Abstract
Mycobacterium tuberculosis and Mycobacterium bovis are responsible for infections that cause a substantial amount of death, suffering, and loss around the world. Still, relatively little is known about the mechanisms of gene expression in these bacteria. Here, we used genome-wide location assays to identify direct target genes for mycobacterial σ factors. Chromatin immunoprecipitation assays were performed with M. bovis BCG for Myc-tagged proteins expressed using an anhydrotetracycline-inducible promoter, and enriched DNA fragments were hybridized to a microarray representing intergenic regions from the M. tuberculosis H37Rv genome. Several putative target genes were validated by quantitative PCR. The corresponding transcriptional start sites were identified for σF, σC, and σK, and consensus promoter sequences are proposed. Our conclusions were supported by the results of in vitro transcription assays. We also examined the role of each holoenzyme in the expression of σ factor genes. Our results revealed that many σ factors are expressed from autoregulated promoters.
Several pathogenic mycobacteria cause infections that lead to tuberculosis or related diseases in a variety of organisms. For example, Mycobacterium tuberculosis infects nearly one-third of the human population (10), while Mycobacterium bovis infections in cattle are responsible for important agricultural losses (12). Advanced knowledge of the biology of these bacteria could significantly contribute to prevention and treatment of the associated diseases. A key step toward this end occurred with the publication of the complete M. tuberculosis and M. bovis genome sequences (8, 12). Several genes have also been interrupted in M. tuberculosis, leading to different outcomes for virulence (for reviews, see references 7 and 38). More recently, in many studies workers have reported gene expression profiles for various growth conditions and during different stages of infection in mice and human macrophages (21, 37, 41), providing new insights into the strategies used by the pathogens to adapt to various environments. Nevertheless, much remains to be discovered about the molecular mechanisms controlling the genetic programs in pathogenic mycobacteria.
Thirteen σ factor genes were predicted based on the genome sequence of M. tuberculosis, and almost identical M. bovis counterparts were also predicted (25, 35). Some of these genes may orchestrate critical responses in the pathogenesis of tuberculosis and therefore may be attractive targets for development of new drugs and vaccines. Indeed, in mouse models infections with sigC, sigD, sigE, sigF, sigh, and sigL mutant strains were all attenuated compared to infections with the wild-type strains (5, 9, 13, 16, 20, 23, 24, 26, 27, 32, 33, 40). Still, the conditions leading to the activities of most σ factors remain elusive (23, 35). Differential gene expression profiles were nonetheless obtained for the σ factor mutant strains, and consensus promoter sequences have been proposed (5, 9, 13, 16, 20, 26, 27, 32, 33, 40). However, direct and indirect effects on the expression of most affected genes are difficult to discriminate. In addition, bioinformatics searches for putative promoters are complicated by the ability of most σ factors to tolerate various mismatches in their consensus sequences. Hence, it is relatively difficult to unambiguously associate a gene with a σ factor that regulates its expression. Consequently, the physiological roles of most mycobacterial σ factors are still poorly understood.
Chromatin immunoprecipitation (ChIP) assays are a powerful technique that allows detection of protein-DNA interactions in vivo (22, 31). Cell components are first cross-linked by using formaldehyde, washed, and used to prepare a whole-cell extract. Next, the DNA is sheared by sonication, and the protein of interest is immunoprecipitated along with the DNA loci to which it is bound. The enriched DNA is then purified and typically analyzed by PCR using specific primers. This method has also been adapted to allow identification of numerous loci in a single experiment using a microarray, a procedure commonly referred to as a genome-wide location assay (4, 34). In this study, we expressed M. tuberculosis σ factors using an inducible promoter, and ChIP assays were performed to identify the corresponding binding sequences. The genome-wide locations of the σ factors σF, σC, and σK were analyzed using the M. bovis BCG-Russia strain. Several candidate promoters were identified and investigated further. Direct target genes were identified for additional σ factors. The roles of various holoenzymes in the transcription of σ factor genes were also studied, and the results revealed that several σ factors regulate their own expression.
MATERIALS AND METHODS
Bacterial strains and media.
