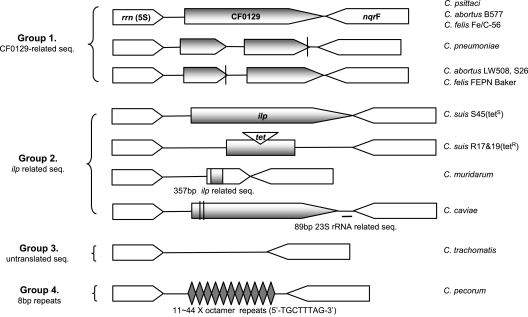
FIG. 1.
Genetic diversity at the 5S rrn-nqrF site. The schematic diagram presents the genetic organization of the rrn-nqrF intergenic segment. Chlamydia species are separated into four groups based on genetic relatedness at the site. Vertical bars indicate mutations that cause frameshifts or translation stops. Homologous ORFs are indicated graphically as shaded arrows. The diagram is drawn approximately to scale. seq., sequence(s).
Species of the Chlamydiaceae can be separated into four groups based on the contents of their rrn-nqrF intergenic segments. Group 1 includes isolates from Chlamydia pneumoniae, Chlamydia psittaci, Chlamydia abortus, and Chlamydia felis, infectious to primate (respiratory tract and eyes), avian (respiratory and digestive tracts and eyes), ovine and bovine (digestive tract and placenta), and feline (respiratory tract and eyes) species, respectively. The rrn-nqrF segment is fully conserved in four C. pneumoniae strains (12, 19, 21) and is only 46 to 49% identical to segments in the three other species. The rrn-nqrF segment of the Japanese C. felis strain Fe/C-56 (1) is 99% identical to that of C. psittaci strain parakeet but only 63 and 61% identical to those of C. psittaci I-10 or 6BC and another C. felis strain, FEPN Baker, respectively. The last strain, however, displays an rrn-nqrF intergenic sequence closely related to those of three C. abortus strains, S26 (24), B577, and LW508. Nucleotide substitutions, insertions, and deletions and hypervariable segments including stretches of C's and T's (Fig. 2) are found in group 1 strains, causing various translational frames. A single open reading frame (ORF) at the rrn-nqrF site, identified as CF0129 in the C. felis strain Fe/C-56 genome (1), is also found in C. psittaci strains 6BC, parakeet, and I-10 and in C. abortus strain B577. However, mutations disrupt this ORF in C. abortus strains S26 and LW508, C. felis strain FEPN Baker, and four C. pneumoniae strains.
FIG. 2.
Multiple alignment of the poly(C) and poly(T) tracts and flanking sequences at the rrn-nqrF sites in three Chlamydia species. Base numbering is from the 3′ end of rrn.
Group 2 includes isolates that carry at least partial ilp sequences at the rrn-nqrF site: C. caviae strain GPIC, Chlamydia muridarum strain MoPn, and eight C. suis strains, which infect guinea pigs (eyes and genital tract), mice (lungs), and pigs (digestive tract), respectively. Of those, only C. suis strain S45 (13) has intact ilp; all others have either fragments of the gene (C. muridarum) or a gene that is interrupted by mutation (C. caviae and Tetr C. suis). The sequence similarity of C. suis Ilp to Yersinia pseudotuberculosis invasin (9) and enteropathogenic Escherichia coli intimin (11) is restricted to the transmembrane and immunoglobulin-like domains (D1). However, the general structure of the predicted Ilp polypeptide is similar to that of the invasin/intimin family and includes an extended rodlike structure consisting of several modular repeat domains (D2 to D7) and a distal domain (D8). Several conserved beta strands in D2 to D7 are consistent with the crystal structures of intimin D0 to D2 and invasin D1 to D3 (8, 16). Secondary-structure predictions, however, reveal no conserved salient features in D8 (http://www.compbio.dundee.ac.uk/∼www-jpred/; domain definitions are per GenBank [accession number DQ076148]).
We reasoned that ilp of the C. caviae reference strain may have been inactivated through mutation during long-term in vitro culture. To address this question, the rrn-nqrF segment was amplified directly from early, archived specimens of C. caviae from the laboratory of Murray (18) (Table 1). Two specimens of the primary isolate that were maintained by passage through live animals, a 1962 specimen corresponding to the first passage and a 1973 specimen, were analyzed. In both cases, two frameshift mutations identical to those obtained from genomic analysis are present.
Group 3 includes only strains of the human pathogen Chlamydia trachomatis. PCR amplicons from six genitotropic strains, including clinical isolates of serovars D, G, H, I, and K and one serovar E reference strain, were compared. Sequences obtained from serovars D, G, and K are identical to the published sequence of serovar D strain UW-3/Cx (23). However, the serovar E sequence contains a 7-nucleotide insertion and 11 additional mutations, 7 of them located within the putative rrn transcription terminator (not shown). The rrn-nrqF intergenic sequence of lymphogranuloma venereum strain L2/434 (12) is identical to that of ocular strain A/HAR-13 (3) but less identical (96 to 99%) to that of urogenital strains (Table 1). The rrn-nrqF intergenic segments of all C. trachomatis strains and serovars contain no recognizable ORF.
