Abstract
An assay modeled on a known polymorphism in the PE_PGRS9 gene of Mycobacterium tuberculosis was designed to assess the mutability of a sequence containing interspersed PGRS repeats. Application of the assay in Mycobacterium smegmatis revealed sequence plasticity: in addition to recapitulating the mutation on which it was based, other mutations likely mediated by replication slippage between PGRS repeats were detected. However, the mutation rates argued against marked hypermutability of such sequences in mycobacteria.
One of the defining features of the Mycobacterium tuberculosis genome is the presence of the large PE and PPE gene families, which together comprise ∼10% of its coding capacity and are characterized by a G+C content of ∼80% (26), which is markedly higher than the average 65.6% G+C content across the genome (8). The PE_PGRS subfamily of the 100-member PE family of M. tuberculosis H37Rv comprises ∼63 members, whose hallmark feature is the presence of multiple PGRS (polymorphic GC-rich sequence) elements in their 3′ coding regions that encode a novel, C-terminal, Gly-Ala-rich “PGRS domain” characterized by numerous repeats of GGA and GGN residues (7). The function of PE_PGRS proteins in M. tuberculosis has been the subject of considerable interest and debate (7). Interaction of PE_PGRS proteins with the immune system is well documented (1, 5, 6, 11, 13), with the most widely studied member, PE_PGRS33 (Rv1818c), being shown to be a cell surface-associated antigen (5). Differential expression of PE_PGRS genes has been observed under a variety of experimental conditions (1, 3, 10, 14, 16, 29, 34), suggesting a possible specialization of function within the family, and studies with M. marinum have provided evidence of the involvement of PE_PGRS genes in mycobacterial virulence (27).
There is substantial evidence implicating PE_PGRS elements in genome plasticity and strain evolution. The earliest report of allelic diversity in a PE_PGRS gene was for the Rv0746 genes (PE_PGRS9) of M. tuberculosis H37Rv and M. bovis BCG Pasteur (8) (Fig. 1A). A subsequent investigation of PE_PGRS variability at the protein level was strongly indicative of allelic diversity within and between species (1). Genome-wide sequence comparisons similarly revealed significant allelic variation in PE_PGRS genes between M. tuberculosis H37Rv and CDC1551 (15). More recently, sequence analysis of PE_PGRS33 in 123 clinical isolates of M. tuberculosis identified 25 different variations, including 9 deletions, 3 insertions, a deletion/insertion, and 12 single-nucleotide polymorphisms, attesting to its polymorphic character (33).
FIG. 1.
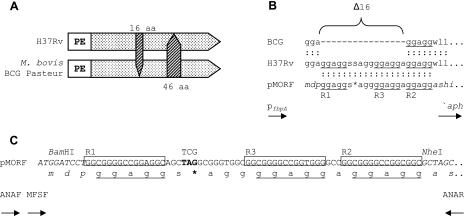
Design of the pMORF reporter system. (A) Schematic representation of the polymorphic PE_PGRS9 genes in M. tuberculosis H37Rv (783 amino acids) and M. bovis BCG (813 amino acids) (not to scale). The arrowheads denote the absence of amino acids (aa) in the respective alleles. (B) Protein sequence alignment of the M. bovis BCG Pasteur and H37Rv homologs of PE_PGRS9 and the reporter construct pMORF, where Δ16 denotes the 16-amino-acid deletion reported by Cole et al. (8). The pentapeptide GGAGG repeats within and immediately adjacent to the 16-amino-acid deletion are denoted by R1, R2, and R3 and are underlined. The DNA sequence of the pMORF reporter spanning from R1 to R2 is identical to that of PE_PGRS9 from M. tuberculosis H37Rv, with the exception of the engineered point mutation (TCG→TAG), which converted the Ser codon to a stop codon. (C) DNA and amino acid sequences of the PGRS-containing regions of the pMORF reporter. DNA sequences flanking the PGRS-containing element are shown in italics. The imperfect 15-bp repeats R1, R3, and R2, which encode the GGAGG pentapeptide repeats, are boxed. Stop codons are shown in bold and are represented by an asterisk in the corresponding protein sequence. The p3STOP vector was designed to monitor slippage events between repeats R1 and R2, which are connected by a synthetic 16-bp linker that contains stop codons in all three reading frames. The locations of the primers used for PCR-based genotyping (ANAF and ANAR) and DNA sequencing (MFSF) are shown below the pMORF reporter sequence.