M. bovis BCG-Russia (ATCC 35740) was grown at 37°C in Middlebrook 7H9 medium supplemented with albumin-dextrose-saline, 0.05% Tween 80, and 10 μg/ml cycloheximide. When appropriate, hygromycin and anhydrotetracycline were added to the media at final concentrations of 50 μg/ml and 50 ng/ml, respectively.
DNA manipulations.
σ factor-encoding genes were amplified by PCR using forward and reverse primers containing PacI and NsiI restriction endonuclease sites, respectively. PCR products were digested and introduced into the corresponding sites of a modified version of pUV15tetORm (11) using standard procedures. The resulting plasmids contained a hygromycin resistance gene, an anhydrotetracycline-inducible promoter, and a fusion between an amino-terminal Myc epitope and the σ factor of interest. Plasmids were introduced into M. bovis BCG-Russia by electroporation as previously described (39).
ChIP.
Cells were first transformed with a plasmid allowing controlled expression of a σ factor of interest, precultured, and used to inoculate 100 ml of medium at an optical density at 600 nm (OD600) of approximately 0.025 to 0.050. Cells were grown to an appropriate OD600 (∼0.200), and expression of the Myc-tagged σ factor was induced with anhydrotetracycline for 48 h. Expression of Myc-tagged σ factor was monitored by immunoblotting using standard procedures. At an OD600 of ∼0.6 to 0.8, formaldehyde was added directly to the medium to a final concentration of 1%. The cultures were incubated with gentle agitation for 20 min at room temperature and then overnight at 4°C. Cells were collected by centrifugation, washed twice with ice-cold Tris-buffered saline buffer (20 mM Tris HCl [pH 7.5], 150 mM NaCl), and resuspended in lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100). Cells were then transferred into 1.5-ml tubes and disrupted by sonication. Samples were centrifuged at the maximum speed, and the supernatant was sonicated to shear the DNA to obtain an average size of approximately 400 bp. Five microliters of this extract was conserved and used as the input material. The rest of the extract was subjected to immunoprecipitation with sheep anti-mouse paramagnetic beads (M-450; Dynal Biotech) coupled to the Myc 9E10 monoclonal antibody. The magnetic beads were then washed twice with 1 ml of lysis buffer, twice with 1 ml of lysis buffer plus 360 mM NaCl, twice with 1 ml of wash buffer (10 mM Tris-HCl [pH 8.0], 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA), and once with 1 ml of Tris-EDTA (TE) (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The bound material was eluted from beads by resuspension in 50 μl of elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% sodium dodecyl sulfate) and incubation for 10 min at 65°C with occasional agitation. Samples were centrifuged briefly, and cross-links were reversed by mixing 45 μl of the supernatant with 150 μl of TE plus 1% sodium dodecyl sulfate and incubating the preparation overnight at 65°C. Samples were treated with proteinase K, extracted twice with phenol-chloroform, precipitated with ethanol, and resuspended in 60 μl of TE. The DNA was then treated with RNase A and purified using a Qiaquick PCR purification kit from QIAGEN.
DNA labeling, hybridization, and data analysis.
Immunoprecipitated and input control DNA were amplified and labeled with Cy5- and Cy3-labeled dCTP, respectively, using ligation-mediated PCR (34). Samples were mixed and hybridized to the M. tuberculosis intergenic microarray using the protocol of Ren et al. (34). For each σ factor, the data from three biological replicates was analyzed using an error model, as previously described (34).
M. tuberculosis intergenic microarray design and preparation.
Intergenic regions (≥30 bp) located upstream of genes or between two divergent genes were used to select 2,018 70-mer oligonucleotides from the Tuberculist R5 annotation of M. tuberculosis H37Rv. Two probes were selected for every intergenic region larger than 600 bp. Most probes had a melting temperature of 82 ± 3°C, contained no more than 15 contiguous nucleotides identical to the nucleotides in any other genome region, and exhibited less than 70% BLASTN identity to other hits in the genome of M. tuberculosis. Oligonucleotides were spotted on Corning GAPSII microarray slides.
qPCR validation of ChIP.
