Abstract
It had been assumed that production of the cytotoxic polyketide mycolactone was strictly associated with Mycobacterium ulcerans, the causative agent of Buruli ulcer. However, a recent study has uncovered a broader distribution of mycolactone-producing mycobacteria (MPM) that includes mycobacteria cultured from diseased fish and frogs in the United States and from diseased fish in the Red and Mediterranean Seas. All of these mycobacteria contain versions of the M. ulcerans pMUM plasmid, produce mycolactones, and show a high degree of genetic relatedness to both M. ulcerans and Mycobacterium marinum. Here, we show by multiple genetic methods, including multilocus sequence analysis and DNA-DNA hybridization, that all MPM have evolved from a common M. marinum progenitor to form a genetically cohesive group among a more diverse assemblage of M. marinum strains. Like M. ulcerans, the fish and frog MPM show multiple copies of the insertion sequence IS2404. Comparisons of pMUM and chromosomal gene sequences demonstrate that plasmid acquisition and the subsequent ability to produce mycolactone were probably the key drivers of speciation. Ongoing evolution among MPM has since produced at least two genetically distinct ecotypes that can be broadly divided into those typically causing disease in ectotherms (but also having a high zoonotic potential) and those causing disease in endotherms, such as humans.
Mycobacterium ulcerans is a significant human pathogen and the causative agent of Buruli ulcer (BU), a disease with severe morbidity characterized by chronic skin ulceration and extensive necrosis of subcutaneous fat (40). Cases of BU have been reported in many parts of the world, with the greatest burden of disease occurring in West and Central Africa (16). However, other than travelers returning from countries where the disease is endemic, no cases of BU have ever been reported in the United States or Europe. M. ulcerans strains are characterized by the presence of a large circular virulence plasmid called pMUM (31, 33). This plasmid harbors three large genes (mlsA1, mlsA2, and mlsB) encoding polyketide synthases that are required for the synthesis of the lipid toxin mycolactone, which is the primary virulence factor for the pathogen (33). Comparisons of multiple plasmid and chromosomal genes among 10 M. ulcerans clinical isolates from diverse origins have suggested that plasmid acquisition was probably the key event that marked and permitted the recent emergence of M. ulcerans from a common Mycobacterium marinum progenitor (31). M. marinum is phenotypically distinct from M. ulcerans, producing photochromogenic pigments and generally growing more quickly. M. marinum causes granulomatous lesions in fish and other ectotherms and can also cause granulomatous skin lesions in humans. Comparisons between the 5.8-Mb genome of the M. ulcerans African epidemic strain Agy99 and the 6.6-Mb genome of M. marinum strain “M” confirmed this hypothesis and showed that M. ulcerans has recently passed through an evolutionary bottleneck, evolving from the generalist M. marinum to become a specialist bacterium, adapted to a more restricted (possibly host-specific) environment (32, 34; http://genolist.pasteur.fr/BuruList/). Like other niche-adapted pathogens, such as Yersinia pestis and Bordetella pertussis, M. ulcerans has undergone extensive gene loss due to DNA deletions, DNA rearrangements, and pseudogene formation. Many of these changes have been mediated by some of the 213 copies of IS2404 and 91 copies of IS2606 (34). Neither of these insertion sequence elements (ISE) is present in M. marinum (32).
It was assumed until recently that mycolactone production was restricted to M. ulcerans. However, in 2004, a previously unreported mycobacterium that contained both IS2404 and IS2606 was recovered from cases of an unusual, lethal edematous disease in laboratory-housed Xenopus laevis and Xenopus tropicalis frogs (37). Limited sequence comparisons of hsp65, the 16S rRNA gene, the 16S-23S rRNA gene internal transcribed sequence spacer, and rpoB gene fragments showed that this mycobacterium shared greater than 98% nucleotide identity with M. ulcerans and M. marinum (37). In a subsequent investigation, the frog mycobacterium was shown to contain a version of the M. ulcerans pMUM plasmid and to produce a new mycolactone, mycolactone E (13, 19). It was also given the unofficial epithet Mycobacterium liflandii (19). In 2005, another new mycobacterium, isolated during an epizootic of mycobacteriosis from diseased striped bass (Morone saxatilis) in Chesapeake Bay, was also shown to contain IS2404 and IS2606 and to share >98% nucleotide sequence identity with M. ulcerans, M. marinum, and Mycobacterium shottsii (28). Based on some phenotypic traits that distinguished it from M. ulcerans, such as photochromogenicity, absence of growth at 37°C, and scant growth at 30°C on Middlebrook 7H10 agar, this mycobacterium was given a new species designation, Mycobacterium pseudoshottsii. Subsequent analysis of the strain showed that it too contained a pMUM-like plasmid with the mycolactone mls genes and that it made yet another mycolactone, called mycolactone F (26). In the same study, a cluster of M. marinum strains that had been isolated from diseased fish in the Red and Mediterranean Seas (38) were also positive for mls gene sequences, and they too produced mycolactone F (26). One of these strains (DL240490) shared identical hsp65 gene sequences with M. pseudoshottsii and another fish pathogen, Mycobacterium seriolae (28).
Thus, the distribution of mycolactone-producing mycobacteria (MPM) appears to be much wider than first appreciated, and this raises several interesting questions regarding the mobility of pMUM and the evolution of M. ulcerans and M. marinum. In this study, we conducted a comprehensive genetic analysis of these strains by DNA-DNA hybridization (DDH), by sequence comparisons of eight chromosomal and four plasmid genes, and by comparing the distributions of pseudogenes and DNA deletions to better define the taxonomic status of all mycolactone-producing strains, including those that cause Buruli ulcer, and to gain insights into their evolution.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Among the isolates used in this study, 18 M. ulcerans strains and 22 M. marinum strains have been described in detail in a previous publication (32). Ten of these strains, each representing a unique multilocus sequence analysis (MLSA) sequence type (ST), are listed in Table 1, because they were used in additional analyses that included DDH, Southern hybridization, and pseudogene and deletion analyses. Table 1 also lists the details of five novel MPM and seven other mycobacteria that were examined in this study. The mycobacteria were cultivated in Middlebrook 7H9 medium or on egg yolk agar slopes at 30°C (32).
TABLE 1.