In spite of the evidence for allelic diversity, at least in some PE and PPE members, it is not known whether the encoding genes are inherently unstable and prone to mutation at significantly elevated rates. To assess the mutability of PGRS-containing sequences in mycobacteria, we developed a reporter assay for detecting mutational events within such sequences, and in this paper we describe the results of its application in Mycobacterium smegmatis.
Forward mutation assay for monitoring rearrangements between PGRS repeats by fluctuation analysis.
We designed a ′aph-based (20) reporter modeled on the sequence of a 16-amino-acid polymorphism in PE_PGRS9 (Δ16) (Fig. 1A) that would allow rearrangements involving neighboring PGRS repeats to be detected by a phenotypic change of the host mycobacterial strain from Kms to Kmr (Fig. 1B and C). The reporter, pMORF, contains a 63-bp region of PE_PGRS9 spanning from the 5′ end of the R1 repeat sequence to the 3′ end of the R2 repeat. This region results in three Gly-Gly-Ala-Gly-Gly (GGAGG) repeats, encoded by R1, R2, and R3. The sequence of this 63-bp element differs from the authentic H37Rv PE_PGRS9 sequence at a single nucleotide position between R1 and R3, which converts a Ser codon to a stop codon (TCG→TAG) (Fig. 1B and C). According to this design, pMORF cannot confer a Kmr phenotype; a gain of kanamycin (Km) resistance would require a mutational event to occur that bypasses the stop codon or converts it to a sense codon.
Luria-Delbrück fluctuation analysis was used to determine the mutation rates of M. smegmatis strains carrying pMORF by analyzing the distribution of Kmr mutant numbers in parallel cultures. Fluctuation tests were performed with between 5 and 13 parallel cultures. Single Gmr (17) colonies from electroporation of pMORF into wild-type M. smegmatis mc2155 (31) or an isogenic recA deletion mutant, ΔrecA (see Table S1 in the supplemental material) (24, 25), were used to generate the inocula, which were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (vol/vol) albumin-dextrose-catalase (Merck), 0.2% glycerol, 0.1% Tween 80, and gentamicin (Gm; 2 μg/ml) to an optical density at 600 nm of approximately 1.0. The inocula were diluted to a cell density of 102 to 103 CFU/ml, as assessed by plating on Middlebrook 7H10 medium (Difco) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (Merck), 0.5% glycerol, and Gm (5 μg/ml), and 2-ml aliquots were grown at 37°C in 15-ml culture tubes for 6 days, with shaking at 300 rpm. To ensure that no preexisting Kmr mutants were present after expansion from the Gmr colonies and dilution to seeding density, inocula were plated directly onto medium containing Km (25 μg/ml). For selection of mutant clones, the entire contents of a culture tube were plated on Km-supplemented medium following the removal of a 50-μl aliquot for CFU determination. Plates were incubated at 32°C and scored for CFU after 3 days of growth. Mutation rates were calculated as follows: μ = ln 2 · m/Nt (19). The m values (number of mutational events per culture) were obtained by mathematical modeling using the Lea-Coulson method of the median (18). For determination of Nt values (final number of cells in the culture) (30), serial dilutions were plated, and CFU were scored after 3 days of growth.
The results of two independent fluctuation tests, with each performed on cultures of mc2155::pMORF and ΔrecA::pMORF grown to stationary phase, are shown in Table 1. These experiments yielded between 62 and 1,504 Kmr mutants from a final number of ∼109 cells per culture (Nt) (see Table S3 in the supplemental material). The overall rate of mutation of M. smegmatis to Km resistance, which is a composite of the rates of the multiple distinct mutational events occurring within parallel cultures, was not affected by deletion of the recA gene. This finding argues against a role for genetic recombination in the mutational events detected in this system, most probably due to the limited size of the stretches of homology between the repeats in the reporter (32).