For selected loci suggested by the intergenic microarray analysis, PCR primers were designed to compare a Myc-tagged σ factor ChIP sample to a no-antibody control sample. Quantitative PCR (qPCR) were performed with a Stratagene MX3000P. Amplicons were detected using SYBR green. Primers used for an invariant control (5′-CACGCAACGTTTGTATCTGC-3′ and 5′-TGACATGTCTGGATTGTGCTC-3′) were selected to amplify a region flanked by two predicted transcription terminators (42) between two convergent genes. The sequences of the primers for the loci tested are available upon request.
RNA extraction and 5′-RACE identification of transcription start sites.
RNA was extracted using an RNeasy kit (QIAGEN) as recommended by the manufacturer, with the modification that 100-μm glass beads and bead beating were used to enhance cell lysis. Rapid amplification of 5′ cDNA ends (5′-RACE) identification of transcription start sites was performed as described previously (14). PCR products were sequenced and analyzed using the M. tuberculosis H37Rv genome sequence as a reference (8).
In vitro transcription assays.
In vitro transcription assays were performed as described elsewhere (19), except that E. coli core RNA polymerase (RNAP) (Epicenter Biotechnologies) was used in combination with M. tuberculosis σ factors.
Preparation of protein extracts and immunoblotting of Myc-tagged σ factors.
Identical amounts of cells, as judged by OD600, were spun, and each pellet was resuspended in 100 μl of buffer C (50 mM Tris-HCl [pH 7.9], 200 mM KCl, 10 μM ZnSO4, 1 mM EDTA, 5 mM 2-mercaptoethanol, 20% glycerol) with 8 M urea. The cells were boiled for 20 min, frozen, and later sonicated for 30 s. Twenty microliters of each extract was electrophoresed on a gel, transferred onto a nitrocellulose membrane, and immunoblotted using the Myc 9E10 monoclonal antibody according to standard procedures.
RESULTS
Inducible expression of σ factors.
σ factors must associate with RNAP in order to direct expression of their target genes. However, the growth conditions leading to the formation of specific holoenzymes are still not known for most M. tuberculosis σ factors (35). Thus, we took advantage of a recently described conditional gene expression system to express σ factors from a plasmid-borne anhydrotetracycline-inducible promoter. All putative M. tuberculosis σ factor-encoding genes were fused to an N-terminus Myc epitope and cloned in a pUV15tetORm derivative (Fig. 1A) (11). The resulting plasmids were used to transform M. bovis BCG-Russia, an attenuated strain whose genome sequence is almost identical to those of M. bovis and M. tuberculosis (3, 12, 29). No significant level of