Mycobacteria used in this study
| ST | Identity no. | Species designation | Yr isolated | Origin | Place of origin | Sourcec | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 99/84a | M. marinum | 1999 | Bilby (Macrotis lagotis) | Western Australia | PC | 32 |
| 2 | JKD2394a | M. marinum | 1994 | Human | Victoria, Australia | VIDRL | 32 |
| 3 | 471a | M. marinum | Human | Norway | UTK | 36 | |
| 4 | 99/87a | M. marinum | 1996 | Human | Western Australia | PC | 32 |
| 5 | 99/89a | M. marinum | 1994 | Human | Tasmania, Australia | PC | 32 |
| 6 | 2000-372 | M. marinum | 2000 | Human | France | IP | 6 |
| 7 | 1726 | M. marinum | 1986 | Armadillo (Dasypus novemcinctus) | Louisiana | ITM | 35 |
| 8 | 1717 | M. marinum | 1986 | Armadillo (Dasypus novemcinctus) | Louisiana | ITM | 35 |
| 9 | DL240490 | M. marinum | 1990 | Fish (Dicentrarchus labrax) | Red Sea, Israel | NCM | 38 |
| 10 | CC240299 | M. marinum | 1999 | Fish (Cyprinus carpio) | Freshwater, Israel | NCM | 38 |
| 11 | DL045 | M. marinum | 2002 | Fish (Dicentrarchus labrax) | Mediterranean Sea, Greece | NCM | 38 |
| 12 | L15 | M. pseudoshottsii | 2002 | Fish (Morone saxatilis) | Chesapeake Bay | VIMS | 28 |
| 14 | 128FXT | M. liflandii | 2005 | Frog (Xenopus sp.) | California | UTK | 37 |
| 15 | 5114a | M. ulcerans | 1953 | Human | Mexico | ITM | 32 |
| 16 | 842a | M. ulcerans | 1986 | Human | Surinam | ITM | 32 |
| 17 | Agy99 | M. ulcerans | 1999 | Human | Ga District, Ghana | VIDRL | 32 |
| 18 | 13822/70a | M. ulcerans | 1971 | Human | North Queensland, Australia | QDRLMD | 32 |
| 18 | 1615 | M. ulcerans | 1964 | Human | Malaysia | UTK | 22 |
| 19 | 98912a | M. ulcerans | 1997 | Human | People's Republic of China | ITM | 9 |
| 19 | 753 | M. ulceransb | 2004 | Human | Japan | RIT | 17 |
| 20 | ATCC 19423a | M. ulcerans | 1948 | Human | Victoria, Australia | ATCC | 32 |
| 22 | M | M. marinum | Human | California | UW | 25 |
These strains used in a previous MLSA study (32).
This isolate is also referred to as Mycobacterium shinshuense.
PC, the Western Australian Centre for Pathology and Medical Research; VIDRL, Victorian Infectious Diseases Reference Laboratory, Victoria, Australia; UTK, Department of Microbiology, University of Tennessee, Knoxville, TN; IP, Institut Pasteur, Paris, France; ITM, Institute for Tropical Medicine, Antwerp, Belgium; NCM, National Centre for Mariculture, Eilat, Israel; VIMS, Virginia Institute of Marine Science, Gloucester Point, VA; QDRLMD,Queensland Diagnostic and Reference Laboratory for Mycobacterial Diseases, Australia; RIT, Research Institute of Tuberculosis, Japan Anti-Tuberculosis Association, Tokyo, Japan; UW, Department of Microbiology, University of Washington, Seattle.
General DNA methods and oligonucleotides used in this study.
Genomic DNA for DDH was extracted from mycobacteria harvested from egg yolk agar slopes. Briefly, 100 mg (wet weight) cells was resuspended in 2 ml of lysis buffer (15% [wt/vol] sucrose, 0.05 M Tris, pH 8.5, 0.05 M EDTA, 1 mg/ml lysozyme, and 2 mg/ml RNase) and incubated at 37°C for 1 hour. The volume was then increased to 5 ml by the addition of 2 ml of 10% (wt/vol) sodium dodecyl sulfate and 1 ml of 1× TE (10 mM Tris, pH 8.5, 1 mM EDTA) containing 5 mg of proteinase K. Following overnight incubation at 37°C, the DNA was separated by successive rounds of extraction with phenol-chloroform (1:1) and chloroform and then precipitated with a 2× volume of absolute ethanol in the presence of 4% NaCl at −20°C overnight. The resulting pellet was resuspended in 400 μl of 1× TE, and the extraction process was repeated. The final DNA pellet was washed twice in 70% (vol/vol) ethanol and resuspended in 100 μl of 1× TE.
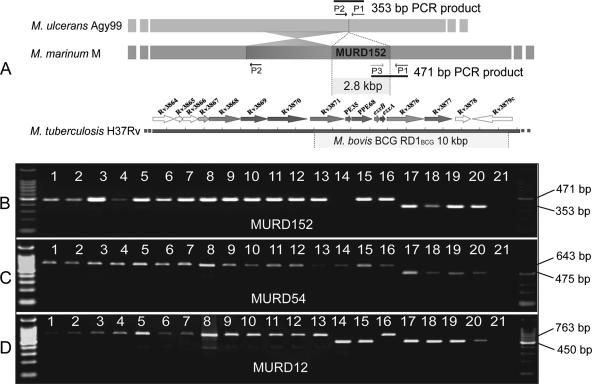
Other methods for PCR, pulsed-field gel electrophoresis, and Southern hybridization were also performed as described previously (32). For the MLSA, sequencing of a 357-bp fragment of the glcB gene was achieved by PCR amplification using glcB-F (5′-GGACTTCACCATCACCACCT-3′) and glcB-R (5′-GACTCCAGGATCACGGTCCTT-3′) (Fig. 1). Other primer pairs used for MLSA have been described before (32). The oligonucleotides used for PCR amplification and sequencing of potential pseudogenes are listed in Table S1 in the supplemental material. Genome comparisons between M. uclerans Agy99 and M. marinum M identified 157 specific DNA differences between the species, referred to as M. ulcerans regions of difference (MURDs) (34). Most of the MURDs are deletions of DNA from M. ulcerans. To investigate the distribution of three deletions (MURD12, -54, and -152) among the strains in this study, we performed PCR with three oligonucleotides for each MURD. These primers were designed based on genome comparisons between M. ulcerans Agy99 and M. marinum M (34), where P1 hybridized to conserved sequences in both strains flanking the region deleted in M. ulcerans, P2 hybridized to sequence adjacent to P1 in M. ulcerans, and P3 hybridized to sequence adjacent to P1 in M. marinum. The oligonucleotide sets used for the detection of MURD12, -54, and -152 are listed in Table S2 in the supplemental material.
FIG. 1.