TABLE 1.
Determination of mutation rate of M. smegmatis carrying pMORF to Km resistance by fluctuation analysis
| Fluctuation expt no. | Strain | No. of cultures | Nt valuea | Lea-Coulson m valueb | Mutation ratec |
|---|---|---|---|---|---|
| 1 | mc2155::pMORF | 13 | 1.0 × 109 ± 0.4 × 109 | 31 | 2.1 × 10−8 |
| 2 | mc2155::pMORF | 6 | 1.7 × 109 ± 0.7 × 109 | 62 | 2.5 × 10−8 |
| 3 | ΔrecA::pMORF | 5 | 1.6 × 108 ± 0.4 × 108 | 76 | 3.3 × 10−8 |
| 4 | ΔrecA::pMORF | 5 | 4.3 × 108 ± 0.4 × 108 | 81 | 1.3 × 10−8 |
Final number of cells in the culture.
Number of mutations per culture.
Probability of mutation per cell per generation.
Genotypic characterization of mutational events detected in fluctuation assays.
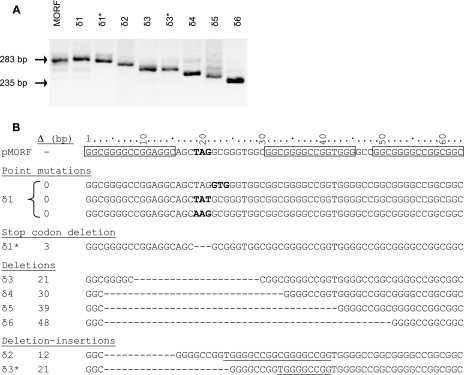
To assess the spectrum of mutations in pMORF leading to Km resistance in the wild-type and recA strains of M. smegmatis, a random selection of Kmr mutants recovered from cultures from the four fluctuation tests was subjected to genotypic characterization. Totals of 100, 71, 39, and 39 individual Kmr clones from experiments 1 to 4, respectively, were genotyped by PCR analysis (Table 2; see Table S3 in the supplemental material). Colony suspensions in water-chloroform (20 μl of each) were boiled (20 min) and centrifuged (10 min), and an aliquot of the supernatant (0.5 μl) was used as a template for PCR amplification of the region of the reporter between the 3′ end of the expression signal cassette (pfbpA) (12) and the 5′ end of the ′aph reporter, using the primers ANAF and ANAR (Fig. 1C; see Table S2 in the supplemental material). The clones thus analyzed yielded a variety of PCR amplicons, ranging in size from 235 bp to the size of the amplicon generated from pMORF itself (283 bp) (Fig. 2A). The genotypes of Kmr clones distinguishable by PCR were designated δ1 to δ6, where δ1 was characterized by PCR amplicons with the same apparent gel electrophoretic mobility as that generated from the pMORF substrate (283 bp) and the remainder (δ2 to δ6) was characterized by PCR amplicons of progressively smaller sizes. The PCR-based genotyping revealed that the distributions of mutants derived from the wild-type versus ΔrecA strains were very similar (Table 2), providing further evidence for the recA independence of the mutations detected using the pMORF reporter.
TABLE 2.
Spectra of pMORF-derived, Km-resistant mutants of M. smegmatis strains isolated from fluctuation assays
| Fluctuation expt no. | Strain | No. of parallel cultures | Total no. of Kmr colonies recovered | No. of Kmr colonies genotyped by PCR analysis | Distribution of mutational events (no. of cultures containing a PCR amplicon representative of the given genotype/ total no. of cultures in the fluctuation test)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| δ1 | δ2 | δ3 | δ4 | δ5 | δ6 | |||||
| 1 | mc2155::pMORF | 13 | 2,270 | 100 | 5/13 | 1/13 | 0/13 | 10/13 | 3/13 | 13/13 |
| 2 | mc2155::pMORF | 6 | 2,950 | 71 | 4/6 | 1/6 | 0/6 | 5/6 | 5/6 | 6/6 |
| 3 | ΔrecA::pMORF | 5 | 2,070 | 39 | 2/5 | 0/5 | 1/5 | 5/5 | 0/5 | 5/5 |
| 4 | ΔrecA::pMORF | 5 | 2,710 | 39 | 1/5 | 0/5 | 1/5 | 5/5 | 1/5 | 5/5 |
FIG. 2.