expression of any Myc-tagged σ factor was detected in the absence of the inducer, as judged by immunoblotting (Fig. 1B and data not shown). However, σ factors were expressed in a time-dependent fashion once anhydrotetracycline was added to the growth medium (Fig. 1B and data not shown). We reasoned that the increase in the cellular concentration of a particular Myc-tagged σ factor should be sufficient to allow a significant amount of the corresponding holoenzymes to form in spite of any possible anti-σ factor antagonism. We next performed ChIP assays to identify novel σ factor binding sites.
FIG. 1.
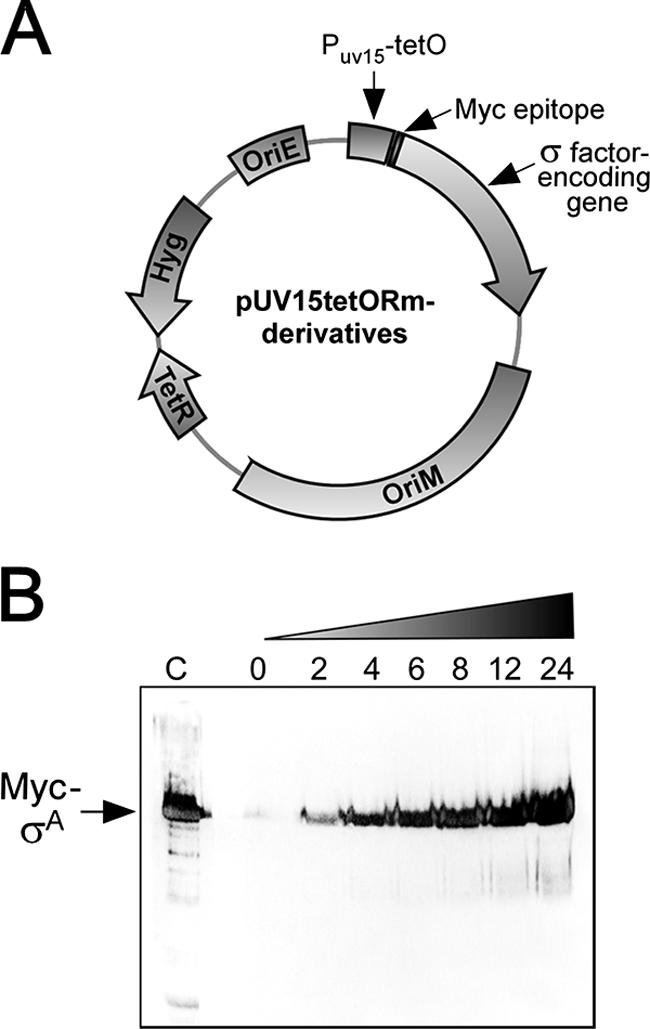
Anhydrotetracycline-inducible expression of Myc-tagged M. tuberculosis σ factors. (A) Structure of the plasmids used for expression of σ factors. OriE, replication origin in E. coli; OriM, pAL5000 mycobacterial replication determinant; Hyg, hygromycin resistance gene; TetR, Tn10 Tet repressor; pUV15-tetOrm, strong TetR-controlled mycobacterial promoter. (B) Time-dependent induction of Myc-tagged σA in M. bovis BCG-Russia. Samples were collected at 2, 4, 6, 8, 12, and 24 h after addition of 50 ng/ml anhydrotetracycline to the growth medium. Protein extracts were prepared and used for immunoblotting with the Myc 9E10 antibody. Lane C contained recombinant Myc-tagged σA.
Genome-wide location of σF.
The σF-dependent usfXP1 promoter is one of the few promoters that have been characterized both in vivo and in vitro (2, 19). Moreover, σF is negatively regulated by the anti-σ factor UsfX (2). In an attempt to validate our experimental scheme, the M. bovis BCG-Russia strain harboring the σF-inducible expression plasmid was used to perform ChIP assays. Direct binding of Myc-tagged σF was observed in the region containing the usfXP1 promoter (Fig. 2B), suggesting that the expression of this protein from the anhydrotetracycline-inducible promoter was sufficient to bypass the negative regulation by UsfX and allow the formation of a competent holoenzyme.
FIG. 2.
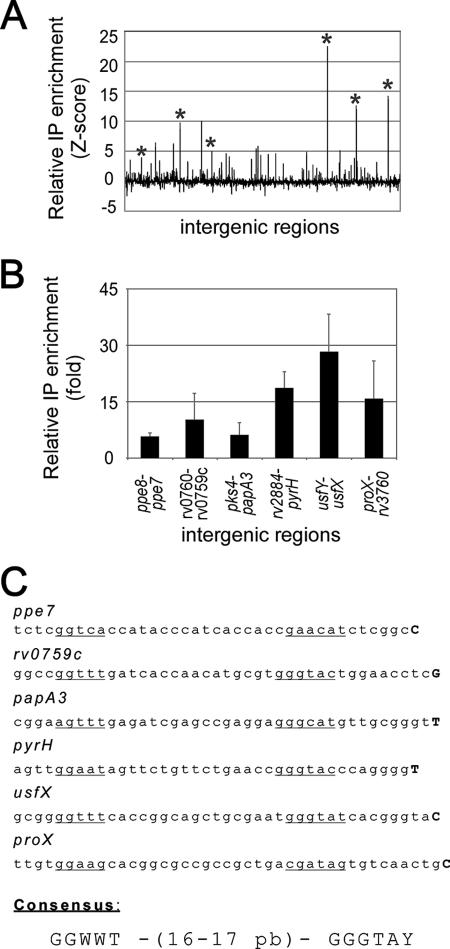
Identification of σF-dependent promoters. (A) Relative ChIP enrichment of Myc-tagged σF for every intergenic region represented on the microarray. The asterisks indicate target genes validated by qPCR. (B) qPCR enrichment of Myc-tagged σF relative to a ChIP assay performed in the absence of the Myc 9E10 antibody for selected intergenic regions. The intergenic regions are in the same order as in panel A. (C) Nucleotide sequences of the putative σF-dependent promoters and proposed consensus sequence. Transcription start sites are indicated by boldface uppercase letters, and proposed promoter boxes are underlined.