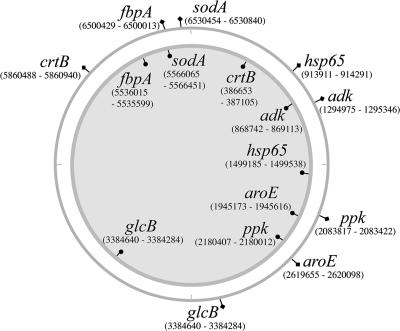
Distributions and nucleotide positions of the eight loci on the 5.6-Mbp chromosome of M. ulcerans Agy99 (inner circle) and the 6.6-Mbp chromosome of M. marinum M (outer circle).
DNA-DNA hybridization.
For each isolate, at least three biological repeats were performed, in which 100 ng of single-stranded genomic DNA was spotted in duplicate onto positively charged nylon membranes using a 96-well vacuum dot blotter (Schleicher & Schuell). DNA was cross-linked to the membranes with UV irradiation (1,200 mJ), washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and probed immediately. Genomic-DNA probes were prepared by incorporating digoxigenin dUTP by random prime labeling, following the manufacturer's instructions (Roche). Blots were hybridized and detected as described previously (15). Blot images were digitized and then analyzed with ImageJ (27). A standard spot area was used to measure the integrated pixel density. The relative binding ratio (RBR), defined as 100% for a blot hybridizing to a probe derived from autologous DNA, was calculated by dividing the mean integrated pixel density for each strain by the mean pixel density of the autologous reaction mixture and multiplying by 100. The mean was calculated from at least three independent hybridization reactions. Analysis of variance in conjunction with Dunnett's two-sided t test was used to determine if differences between RBRs were significant (P < 0.05). Where variance was not homogeneous (as determined by Levene's statistic), Dunnett's T3 post hoc test was employed. All calculations were performed using SPSS (version 14.0.1; LEAD Technologies, Inc.).
MLSA.
The methods used for MLSA were as previously described (32), with the addition of an eighth locus, a 357-bp fragment of the glcB gene that encodes malate synthase, the enzyme involved in the second step of the glyoxylate bypass. The distribution and nucleotide positions of the eight protein-coding DNA sequences (CDS) on the 5.6-Mbp chromosome of M. ulcerans Agy99 and the 6.6-Mbp chromosome of M. marinum M are shown in Fig. 1. Double-stranded DNA sequences were determined from the PCR products of each locus using dye terminator sequencing as described previously (32). Sequence assembly was managed with Gap4 (3). An allelic profile was constructed for each strain, and the unique combination of alleles defined a sequence type (Table 1; see Table S3 in the supplemental material). The DNA sequences of the alleles are listed in Table S3 in the supplemental material. The allelic profiles for some isolates differed at more than three of the eight loci, so phylogeny was inferred by using a distance method rather than a pairwise comparison of the allelic profiles (29); the latter approach is referred to as multilocus sequence typing (10). For each ST, the nucleotide sequences of the alleles were concatenated in frame to generate an artificial CDS. Using the same approach, all MPM were subjected to further MLSA analysis by determining the partial DNA sequences of four pMUM plasmid CDS (repA, parA, MUP045, and the mls loading domain) as described previously (31).
Phylogenetic methods.
Phylogenetic analysis was performed with MEGA software version 2.1.2 (18) and Splits Tree version 4 (14). “P” distances were used throughout, as the overall level of sequence divergence was small. Values for synonymous (dS) and nonsynonymous mutation frequencies were calculated by Nei and Gojobori's method (20), and standard errors (SE) for the means of these values were estimated by the method of Nei and Jin (21) using dSdNqw (7).
VNTR analysis.
The primers and conditions used for PCR amplification of the nine variable-number tandem repeat (VNTR) loci 1, 4, 6, 8, 9, 14, 15, 18, and 19 were as described previously (2). Amplicon sizes were estimated following electrophoresis of the PCR products through a 2% agarose gel and comparison with a 100-bp DNA ladder (Promega).
Nucleotide sequence accession numbers.
The DNA sequences of the alleles have been assigned GenBank accession numbers DQ985290 to DQ985355.
RESULTS
DNA-DNA hybridization.
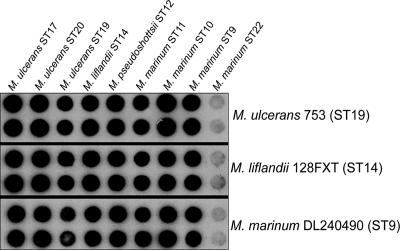
Whole-genome probes derived from the five novel MPM strains with MLSA ST9, -10, -11, -12, and -14 (see below); three M. ulcerans strains (ST17, -19, and -20); and a representative non-MPM M. marinum strain (ST22) were hybridized to each other in a reciprocal fashion. The five novel MPM strains were then tested against genomic DNAs from three additional M. marinum strains (ST1, -2, and -5). The results are summarized in Table 2, and an example is given in Fig. 2, showing that by this technique the five new MPM are indistinguishable from strains of M. ulcerans, with an overall mean RBR of 98% (range, 79 to 117%). In contrast, the mean RBR between the MPM and non-MPM was significantly lower at 40% (range, 13 to 68%; P < 0.05).
TABLE 2.
RBR for hybridization of mycobacteria using Digoxigenin-labeled genomic DNA probes
| DNA source | Mean (range) RBR with DNA probe:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
M. ulcerans
|
M. liflandii ST14 | M. pseudoshottsii ST12 |
M. marinum
|
||||||
| ST17 | ST20 | ST19 | ST11 | ST10 | ST9 | ST22 | |||
| M. ulcerans ST17 | 100 | ||||||||
| M. ulcerans ST20 | 97 (88-117) | 100 | |||||||
| M. ulcerans ST19 | 94 (84-101) | 104 (92-115) | 100 | ||||||
| M. liflandii ST14 | 100 (89-109) | 107 (95-105) | 99 (94-106) | 100 | |||||
| M. pseudoshottsii ST12 | 98 (80-110) | 102 (96-115) | 101 (95-107) | 102 (94-112) | 100 | ||||
| M. marinum ST11 | 94 (86-103) | 100 (72-113) | 88 (71-95) | 98 (86-107) | 102 (83-110) | 100 | |||
| M. marinum ST10 | 103 (93-108) | 100 (89-107) | 95 (82-100) | 100 (79-107) | 100 (82-105) | 96 (88-105) | 100 | ||
| M. marinum ST9 | 91 (80-105) | 101 (90-109) | 88 (79-98) | 96 (79-104) | 99 (88-108) | 100 (90-116) | 98 (88-112) | 100 | |
| M. marinum ST22 | 52 (40-59) | 60 (41-68) | 29 (21-44) | 33 (24-49) | 41 (32-52) | 32 (24-43) | 39 (26-50) | 37 (21-51) | 100 |
| M. marinum ST1 | NDa,b | NDb | ND | 19 (18-21) | 15 (13-15) | 27 (22-34) | 21 (18-24) | 23 (21-24) | ND |
| M. marinum ST2 | NDb | NDb | ND | 26 (24-27) | 15 (15-17) | 25 (20-34) | 23 (21-25) | 21 (18-25) | ND |
| M. marinum ST5 | NDb | NDb | ND | 32 (27-34) | 21 (20-22) | 30 (19-38) | 33 (28-38) | 23 (19-27) | ND |
ND, not done.