Genotypic characterization of mutational events detected using the pMORF reporter assay. (A) PCR analysis of Kmr mutants using the ANAF and ANAR primers (Fig. 1C). (B) DNA sequence analysis of the 63-bp PGRS-containing pMORF region in Kmr mutants. The sizes of the deletions (Δ) in the various genotypes are indicated. The R1, R2, and R3 repeats in pMORF are boxed, and the stop codon is shown in bold. Codons that carry point mutations in clones with a δ1 genotype are also shown in bold. The sequences that are duplicated in the deletion-insertion mutants δ2 and δ3* are underlined. The asterisks in δ1* and δ3* denote genotypes that were indistinguishable in size (as determined by gel electrophoretic mobility) but differed in sequence from δ1 and δ3, respectively. The 48-bp deletion in δ6 is identical to the product of p3STOP rearrangement and recapitulates the Δ16 polymorphism in PE_PGRS9.
Eighteen of the 249 Kmr clones from the four fluctuation tests analyzed by PCR were then selected for genotyping by DNA sequence analysis using the primer MFSF (see Table S2 in the supplemental material). The fluctuation test cultures from which the 18 clones originated are indicated in Table S3 in the supplemental material. Preliminary observations had suggested that the PCR amplicon size correlated closely with the DNA sequence in the case of the δ2 to δ6 genotypes (data not shown). The selection of clones for sequence analysis was therefore biased towards those carrying a δ1 genotype, since the amplicon size was not informative of the sequence in such cases. The results of the sequence analysis are summarized in Fig. 2B. Of 11 δ1 clones that were sequenced, 10 displayed point mutations that converted the stop codon to a sense codon (TAG→TAT or AAG) or generated an alternate start codon from that immediately adjacent to the stop codon (GCG→GTG). In contrast, the δ1* clone was found to contain an in-frame deletion of the stop codon. The seven remaining clones contained deletions of between 12 and 48 bp. Interestingly, the two δ3 clones selected for DNA sequence analysis contained deletions of the same size (21 bp) but with different sequences (δ3 and δ3*) (Fig. 2B).
Rates and spectra of mutational events.
In all four fluctuation tests, a multiplicity of mutant genotypes were detected in every culture. Two to four distinct amplicon sizes were identified by PCR screening, with at least one being smaller than 283 bp (δ2 to δ6) (Table 2 and data not shown). A significant number of cultures yielded no amplicons characteristic of the δ1 genotype (8/13, 2/6, 3/5, and 4/5 in experiments 1 to 4, respectively) (Table 2). This bias towards products of deletion rather than point mutation suggests that the former occurred at higher rates than the latter, albeit not to the extent that point mutations would be overwhelmed and hence escape detection. Based on the data obtained from experiment 1, the mutation rate for point mutations alone was calculated to be 3.4 × 10−10 mutations per cell per generation (Lea-Coulson m = 0.5; Nt = 1.0 × 109). This rate is comparable to the rate of 2.3 × 10−10 for mutation of M. tuberculosis to rifampin resistance (9), which occurs by base substitution mutagenesis and provides an important benchmark against which the rate of mutations leading to deletion products can be compared.