A valuable approach for identifying novel σF binding sites is to perform ChIP assays in combination with analysis of a microarray representing various regions of the M. tuberculosis genome. Since no commercially available microarray proved to be useful for these assays, we designed a microarray consisting of 2,018 70-mer isothermal oligonucleotides. Probes were selected from ≥30-bp intergenic regions located upstream of all annotated genes and were printed on appropriate glass slides (see Materials and Methods for a detailed description of the microarray). DNA fragments obtained from Myc-tagged σF ChIP assays were amplified and simultaneously labeled with fluorescent dCTP by ligation-mediated PCR (34). A control sample was treated similarly, and identical amounts of the resulting products were mixed and hybridized to the M. tuberculosis intergenic microarray. Figure 2A shows the relative enrichment of Myc-tagged σF for all probes on the microarray. Using a single-array error model (34), we found that 28 intergenic regions were significantly enriched (P ≤ 0.001) compared to the control (Table 1). The six most enriched loci were selected for validation by qPCR. In the latter assays, Myc-tagged σF ChIP reactions were compared to a control reaction in which the Myc 9E10 antibody was omitted, and the results were normalized to a region where no σ factor binding should have been detected (Fig. 2B). The genome region corresponding to the previously identified σF-dependent promoter usfXP1 showed one of the strongest enrichment signals in the genome-wide location assays and in the qPCR validation experiments (Fig. 2A and B). The binding of Myc-tagged σF to five additional intergenic regions was also validated by the qPCR approach (Fig. 2B). Two loci that were not enriched in the microarray experiments were also tested, and no enrichment was detected (data not shown).
TABLE 1.
Intergenic regions predicted to be directly bound by σF, σC, and σK using a single-array error model (P ≤ 0.001)a
| σ factor | Regionb |
|---|---|
| σF | Rv0213c-fadD4 |
| ppe7-ppe8 | |
| ctpH-Rv0424c | |
| hbhA-Rv0476 | |
| Rv0547c-Rv0546c | |
| Rv0760c-Rv0759c | |
| moaD2-rpfA | |
| Rv1054-Rv1053c | |
| pks4-papA3 | |
| pe_pgrs25-Rv1397c | |
| pe_pgrs27-tkt | |
| gnd1-guaB1 | |
| Rv1870c-Rv1869c | |
| Rv1871c-Rv1870c | |
| furA-katG | |
| Rv2023Ac-Rv2023c | |
| dapE2-tb18.6 | |
| Rv2576c-Rv2577 | |
| ribD-Rv2672 | |
| pyrH-Rv2884 | |
| nrdH-nrdI | |
| Rv3210c-rhlE | |
| Rv3253c-Rv3254 | |
| usfY-usfX | |
| glpD2-phoY1 | |
| Rv3605c-Rv3604c | |
| proX-Rv3760 | |
| σC | Rv0095c-ppe1 |
| ctpB-Rv0104 | |
| Rv0311-Rv0312 | |
| Rv0330c-Rv0331 | |
| ansP2-Rv0347 | |
| grcC1-htpX | |
| Rv0575c-Rv0574c | |
| secE1-nusG | |
| rplJ-rplL | |
| Rv0680c-Rv0681 | |
| rplW-rplB | |
| rpsQ-atsA | |
| rplO-sppA | |
| mbtI-Rv2387 | |
| Rv3483c-Rv3482c | |
| σK | gmhA-gmhB |
| Rv0245-Rv0246 | |
| Rv0281-Rv0282 | |
| Rv0699-rpsJ | |
| rplW-rplB | |
| Rv1069c-pe_pgrs20 | |
| Rv1115-Rv1116 | |
| rrl-rrf | |
| Rv1501-Rv1502 | |
| Rv1534-Rv1535 | |
| Rv2106-pe22 | |
| Rv2872-mpt83 | |
| dipZ-mpt70 | |
| mpt70-Rv2876 | |
| Rv3889c-Rv3888c | |
| Rv3920c-gid |
The single-array error model used has been described previously (34).