Previous DDH experiments reported a mean RBR of 38 (range, 25 to 47) between these strains of M. ulcerans and M. marinum (36).
FIG. 2.
Representative DNA-DNA hybridization result showing using whole-genome probes from three MPM against a panel of nine strains representing different MLSA sequence types.
MLSA of chromosomal genes.
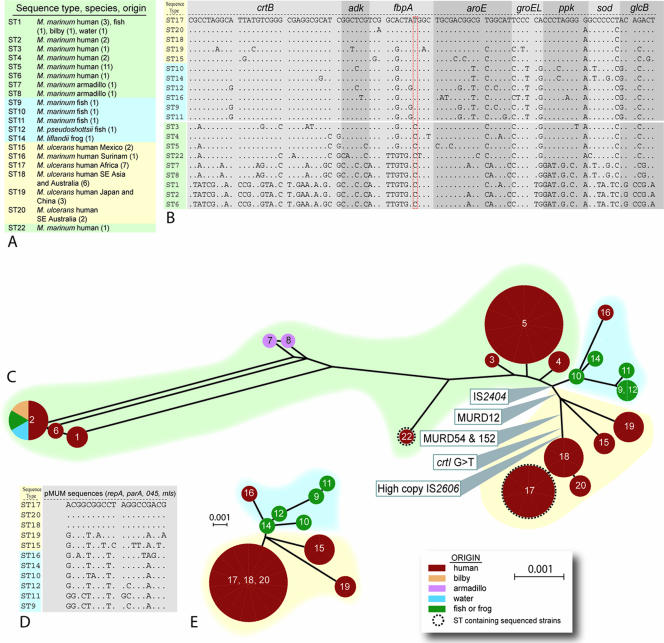
To improve chromosome coverage, we expanded our original MLSA method and added a 357-bp fragment of the glcB gene to the seven loci we had previously sequenced from 18 isolates of M. ulcerans and 22 isolates of M. marinum (Fig. 1) (32). A large difference in chromosomal location between the species was noted for some genes, and this was due to the extensive genome rearrangements that have occurred in M. ulcerans (34) (http://genolist.pasteur.fr/BuruList/). All eight loci were then sequenced for the 12 new isolates used in this study, which included five MPM recovered from diseased fish and frogs. Multiple alleles were obtained for each locus among the total of 52 mycobacteria tested. For example, 13 different crtB alleles, 7 different adk alleles, and 10 different fbpA alleles were obtained. A unique combination of alleles is referred to as an ST, and 20 STs were obtained (see Table S3 in the supplemental material). The eight alleles for each ST were concatenated in frame in the order crtB, adk, fbpA, aroE, groEL, ppk, sodA, and glcB to produce an artificial 3,210-bp CDS (Fig. 3A and B). DNA sequence alignments and comparisons of this CDS across all 20 STs revealed only 95 variable sites, representing a shared nucleotide identity of 97%, highlighting the very close genetic relationship between all the mycobacteria analyzed in this study (Fig. 3A and B). The MPM were represented by 11 STs, and they exhibited a very high level of nucleotide identity (>99%) (Fig. 3B). Significantly, all MPM contain the T-for-C transition within the fbpA gene, a polymorphic site suggested previously as a key discriminator between M. ulcerans and M. marinum (Fig. 3B) (32). A phylogeny inferred by split-decomposition analysis of the MLSA data shows a treelike structure among the MPM and divergence from other, non-MPM strains at a single node (Fig. 3C). Within the MPM, there is a second split into at least two distinct lineages that broadly correlate with those MPM causing disease in fish and frogs and those identified as M. ulcerans that cause Buruli ulcer in humans. Each of the novel MPM had related but distinct STs that clustered with a human M. ulcerans isolate from Surinam.
FIG. 3.
Multilocus sequence analysis. (A) Species designations for the 20 sequence types, their origins, and (in parentheses) the numbers of isolates tested. (B) Alignment of the 3,210-bp sequences derived from the eight concatenated chromosomal CDS fragments among 20 different sequence types. Only variable nucleotides are shown. A period indicates identity with M. ulcerans ST17 (African type). (C) Split-decomposition representation of the phylogenetic relationships among M. ulcerans, other mycolactone-producing mycobacteria, and M. marinum strains. The circles at the vertices are labeled with the sequence types, and their sizes are proportional to the numbers of isolates within the groups, following a layout described previously (8). M. marinum ST1, -2, -3, -4, and -5 correspond to STI, -II, -III, -IV, and -V as previously reported (32). All edges had 100% bootstrap support (1,000 replicates). (D) Alignment of the 1,266-bp sequences derived from the four concatenated pMUM CDS fragments among 11 different sequence types. Only variable nucleotides are shown. A period indicates identity with M. ulcerans ST17 (African type). (E) Split-decomposition graph of the phylogenetic relationships among M. ulcerans and other mycolactone-producing mycobacteria, derived from the plasmid gene sequences. All edges had >60% bootstrap support (1,000 replicates).
Among the nine non-MPM M. marinum sequence types, only ST2 contained isolates from a variety of sources, including humans, fish, and water. The M. marinum strain 2000-372 (ST6) has been proposed as a strain that represents an evolutionary “missing link” between M. ulcerans and M. marinum, because it does not make mycolactone but returns a positive result in a PCR test for IS2404 (6). MLSA unambiguously grouped this strain within the STs most distant from M. ulcerans, and hence, it is very unlikely that this strain is an intermediate between the species (Fig. 3C). Sequence analysis of the IS2404 PCR product from this strain showed that it contains an ISE that is distinct from IS2404, as it is only 86% identical at the nucleotide level.