In addition to point mutations in the vicinity of the stop codon and deletion of the entire stop codon, six distinct genotypes leading to deletions within the PGRS-containing sequence were also detected (Fig. 2B). Of these, two were most common (δ4 and δ6), one was less common (δ5), and the remaining three were rare (δ2, δ3, and δ3*) (Table 2; see Table S3 in the supplemental material). The δ6 genotype, in which 48 bp of the PGRS-containing sequence in pMORF was deleted, recapitulates precisely the 48-bp (Δ16) deletion polymorphism observed in PE_PGRS9 (10) (Fig. 1). The formation of this deletion mutation confirms the ability of the pMORF reporter to accurately detect naturally occurring M. tuberculosis and M. bovis polymorphisms when applied in the surrogate host, M. smegmatis. This mutation likely resulted from replication slippage between the distal 11-bp GGCGGGGCCGG repeats spanning positions 1 to 11 and 49 to 59 in the PGRS-containing region of pMORF (Fig. 2B). By analogy, we propose that δ4 resulted from replication slippage directed by the proximal 11-bp GGCGGGGCCGG repeats at positions 1 to 11 and 31 to 41, δ5 resulted from slippage between 8-bp GGGGCCGG repeats (positions 4 to 11 and 43 to 50), and δ3 resulted from slippage between 3-bp CGG repeats (positions 9 to 11 and 30 to 32).
The predominance of the δ4 and δ6 genotypes is consistent with the notion that deletion events caused by replication slippage are driven more effectively by longer repeats. Two genotypes consistent with deletion-insertion events were also recovered, namely, δ2 and δ3*. Their formation can be explained by a combination of “forward” slippage (which eliminates the intervening stop codon, as in the δ4 genotype) and “reverse” slippage events, leading to the duplication of either 9 (positions 42 to 50) or 18 nucleotides (positions 42 to 59) to generate δ3* or δ2, respectively.
Conclusions.
The results of this study clearly confirm the utility of the pMORF reporter for assessing the mutability of PGRS-containing sequences in mycobacteria. However, the conclusions that can be drawn are limited by a number of factors. (i) The gain-of-function design of the pMORF-based assay restricted the detectable mutational events to those that allowed readthrough from the expression signals to the ′aph reporter through bypass of the stop codon engineered in the 63-bp PGRS-containing sequence. Of the mutational events resulting in a Kmr phenotype, the spectrum was further limited to those that occurred in sufficiently large numbers to ensure their selection for PCR-based genotyping among the mutants sampled for analysis. Since mutational events that did not lead to a Kmr phenotype were not detected, this assay system likely underestimated the extent of plasticity of the PGRS-containing sequence. Thus, “backward” slippage events involving sequences within the R2 or R3 element and corresponding regions of identity in R1 which duplicated the stop codon and those involving R2 and R3 which did not bypass the stop codon would not have been detected. Similarly, out-of-frame rearrangements would not have been detected. (ii) The mutational analysis was based on a polymorphic region of only one PE_PGRS gene (PE_PGRS9); as such, the extent to which the findings can be generalized to other PGRS genes is unknown. (iii) Similarly, the extent to which the findings of this study can be applied to M. tuberculosis is unclear. Although M. smegmatis has been used successfully as a surrogate for M. tuberculosis in other studies of DNA metabolism (4), this mycobacterium has no naturally occurring PE_PGRS genes, and we cannot rule out the possibility that it may be less permissive to replication slippage involving PGRS repeats than M. tuberculosis and its slow-growing relatives. Resolution of this issue awaits application of the pMORF reporter assay to M. tuberculosis. (iv) Given the limited size of the PGRS-containing sequence included in the reporter (63 bp), it is not possible to predict how the proximal versus distal distributions of slippage events within (or between) PE_PGRS genes may be affected by the multiplicity of repeats and/or by the spacing between the repeats. Larger slippage events are evidently possible in M. tuberculosis, as illustrated by the 46-amino-acid deletion documented for PE_PGRS9, in which 138 bp have either been deleted or inserted in one of the homologues (8). PE_PGRS9 contains a total of 31 identical 8-bp GGGGCCGG repeats in its PGRS region, creating multiple possibilities for deletions and insertions. Interestingly, the identical 11-bp repeats flanking the 16-amino-acid deletion were also found to flank the 46-amino-acid deletion in this gene (8), attesting to the ability of this repeat to serve as a driver of replication slippage events in members of the M. tuberculosis complex. The largest reported deletion in PE_PGRS33 is 273 bp (33). In this case, the flanking sequences are identical 7-bp CGGCGGA repeats, suggesting that repeats of this length are also able to direct replication slippage. (v) Since the results of this study were derived from M. smegmatis recombinants harboring the pMORF reporter cloned in one orientation only at a single site in the chromosome (attB), we were unable to assess whether the mutability is affected by chromosomal orientation (i.e., on the leading versus lagging strand) (23) or location.