The gene annotations are from Tuberculist (http://genolist.pasteur.fr/TubercuList/).
Next, RNA was extracted from M. bovis BCG cultures grown in the same conditions that were used for the ChIP assays, and 5′-RACE assays were performed to identify the transcriptional start sites at the loci where clear binding of Myc-tagged σF was detected (Fig. 2C). Putative promoter boxes were proposed by analyzing the corresponding upstream region of the M. tuberculosis H37Rv genome (Fig. 2C). In these experiments, transcription of the usfX-sigF operon was found to originate from the same nucleotide with respect to the σF-dependent usfXP1 promoter (2). Moreover, analysis of transcription start sites for several regions revealed conserved nucleotides that adequately matched the sequence of the usfXP1 promoter at specific positions relative to the putative −35 and −10 boxes. Taken together, these results suggest that the promoters are directly recognized by σF and that genome-wide location assays identify bona fide direct σ factor binding sites.
σC and σK target genes.
Genome-wide location assays were then carried out for other M tuberculosis σ factors, most of which are poorly characterized. A similar procedure was used for the other σ factors, including σC and σK. Two significant Myc-tagged σC binding signals were detected in the intergenic regions between the genes corresponding to M. tuberculosis Rv0095c-ppe1 and ctpB-Rv0104 using genome-wide location assays (Fig. 3A). Although their relative enrichment was weaker, 13 additional putative target genes (P ≤ 0.001) were also predicted from the single-array error model analysis (Table 1). ChIP enrichment for the Rv0095c-ppe1 and ctpB-Rv0104 loci was also detected by qPCR assays (Fig. 3B). Since these two intergenic regions should support transcription initiation of divergent genes, 5′-RACE was conducted for both orientations with RNA extracted from anhydrotetracycline-induced cultures. Transcription start sites were detected in regions upstream of Rv0095c and ctpB, revealing nearly identical putative promoter boxes (Fig. 3C). Moreover, in vitro transcription assays were performed using E. coli RNAP and M. tuberculosis σ factors, and a transcript was obtained from σC-containing holoenzymes only in the Rv0095c and ctpB upstream regions (Fig. 3D and data not shown).
FIG. 3.
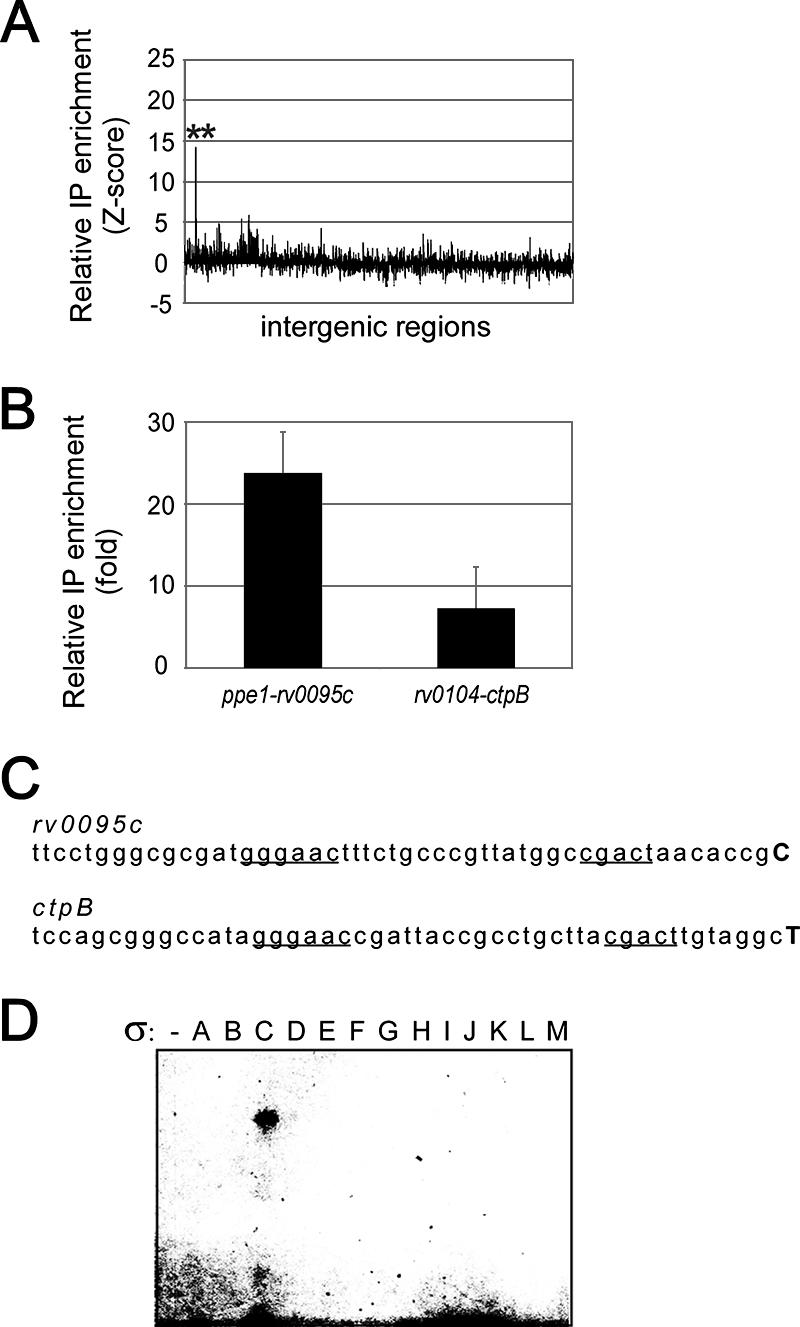