MLSA of pMUM plasmid genes.
Sequence comparisons and split-decomposition analysis of the 1,266-bp concatenated DNA sequences from four pMUM gene sequences among the MPM produced a tree topology very similar to that obtained from the eight chromosomal loci (Fig. 3D and E). There was clear separation between the fish and frog MPM and M. ulcerans. dS is a measure of the time a given coding sequence has been extant relative to another. Sequences with similar dS are likely to have been acquired at the same time. The parameter dS was not significantly different for plasmid (dS mean = 0.00639; SE = 0.00234) and chromosome (dS mean = 0.00515; SE = 0.00135) sequences, consistent with the idea that the plasmid has coevolved with all MPM from a common progenitor.
Comparison of plasmid sizes and distributions of IS2404 and IS2606.
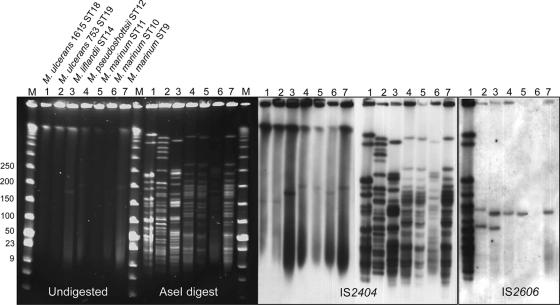
Pulsed-field gel electrophoresis and Southern hybridization were used to compare the sizes of the pMUM plasmids among the novel MPM and to explore ISE copy numbers. M. ulcerans strains from Malaysia and Japan had plasmids of approximately 170 kb, as previously reported for strains from Africa and Australia (Fig. 4) (31, 33). The novel MPMs had plasmids of larger sizes. M. liflandii (ST14) and M. marinum strain CC240299 (ST10) harbor plasmids of approximately 180 kbp, while M. pseudoshottsii (ST12), M. marinum DL240490 (ST9), and M. marinum DL045 (ST11) all harbor plasmids of approximately 200 kbp (Fig. 4). The plasmid size correlated with restriction fragment polymorphism, as shown after digestion of DNAs from these strains with AseI and hybridization with probes derived from IS2404 and IS2606. Of particular note are the patterns of bands, which are indistinguishable from each other for the strains with 200-kbp plasmids (ST9, -11, and -12). MLSA supported this finding with 99.9% shared nucleotide identity and only three variable nucleotides from the 3,210 bp sequenced among these strains (Fig. 3). Southern hybridization confirmed that all MPM contain many copies of IS2404, suggesting that this insertion sequence and possibly its expansion to a high copy number were characteristics of the common progenitor. The same is not true for IS2606. It was not detected in M. marinum CC240299, a finding confirmed by PCR (data not shown), and was present in low copy numbers in the M. ulcerans strain from Japan (ST19) and the novel MPM (Fig. 4), suggesting that it was acquired independently by the two different lineages of MPM.
FIG. 4.
Pulsed-field gel electrophoresis and Southern hybridization analysis of MPM showing the presence in all isolates of pMUM-like plasmids and the distributions of IS2404 and IS2606.
Distribution of pseudogenes among novel MPM.
Approximately 14% of the original coding potential of M. ulcerans Agy99 has been lost due to point mutations or insertion sequences that have disrupted CDS and led to the formation of pseudogenes. To explore the distribution of pseudogenes in the novel MPM, we used PCR to amplify and then sequence the region spanning different mutations. These mutations were nucleotide changes that changed the reading frame or introduced a premature stop codon or disruptions by insertion of IS2404. Twenty of the 743 pseudogenes (including 21 mutations) identified in M. ulcerans Agy99 were selected for comparison with novel MPM of ST9, -10, and -11. These sequences were selected as a random sample of the total pseudogene complement with potential to explain phenotypic differences between strains. The results are summarized in Table S4 in the supplemental material and show that 15 of these pseudogenes are still intact in the novel MPM; of the five that were pseudogenes, only one (an insertion of 5 nucleotides in arsC) was caused by the same mutation as in M. ulcerans Agy99. The frequency of the G-to-T transition in crtI, encoding phytoene dehydrogenase, which is required for carotenoid pigment production, was more widely investigated by testing all 52 mycobacteria. This mutation was found to be restricted to M. ulcerans strains belonging to ST17, -18, and -20 (Fig. 3C).
Distribution of DNA deletions among MPM.
Deletion of DNA fragments occurs frequently in mycobacteria, and their distribution among strains can be used to reconstruct evolutionary pathways (5). Comparative genomics revealed 157 between M. ulcerans strain Agy99 (ST17) and M. marinum M (ST22) (34). Three of these regions (MURD12, MURD54, and MURD152) that were shown to be deletions in M. ulcerans Agy99 were selected for further study. Using PCR, MPM and M. marinum were screened for the presence or absence of each MURD, using the approach developed for studying members of the Mycobacterium tuberculosis complex (5) (Fig. 5A). Analysis of strains representing each of the 20 STs revealed that MURD12 had been lost from all strains that constituted the endotherm lineage, represented by ST15, -17, -18, -19, and -20 (Fig. 3C and 5). M. ulcerans strains of the subcluster within the endotherm lineage (ST17, -18, and -20) have lost MURD54 and MURD152 (Fig. 3C and 5). M. ulcerans strain 5114 from Mexico (ST15) did not produce a product in the MURD152 assay, suggesting that this strain has undergone a different chromosomal modification in this region.
FIG. 5.
PCR deletion analysis for MURD12, MURD55, and MURD152 of strains representing each MLSA sequence type. (A) Arrangement of the oligonucleotides used for PCR of MURD152, showing the 471-bp product predicted from the genome sequence of M. marinum strain M and the 353-bp product predicted from the genome sequence of M. ulcerans Agy99. Also shown for reference is the alignment of MURD152 with the RD1 region absent from Mycobacterium bovis BCG. (B, C, and D) Results of deletion PCR analysis for MURD152, MURD54, and MURD12, respectively. Lanes 1 to 11, M. marinum (1, 99/84; 2, JKD2394; 3, 471; 4, 99/87; 5, 99/89; 6, 2000-372; 7, 1717; 8, M; 9, DL240490; 10, CC240299; 11, DL045); lane 12, M. pseudoshottsii L15; lane 13, M. liflandii 128FXT; lane 14, M. ulcerans Mexico 5114; lane 15, M. ulcerans Japan 753; lane 16, M. marinum Surinam 842; lane 17, M. ulcerans Agy99; lane 18, M. ulcerans 1615; lane 19, M. ulcerans 13822/70; lane 20, M. ulcerans 19423; lane 21, no template control. On the left is a 100-bp DNA molecular size ladder (Promega).