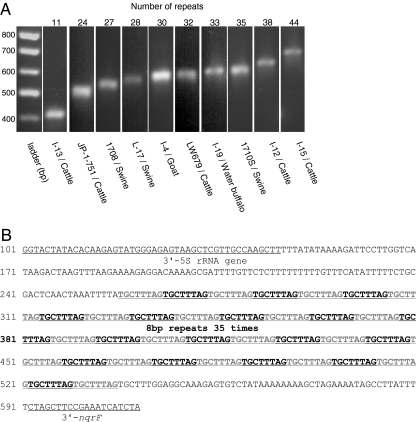
Group 4 includes 10 isolates of Chlamydia pecorum obtained from mammalian species on two continents. Amplicons of the segments from these isolates range between 400 and 700 bp (Fig. 3A). Sequence analysis reveals multiple copies (between 11 and 44) of the simple sequence repeat (SSR) 5′-TGCTTTAG-3′ (Fig. 3B). BLAST searches reveal no matches, suggesting that this SSR is unique to C. pecorum.
FIG. 3.
Multiple repeats at the rrn-nqrF site of C. pecorum. (A) Gel electrophoresis of amplicons generated with primers 5S rRNA F and nqrF R (data available upon request) for 10 C. pecorum isolates (Table 1). The numbers of octamer repeats are indicated above the lanes. Numbers at the left are molecular sizes. (B) Nucleotide sequence of the rrn-nqrF site of C. pecorum strain 1710S, which contains 35 octamer repeats.
Taken together, our results indicate an unusual degree of genetic plasticity at the rrn-nqrF locus in the family Chlamydiaceae that transcends phylogenetic boundaries. For instance, the ilp sequence, albeit degenerate or mutated, is common to C. suis, C. muridarum, and C. caviae, i.e., three species representing three different hosts and two phylogenetic lineages of the Chlamydiaceae (7). The apparent genetic plasticity of this site stands in sharp contrast with that of the neighboring rRNA operon, whose functional constancy and genetic stability are a basis of current phylogenetic analysis. It is possible that the plasticity of the rrn-nqrF segment stems from its being frequently exposed during chlamydial development. There are only one or two copies of rrn in the Chlamydia genomes. Elevated transcription activity of the rrn operon would require frequent unwinding of supercoiled DNA, which might promote physical damage (15), consistent with the site's being a preferred target for recombination. The presence of an 89-bp sequence nearly identical to a chlamydial 23S rRNA domain 1 sequence, flanking ilp in C. caviae, may indeed be a vestige of a previous horizontal ilp transfer event (Fig. 1). The identification of four genomic islands within ilp in seven Tetr C. suis strains further supports this hypothesis. Although the Tetr cassettes are all recombined at the same position within ilp (5), strain-specific differences in the sequences of ompA or the Tetr islands suggest that these recombinations result from independent integration events (2, 5). Similarly, in group 1, although rRNA sequences of C. felis strains Fe/C-56 and FEPN Baker are almost identical (>99.9%), their rrn-nqrF intergenic segments are nearly 40% divergent and align with those of C. psittaci and C. abortus, respectively. Our results suggest that genetic exchange has occurred at this site between C. felis, C. psittaci, and C. abortus. Apart from recombination events, modifications of the rrn-nqrF segment include frameshifts in ilp of C. caviae; truncation of ilp in C. muridarum; ORF fusion and gene decay in CF0129-related sequences in C. pneumoniae, C. psittaci, C. abortus, and C. felis; and unique SSR duplications and deletions in C. pecorum.
Sequence variation in pathogens often occurs in genes encoding surface components. Among ORFs of the rrn-nqrF locus, CF0129-related ORFs and ilp encode predicted membrane proteins. The predicted product of the CF0129-related ORF CAB852 of C. abortus S26 includes three transmembrane domains (24) that are conserved in all tested strains of C. abortus, C. felis, and C. psittaci. Slipped mispairing at poly(C) and poly(T) tracts (Fig. 2), however, results in various translational stops and frameshifts. Likewise, although undisrupted ilp isolated from C. suis strain S45 is transcribed in vitro (not shown), ilp sequence interruption by Tetr cassettes in seven C. suis strains (5), frameshift mutations in C. caviae, and a sequence truncation in C. muridarum (19) indicate that ilp is not required for infection. The roles of other members of the invasin/intimin family in pathogenesis also vary. The yersinial invasin confers an advantage to the bacterium early in infection, but loss of invasin function does not prevent infection (10, 17).
Sequence analysis reveals that the 5′-TGCTTTAG-3′ octamer occurs four to six times more than a random sequence octamer in all chlamydial genomes and is oriented relative to ori. These properties are similar to previously reported properties of the genus-specific “Chi” recombination hot spot of Escherichia coli (14) and other bacteria (4, 6, 22). However, the function of multiple repeats of the octamer at the rrn-nqrF site of C. pecorum is unknown.