Expression of Rv0095c and ctpB is regulated by σC. (A) Genome-wide enrichment of σC by ChIP. The asterisks indicate target genes validated by qPCR. (B) ChIP qPCR validation for the region upstream of Rv0095c and ctpB. (C) Detected transcription start sites and putative σC recognized boxes. Transcription start sites are indicated by boldface uppercase letters, and proposed promoter boxes are underlined. (D) In vitro transcription using the 13 M. tuberculosis σ factors in the ctpB upstream region.
In the case of Myc-tagged σK, three genome regions located upstream of mpt83, mpt70, and Rv2876 were particularly enriched in genome-wide location experiments (Fig. 4A). Interestingly, these genes, along with an operon starting with Rv0449c, were recently reported to be specifically downregulated in some M. bovis BCG strains by a mutation in the σK translation start codon (6). However, the Rv0449c upstream region was not represented on the intergenic microarray. Therefore, we performed qPCR assays for this putative σK-dependent promoter-containing region, as well as for the regions upstream of mpt83, mpt70, and Rv2876. These four genome regions were found to be significantly enriched compared to the results of a control ChIP reaction (Fig. 4B). 5′-RACE resulted in prediction of nearly identical −35 and −10 promoter boxes upstream of mpt83 and mpt70 (Fig. 4C). Moreover, DNA motifs that could be recognized by SigK were also found upstream of Rv2876 and Rv0449c, although we did not find a clear transcription start site upstream of the latter genes (Fig. 4C and data not shown). In vitro transcription experiments using the mpt70 intergenic region allowed transcription only by a σK holoenzyme, suggesting that σK recognized the previously identified promoter (Fig. 4D). Interestingly, the corresponding transcription start site perfectly matched the transcription start site reported by Matsuo and colleagues (28), although the proposed promoter boxes were slightly shifted.
FIG. 4.
Identification of σK-dependent promoters. (A) Genome-wide location analysis of Myc-tagged σK. The asterisks indicate target genes validated by qPCR. (B) qPCR enrichment of Myc-tagged σK in the regions upstream of mpt83, mpt70, and Rv2876 relative to a negative ChIP control reaction. (C) Transcription start sites (indicated by boldface uppercase letters) identified upstream of the mpt83 and mpt70 genes. Proposed promoter boxes and DNA motifs possibly recognized by SigK upstream of Rv2876 and Rv0449c are underlined. (D) In vitro transcription with every M. tuberculosis σ factor in the mpt70 upstream region.