VNTR analysis distinguishes different MPM.
Variable-number tandem repeat loci are widespread in mycobacterial genomes, and their varied distribution has been exploited for strain differentiation. A system of PCR and sequence-based VNTR typing has been described for high-resolution differentiation among M. marinum and M. ulcerans strains (1, 2, 12, 35), and we applied this technique to the MPM strains in this study. Comparisons of fragment sizes at VNTR loci 1, 4, 6, 8, 9, 14, 15, 18, and 19 permitted the discrimination of the novel MPM from other strains of M. marinum and M. ulcerans. However, M. pseudoshottsii L15 (ST12), M. marinum DL240490 (ST9), and M. marinum DL045 (ST11) shared the same VNTR profile (data not shown).
DISCUSSION
Initial MLSA analyses and subsequent whole-genome comparisons have shown that M. ulcerans has recently evolved from M. marinum by acquisition of the pMUM plasmid and reductive evolution (31-34). The recent discovery of MPM that are phenotypically distinct from M. ulcerans in diseased fish and frogs has highlighted the possibility that pMUM is being transferred among different mycobacterial species (19, 26). In this report, we show by a systematic genetic approach that all MPM are very closely related to each other and have evolved, not by multiple exchanges of pMUM, but from a common M. marinum progenitor that acquired the plasmid.
MLSA is widely used to understand the taxonomic relationships among bacterial populations (8, 10, 11), and it was the method we employed in an earlier study to suggest that M. ulcerans recently evolved from M. marinum (32). To improve genome coverage and increase resolution of the MLSA method for the present investigation, we added an eighth locus to create a 3,210-bp semantide (a large information-bearing molecule). We reanalyzed our original data set and then added sequences from 12 additional isolates that included 5 mycobacteria recently shown to contain pMUM and to produce mycolactones. There was significant sequence diversity among non-mycolactone-producing M. marinum isolates, and as shown by others (32, 39), the majority of isolates fell into two distinct clusters represented by ST1, -2, and -6 and ST 3, -4, and -5. The sample size in this study was too small to draw conclusions linking specific M. marinum genotypes to virulence in humans, as has been proposed (39); however, such a correlation seems unlikely, given that M. marinum isolates of human origin spanned the spectrum of sequence diversity revealed by MLSA. Two M. marinum isolates recovered from armadillos showed intermediate sequence types (ST7 and -8), as did M. marinum strain “M” (ST22), whose genome has recently been sequenced (http://www.sanger.ac.uk/Projects/M_marinum).
MLSA unambiguously showed that, despite their varying phenotypes, all MPM have evolved from a single M. marinum clone that has since expanded into at least two distinct lineages, and this was supported by the congruent tree topology derived from the four pMUM plasmid loci (Fig. 3). Together with equivalent levels of synonymous nucleotide substitution frequency between chromosome and episome sequences, these data suggest that plasmid acquisition was probably the principal event that enabled an M. marinum progenitor to survive in a new environment.
The genetic homogeneity of MPM was also reflected in their high DDH values. DDH has been widely used for over 30 years in bacterial taxonomy to infer relatedness between genomes, and a DDH value greater than 70% is one criterion used to help define a bacterial species. The high DDH values among M. ulcerans and the other MPM were further evidence of their common origin and contrasted with the low (<55% RBR) values when the same MPM were tested against non-mycolactone-producing M. marinum strains. These data are consistent with an earlier investigation of M. ulcerans and M. marinum that showed intraspecies DDH values of >90% and interspecies DDH values of <50% (36). The presence of pMUM-like plasmids and the multiple copies of IS2404 in all MPM may explain, at least in part, the striking DDH results. These data also indicate that IS2404 acquisition (and possibly its expansion to high copy numbers) occurred before radiation of MPM around the world. In contrast, IS2606 is present in high copy numbers only in the lineage of M. ulcerans strains that contain isolates from Africa, Australia, and Malaysia (ST17, -18, and -20) (30, 32). The varied distribution of this ISE among other MPM and its absence from the MPM M. marinum strain CC240299 (ST10) suggest it has been transferred independently to at least two different populations of MPM subsequent to IS2404 and pMUM acquisition.
The M. ulcerans genome project and a recent microarray-based study have both revealed extensive DNA deletion polymorphism among M. ulcerans strains (34; M. Käser, S. Rondini, T. Stinear, M. Tessier, C. Mangold, G. Dernick, M. Naegeli, F. Portaels, U. Certa, and G. Pluschke, submitted for publication). In the current work, the pattern of DNA deletion observed among MPM for three deletions was in good agreement with MLSA (Fig. 5). MURD12, a 10-kbp fragment containing CDS involved in secondary metabolism, was a marker for distinguishing between the ectotherm and endotherm lineages, as it was absent from M. ulcerans isolates from both Mexico and Japan (ST15 and -19) and the African, Malaysian, and Australian cluster (ST17, -18, and -20) (Fig. 3C and 5C). The MURD54 and MURD152 deletions differentiate the ST17, -18, and -20 subcluster from other MPM and are indicative of more advanced genome reduction in these strains. MURD152 is a 2.8-kb DNA fragment deleted from M. ulcerans strain Agy99 (ST17) that spans esxA and esxB, genes encoding key components of the ESX-1 secretion apparatus and virulence factor (4, 34). The MURD152 deletion PCR assay confirmed earlier findings that showed that esxA and esxB are absent from M. ulcerans strains from Africa, Australia, and Malaysia but present in other MPM (19, 26). Both the MURD54 and MURD152 assays may have diagnostic applications in countries where Buruli ulcer is endemic, such as Africa and Australia, where it will be useful, particularly when screening environmental samples, to distinguish between M. ulcerans strains that cause Buruli ulcer and other MPM.
The split of MPM into two distinct lineages, which include strains with different species names (e.g., M. marinum, M. liflandii, and M. pseudoshottsii) that typically cause disease in ectotherms but also have a high zoonotic potential and strains of M. ulcerans (ST17, -18, and -20) that cause Buruli ulcer in humans and target other endotherms, is an important finding. Some insights into the genetic basis for this separation have been gleaned from this study and, combined with previous research showing that these strains have different optimal growth temperatures and produce mycolactones with varying potencies (19, 26), they suggest MPM have evolved to occupy different ecological niches. M. ulcerans is not known to infect fish, while the diseases caused by M. marinum and M. ulcerans in humans differ greatly in their clinical, histopathologic, and epidemiologic aspects (32).