These limitations notwithstanding, the results presented herein have nonetheless provided some important insights into the mutagenic potential of PGRS-containing sequences in mycobacteria and the molecular mechanisms operating therein. The recA-independent replication slippage occurring between neighboring PGRS repeats is reminiscent of the mutational mechanism underlying the hypermutability of repeat sequences, such as the microsatellite contingency loci—simple repeat sequences comprising tandem repeating motifs of 1 to 8 bp in length—found in bacterial pathogens such as Haemophilus influenzae and Neisseria meningitidis (2). However, since mutagenesis by replication slippage between neighboring PGRS repeats was only moderately favored over base substitution mutagenesis at the same locus, our findings were not consistent with marked hypermutability of the PE_PGRS9-based sequence under the conditions tested. Allelic diversification through slippage between repeats appears to be constrained by the local DNA sequence and the context of the repeats that direct the rearrangement. In this respect, the length and unusually high G+C content of PGRS-containing sequences likely play a crucial role and may further differentiate the PGRS repeats interspersed in PE_PGRS genes from the hypermutable tandem repeats in the contingency genes of other bacterial pathogens (2, 21). On the one hand, the high G+C content of the PGRS repeats may stabilize duplex structures and hence disfavor slippage, but on the other hand, it may permit slippage between relatively short stretches of homology. These factors could temper the mutability of PGRS-containing sequences in a manner that balances the needs of allowing allelic diversification to occur—which may be associated with a selective advantage—while maintaining genome stability. This conclusion is consistent with the stability observed in the PGRS restriction fragment length polymorphism banding pattern for M. tuberculosis cultures from patients for whom multiple cultures from a single disease episode were available (28). Although not tested in our experimental system, the same conclusion may also apply to PPE-MPTR loci, considering the low frequency of size variants in PPE50 (Rv3135) in isolates of M. tuberculosis recovered over time from an individual patient (22).
Supplementary Material
Acknowledgments
This work was supported by grants from the Wellcome Trust (065578 to Neil G. Stoker and Valerie Mizrahi), the Medical Research Council of South Africa, and the National Research Foundation and by an international research scholar's grant from the Howard Hughes Medical Institute (to V.M.).
Preliminary sequence data were obtained from the Institute for Genomic Research. We thank Erik Böttger, Digby Warner, and Neil Stoker for advice and assistance and Nico Gey van Pittius, Stephanie Dawes, and Bavesh Kana for constructively reviewing the manuscript.
Footnotes
Published ahead of print on 15 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Banu, S., N. Honoré, B. Saint-Joanis, D. Philpott, M. C. Prevost, and S. T. Cole. 2002. Are the PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 44:9-19. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss, C. D., D. Field, and E. R. Moxon. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Investig. 107:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff, H. I., M. B. Reed, C. E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, M. J., G. Delogu, Y. Chen, S. Bardarov, J. Kriakov, M. Alavi, and W. R. Jacobs, Jr. 2001. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect. Immun. 69:7326-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, M. J., and G. Delogu. 2002. The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol. 10:246-249. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, M. J., N. Gey van Pittius, and C. Espitia. 2005. The PE and PPE multigene families of mycobacteria, p. 513-525. In S. T. Cole, D. McMurray, B. Gicquel, and W. R. Jacobs, Jr. (ed.), Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC.