In summary, our analysis has revealed highly polymorphic sequences at the rrn-nqrF site across 45 chlamydial strains from nine species of diverse geographical and pathological origins. Although the underlying mechanism(s) is not clear, the data presented here support the notion that this site is a hot spot for gene recombination in the Chlamydiaceae.
Acknowledgments
We thank Pier Giorgio Vigo, Iris Labalestra, and Kyle Burall for technical assistance and an anonymous reviewer for his assistance in the analysis.
This work was supported by a subcontract to P. M. Bavoil of National Institutes of Health (NIH) grant RO1 AI51472 to G. Myers (The Institute for Genomic Research). Z. Liu was supported by Chongqing Southwest Hospital, China, and NIH grant RO1 AI4310 to P. M. Bavoil. L. Burall was supported by training grant NIDCR T32 DE07309-08. B. Kaltenboeck was supported by NIH grant AI47202.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Azuma, Y., H. Hirakawa, A. Yamashita, Y. Cai, M. A. Rahman, H. Suzuki, S. Mitaku, H. Toh, S. Goto, T. Murakami, K. Sugi, H. Hayashi, H. Fukushi, M. Hattori, S. Kuhara, and M. Shirai. 2006. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 13:15-23. [DOI] [PubMed] [Google Scholar]
- 2.Bush, R. M., and K. D. Everett. 2001. Molecular evolution of the Chlamydiaceae. Int. J. Syst. Evol. Microbiol. 51:203-220. [DOI] [PubMed] [Google Scholar]
- 3.Carlson, J. H., S. F. Porcella, G. McClarty, and H. D. Caldwell. 2005. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 73:6407-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chedin, F., S. D. Ehrlich, and S. C. Kowalczykowski. 2000. The Bacillus subtilis AddAB helicase/nuclease is regulated by its cognate Chi sequence in vitro. J. Mol. Biol. 298:7-20. [DOI] [PubMed] [Google Scholar]
- 5.Dugan, J., D. D. Rockey, L. Jones, and A. A. Andersen. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 48:3989-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.el Karoui, M., D. Ehrlich, and A. Gruss. 1998. Identification of the lactococcal exonuclease/recombinase and its modulation by the putative Chi sequence. Proc. Natl. Acad. Sci. USA 95:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 8.Hamburger, Z. A., M. S. Brown, R. R. Isberg, and P. J. Bjorkman. 1999. Crystal structure of invasin: a bacterial integrin-binding protein. Science 286:291-295. [DOI] [PubMed] [Google Scholar]
- 9.Isberg, R. P., H. L. Voorhis, and S. Falkow. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769-778. [DOI] [PubMed] [Google Scholar]
- 10.Jepson, M. A., and M. A. Clark. 1998. Studying M cells and their role in infection. Trends Microbiol. 6:359-365. [DOI] [PubMed] [Google Scholar]
- 11.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 13.Kaltenboeck, B., and J. Storz. 1992. Biological properties and genetic analysis of the ompA locus in chlamydiae isolated from swine. Am. J. Vet. Res. 53:1482-1487. [PubMed] [Google Scholar]
- 14.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 15.Kuraoka, I., M. Endou, Y. Yamaguchi, T. Wada, H. Handa, and K. Tanaka. 2003. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase. II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 278:7294-7299. [DOI] [PubMed] [Google Scholar]
- 16.Luo, Y., E. A. Frey, R. A. Pfuetzner, A. L. Creagh, D. G. Knoechel, C. A. Haynes, B. B. Finlay, and N. C. Strynadka. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073-1077. [DOI] [PubMed] [Google Scholar]
- 17.Marra, A., and R. R. Isberg. 1997. Invasin-dependent and invasin-independent pathways for translocation of Yersinia pseudotuberculosis across the Peyer's patch intestinal epithelium. Infect. Immun. 65:3412-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray, E. S. 1964. Guinea pig inclusion conjunctivitis. I. Isolation and identification as a member of the Psittacosis-Lymphogranuloma-Trachoma group. J. Infect. Dis. 114:1-12. [DOI] [PubMed] [Google Scholar]
- 19.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sourice, S., V. Biaudet, M. El Karoui, S. D. Ehrlich, and A. Gruss. 1998. Identification of the Chi site of Haemophilus influenzae as several sequences related to the Escherichia coli Chi site. Mol. Microbiol. 27:1021-1029. [DOI] [PubMed] [Google Scholar]
- 23.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 24.Thomson, N. R., C. Yeats, K. Bell, M. T. Holden, S. D. Bentley, M. Livingstone, A. M. Cerdeno-Tarraga, B. Harris, J. Doggett, D. Ormond, K. Mungall, K. Clarke, T. Feltwell, Z. Hance, M. Sanders, M. A. Quail, C. Price, B. G. Barrell, J. Parkhill, and D. Longbottom. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15:629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]