The large number of gene deletions and pseudogenes in the M. ulcerans Agy99 genome compared with M. marinum M is indicative of a bacterium adapting to a restricted and privileged environment, where mutations are tolerated in genes that are no longer required for survival. However, testing three MPM from the ectotherm lineage for 20 of these pseudogenes found only five inactivated CDS, indicating a much less advanced level of genome decay and metabolic streamlining, consistent with the hypothesis that they occupy different environments. Only one of the five mutations (an insertion in arsC) was conserved between the two MPM lineages (see Table S4 in the supplemental material), suggesting that a certain level of genome decay had begun before divergence. The other four mutations occurred at different positions in the same genes (sigJ, echA13, and accD1), and this may indicate that the products of these CDS are not only redundant but perhaps deleterious for survival of MPM and so have been subjected to independent, purifying selection.
The difference in pseudogene profiles may also help explain the phenotypic variation observed among MPM. For example, crtI encodes phytoene dehydrogenase, an enzyme essential for the production of carotenoid pigments in M. marinum (25). M. ulcerans Agy99 has the same crt locus as M. marinum, but it is nonpigmented, and this has been explained by a point mutation in crtI that introduces a premature stop codon and truncates the gene (see Table S4 in the supplemental material) (34). There was complete correlation between lack of pigment production and the disrupted crtI gene, as only MPM of the ST17, -18, and -20 cluster are nonpigmented (24, 26), and it was only this cluster that contained the mutation (Fig. 3C).
In this report, we have sought to clarify the genetic relationships among mycolactone- and non-mycolactone-producing mycobacteria, but this has in turn highlighted the recurring problem of assigning species status to highly related bacteria, as the question remains how much diversity is permissible in a genetically discrete cluster for it to be regarded as a distinct taxon. From a population genetics standpoint, the data presented here do not support the separation of MPM into different species. Employing a subspecies nomenclature might allow a more meaningful naming system that accurately reflects the common origin of MPM. A comprehensive polyphasic and multicenter study of MPM, as performed by Wayne et al. (41), would help decide their taxonomic positions.
Defining the host specificity and natural ecology of MPM is also a research priority. It may be that there are many different MPM but the only strains isolated are those producing mycolactones with sufficient potency to cause disease in humans, fish, frogs, possums, and koalas (23). A better understanding of MPM in the environment will be crucial to halting the spread of the diseases they cause, in particular Buruli ulcer.
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support of the National Health and Medical Research Council of Australia. The collection of the Israeli mycobacterial isolates is maintained at NCM, Eilat, Israel, thanks to the generosity of Dan and Florence Green (North American Friends of IOLR).
We thank Amy Shields for statistical advice, Y. Kazumi and P. L. C. Small for providing M. ulcerans strains, and J. Parkhill for the use of unpublished M. marinum genome sequence data produced by the Pathogen Sequencing Group at the Sanger Institute (ftp://ftp.sanger.ac.uk/pub/pathogens/mm).
Footnotes
Published ahead of print on 15 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ablordey, A., M. Hilty, P. Stragier, J. Swings, and F. Portaels. 2005. Comparative nucleotide sequence analysis of polymorphic variable-number tandem-repeat loci in Mycobacterium ulcerans. J. Clin. Microbiol. 43:5281-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablordey, A., J. Swings, C. Hubans, K. Chemlal, C. Locht, F. Portaels, and P. Supply. 2005. Multilocus variable-number tandem repeat typing of Mycobacterium ulcerans. J. Clin. Microbiol. 43:1546-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonfield, J. K., K. F. Smith, and R. Staden. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 24:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodin, P., I. Rosenkrands, P. Andersen, S. T. Cole, and R. Brosch. 2004. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 12:500-508. [DOI] [PubMed] [Google Scholar]
- 5.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chemlal, K., G. Huys, F. Laval, V. Vincent, C. Savage, C. Gutierrez, M. A. Laneelle, J. Swings, W. M. Meyers, M. Daffe, and F. Portaels. 2002. Characterization of an unusual Mycobacterium: a possible missing link between Mycobacterium marinum and Mycobacterium ulcerans. J. Clin. Microbiol. 40:2370-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Silva, J., and A. L. Hughes. 1998. dSdNqw, 1.0 ed. Pennsylvania State University, University Park, PA.
- 8.Diavatopoulos, D. A., C. A. Cummings, L. M. Schouls, M. M. Brinig, D. A. Relman, and F. R. Mooi. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathogens 1:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faber, W. R., L. M. Arias-Bouda, J. E. Zeegelaar, A. H. Kolk, P. A. Fonteyne, J. Toonstra, and F. Portaels. 2000. First reported case of Mycobacterium ulcerans infection in a patient from China. Trans. R. Soc. Trop. Med. Hyg. 94:277-279. [DOI] [PubMed] [Google Scholar]
- 10.Gevers, D., F. M. Cohan, J. G. Lawrence, B. G. Spratt, T. Coenye, E. J. Feil, E. Stackebrandt, Y. Van de Peer, P. Vandamme, F. L. Thompson, and J. Swings. 2005. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733-739. [DOI] [PubMed] [Google Scholar]
- 11.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilty, M., D. Yeboah-Manu, D. Boakye, E. Mensah-Quainoo, S. Rondini, E. Schelling, D. Ofori-Adjei, F. Portaels, J. Zinsstag, and G. Pluschke. 2006. Genetic diversity in Mycobacterium ulcerans isolates from Ghana revealed by a newly identified locus containing a variable number of tandem repeats. J. Bacteriol. 188:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong, H., T. Stinear, P. Skelton, J. B. Spencer, and P. F. Leadlay. 2005. Structure elucidation of a novel family of mycolactone toxins from the frog pathogen Mycobacterium sp. MU128FXT by mass spectrometry. Chem. Commun. 14:4306-4308. [DOI] [PubMed] [Google Scholar]