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. (Erratum, 396:190.) [DOI] [PubMed] [Google Scholar]
- 9.David, H. L. 1970. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl. Microbiol. 20:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delogu, G., M. Sanguinetti, C. Pusceddu, A. Bua, M. J. Brennan, S. Zanetti, and G. Fadda. 2006. PE_PGRS proteins are differentially expressed by Mycobacterium tuberculosis in host tissues. Microbes Infect. 8:2061-2067. [DOI] [PubMed] [Google Scholar]
- 11.Dheenadhayalan, V., G. Delogu, M. Sanguinetti, G. Fadda, and M. J. Brennan. 2006. Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J. Bacteriol. 188:3721-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downing, K. J., R. A. McAdam, and V. Mizrahi. 1999. Staphylococcus aureus nuclease is a useful secretion reporter for mycobacteria. Gene 239:293-299. [DOI] [PubMed] [Google Scholar]
- 13.Espitia, C., J. P. Laclette, M. Mondragon-Palomino, A. Amador, J. Campuzano, A. Martens, M. Singh, R. Cicero, Y. Zhang, and C. Moreno. 1999. The PE_PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology 145:3487-3495. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, M. A., B. Plikaytis, and T. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weiman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores, J., and C. Espitia. 2003. Differential expression of PE and PE_PGRS genes in Mycobacterium tuberculosis strains. Gene 318:75-81. [DOI] [PubMed] [Google Scholar]
- 17.Labes, M., A. Puhler, and R. Simon. 1990. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene 89:37-46. [DOI] [PubMed] [Google Scholar]
- 18.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 19.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machowski, E. E., R. A. McAdam, K. M. Derbyshire, and V. Mizrahi. 2000. Construction and application of mycobacterial reporter transposons. Gene 253:67-75. [DOI] [PubMed] [Google Scholar]
- 21.Moxon, R. E., P. B. Rainey, M. A. Nowak, and R. E. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4:24-33. [DOI] [PubMed] [Google Scholar]
- 22.Musser, J. M., A. Amin, and S. Ramaswamy. 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagata, Y., G. Kawaguchi, Y. Tago, M. Imai, T. Watanabe, S. Sakurai, M. Ihara, M. Kawata, and K. Yamamoto. 2005. Absence of strand bias for deletion mutagenesis during chromosomal leading and lagging strand replication in Escherichia coli. Genes Genet. Syst. 80:1-8. [DOI] [PubMed] [Google Scholar]
- 24.O'Gaora, P., S. Barnini, C. Hayward, E. Filley, G. Rook, D. Young, and J. Thole. 1997. Mycobacteria as immunogens: development of expression vectors for use in multiple mycobacterial species. Med. Princ. Pract. 6:91-96. [Google Scholar]
- 25.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 26.Poulet, S., and S. T. Cole. 1995. Characterization of the highly abundant polymorphic GC-rich-repetitive sequence (PGRS) present in Mycobacterium tuberculosis. Arch. Microbiol. 163:87-95. [DOI] [PubMed] [Google Scholar]
- 27.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE_PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, M., G. D. van der Spuy, S. L. Sampson, N. Beyers, P. D. van Helden, and R. M. Warren. 2004. Stability of polymorphic GC-rich repeat sequence-containing regions of Mycobacterium tuberculosis. J. Clin. Microbiol. 42:1302-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 20:4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snapper, S. B., R. E. Melton, T. Mustafa, T. Keiser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 32.Springer, B., P. Sander, L. Sedlacek, W. D. Hardt, V. Mizrahi, P. Schar, and E. C. Böttger. 2004. Lack of mismatch correction facilitates genome evolution in mycobacteria. Mol. Microbiol. 53:1601-1609. [DOI] [PubMed] [Google Scholar]
- 33.Talarico, S., M. D. Cave, C. F. Marrs, B. Foxman, L. Zhang, and Z. Yang. 2005. Variation of the Mycobacterium tuberculosis PE_PGRS33 gene among clinical isolates. J. Clin. Microbiol. 43:4954-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voskuil, M. I., D. Schnappinger, R. Rutherford, Y. Liu, and G. K. Schoolnik. 2004. Regulation of the Mycobacterium tuberculosis PE/PPE genes. Tuberculosis (Edinburgh) 84:256-262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.