- 14.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 15.Jenkin, G. A., T. P. Stinear, P. D. Johnson, and J. K. Davies. 2003. Subtractive hybridization reveals a type I polyketide synthase locus specific to Mycobacterium ulcerans. J. Bacteriol. 185:6870-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, P. D., T. Stinear, P. L. Small, G. Pluschke, R. W. Merritt, F. Portaels, K. Huygen, J. A. Hayman, and K. Asiedu. 2005. Buruli ulcer (M. ulcerans infection): new insights, new hope for disease control. PLoS Med. 2:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazumi, Y., K. Ohtomo, M. Takahashi, S. Mitarai, I. Sugawara, J. Izumi, A. Andoh, and H. Hasegawa. 2004. Mycobacterium shinshuense isolated from cutaneous ulcer lesion of right lower extremity in a 37-year-old woman. Kekkaku 79:437-441. [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Mve-Obiang, A., R. E. Lee, E. S. Umstot, K. A. Trott, T. C. Grammer, J. M. Parker, B. S. Ranger, R. Grainger, E. A. Mahrous, and P. L. Small. 2005. A newly discovered mycobacterial pathogen isolated from laboratory colonies of Xenopus species with lethal infections produces a novel form of mycolactone, the Mycobacterium ulcerans macrolide toxin. Infect. Immun. 73:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 21.Nei, M., and L. Jin. 1989. Variances of the average numbers of nucleotide substitutions within and between populations. Mol. Biol. Evol. 6:290-300. [DOI] [PubMed] [Google Scholar]
- 22.Pettit, J. H., N. J. Marchette, and R. J. Rees. 1966. Mycobacterium ulcerans infection. Clinical and bacteriological study of the first cases recognized in South East Asia. Br. J. Dermatol. 78:187-197. [DOI] [PubMed] [Google Scholar]
- 23.Portaels, F., K. Chemlal, P. Elsen, P. D. Johnson, J. A. Hayman, J. Hibble, R. Kirkwood, and W. M. Meyers. 2001. Mycobacterium ulcerans in wild animals. Rev. Sci. Technol. 20:252-264. [DOI] [PubMed] [Google Scholar]
- 24.Portaels, F., P. A. Fonteyene, H. de Beenhouwer, P. de Rijk, A. Guedenon, J. Hayman, and M. W. Meyers. 1996. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates. J. Clin. Microbiol. 34:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramakrishnan, L., H. T. Tran, N. A. Federspiel, and S. Falkow. 1997. A crtB homolog essential for photochromogenicity in Mycobacterium marinum: isolation, characterization, and gene disruption via homologous recombination. J. Bacteriol. 179:5862-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranger, B. S., E. A. Mahrous, L. Mosi, S. Adusumilli, R. E. Lee, A. Colorni, M. Rhodes, and P. L. Small. 2006. Globally distributed mycobacterial fish pathogens produce a novel plasmid-encoded toxic macrolide, mycolactone F. Infect. Immun. 74:6037-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasband, W. S. 2006. ImageJ, 1.36b ed. National Institutes of Health, Bethesda, MD.
- 28.Rhodes, M. W., H. Kator, A. McNabb, C. Deshayes, J. M. Reyrat, B. A. Brown-Elliott, R. Wallace, Jr., K. A. Trott, J. M. Parker, B. Lifland, G. Osterhout, I. Kaattari, K. Reece, W. Vogelbein, and C. A. Ottinger. 2005. Mycobacterium pseudoshottsii sp. nov., a slowly growing chromogenic species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int. J. Syst. Evol. Microbiol. 55:1139-1147. [DOI] [PubMed] [Google Scholar]
- 29.Spratt, B. G., and M. C. Maiden. 1999. Bacterial population genetics, evolution and epidemiology. Phil. Trans. R. Soc. Lond B 354:701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stinear, T., B. C. Ross, J. K. Davies, L. Marino, R. M. Robins-Browne, F. Oppedisano, A. Sievers, and P. D. Johnson. 1999. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 37:1018-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stinear, T. P., H. Hong, W. Frigui, M. J. Pryor, R. Brosch, T. Garnier, P. F. Leadlay, and S. T. Cole. 2005. Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J. Bacteriol. 187:1668-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stinear, T. P., G. A. Jenkin, P. D. R. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stinear, T. P., A. Mve-Obiang, P. L. Small, W. Frigui, M. J. Pryor, R. Brosch, G. A. Jenkin, P. D. Johnson, J. K. Davies, R. E. Lee, S. Adusumilli, T. Garnier, S. F. Haydock, P. F. Leadlay, and S. T. Cole. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA 101:1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stinear, T. P., T. Seemann, S. Pidot, W. Frigui, G. Reysset, T. Garnier, G. Meurice, D. Simon, C. Bouchier, L. Ma, M. Tichit, J. L. Porter, J. Ryan, P. D. R. Johnson, J. K. Davies, G. A. Jenkin, P. L. C. Small, L. M. Jones, T. F. F. Laval, M. Daffé, J. Parkhill, and S. T. Cole. 8 January 2007. Reductive evolution and niche-adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed]
- 35.Stragier, P., A. Ablordey, W. M. Meyers, and F. Portaels. 2005. Genotyping Mycobacterium ulcerans and Mycobacterium marinum by using mycobacterial interspersed repetitive units. J. Bacteriol. 187:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tønjum, T., D. B. Welty, E. Jantzen, and P. L. Small. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trott, K. A., B. A. Stacy, B. D. Lifland, H. E. Diggs, R. M. Harland, M. K. Khokha, T. C. Grammer, and J. M. Parker. 2004. Characterization of a Mycobacterium ulcerans-like infection in a colony of African tropical clawed frogs (Xenopus tropicalis). Comp. Med. 54:309-317. [PubMed] [Google Scholar]
- 38.Ucko, M., and A. Colorni. 2005. Mycobacterium marinum infections in fish and humans in Israel. J. Clin. Microbiol. 43:892-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Sar, A. M., A. M. Abdallah, M. Sparrius, E. Reinders, C. M. Vandenbroucke-Grauls, and W. Bitter. 2004. Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infect. Immun. 72:6306-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Werf, T. S., T. Stinear, Y. Stienstra, W. T. van der Graaf, and P. L. Small. 2003. Mycolactones and Mycobacterium ulcerans disease. Lancet 362:1062-1064. [DOI] [PubMed] [Google Scholar]
- 41.Wayne, L. G., R. C. Good, E. C. Bottger, R. Butler, M. Dorsch, T. Ezaki, W. Gross, V. Jonas, J. Kilburn, P. Kirschner, M. I. Krichevsky, M. Ridell, T. M. Shinnick, B. Springer, E. Stackebrandt, I. Tarnok, Z. Tarnok, H. Tasaka, V. Vincent, N. G. Warren, C. A. Knott, and R. Johnson. 1996. Semantide- and chemotaxonomy-based analyses of some problematic phenotypic clusters of slowly growing mycobacteria, a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 46:280-297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.