We also attempted to find direct targets for the remaining 10 M. tuberculosis σ factors. However, no significant enrichment was observed for Myc-tagged σE, σG, σI, σJ, and σM in any region of the intergenic microarray (data not shown). Moreover, relatively few binding sites were detected for the remaining σ factors, and the relative level of enrichment monitored by genome-wide location assays remained close to the background level (data not shown). Still, novel σ factor target genes were discovered using the qPCR assays, as shown in Fig. 5.
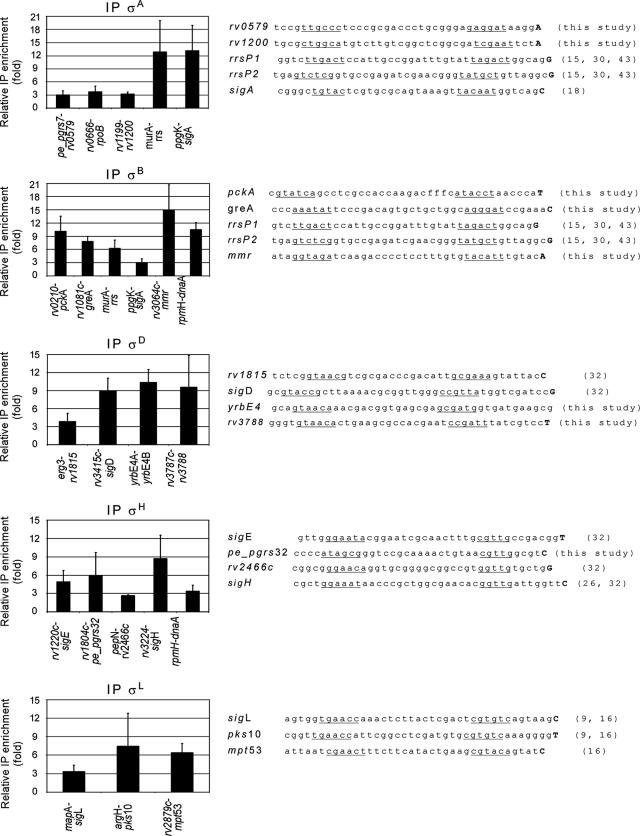
FIG. 5.
Additional σ factor binding sites identified by ChIP and promoters observed in the corresponding intergenic regions. The graphs on the left show the relative enrichment of specific σ factors for various intergenic regions, as monitored by the qPCR assay. The sequences on the right are promoter sequences found in intergenic regions where binding of the corresponding σ factors was detected. Proposed promoter boxes are underlined, and the first transcribed nucleotide is indicated by a boldface uppercase letter where it is known. The numbers in parentheses are references.
σ factor interplay.
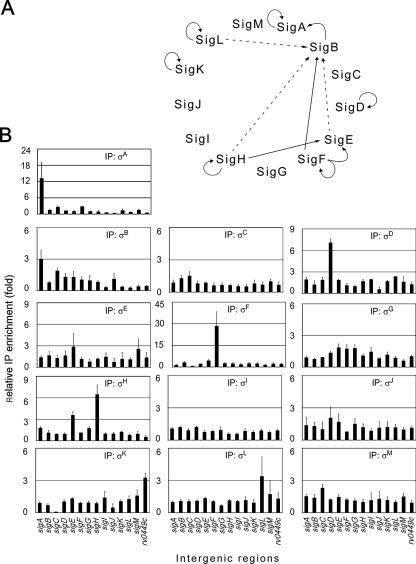
We were also interested in determining how the expression of σ factors is regulated by various holoenzymes, which could reveal regulatory cascades or other network motifs. ChIP assays were performed as described above for every predicted M. tuberculosis σ factor, and the intergenic regions located upstream of the encoding genes were interrogated using specific qPCR primers. ChIP reactions were next compared to a control reaction, and enrichment ratios greater than threefold were considered ratios that indicated relevant binding sites (Fig. 6). Figure 6A shows the regulatory patterns that were observed. Interestingly, most σ factors for which a signal was detected appeared to be expressed from an autoregulated promoter. Indeed, σA, σD, σH, and σL were found to localize in the region located upstream of the corresponding encoding genes, while σF and σK bound upstream of a putative operon comprising the encoding gene (Fig. 6B). Thus, our results are consistent with previously published suggestions for σD, σF, σH, σK, and σL, which were all proposed to regulate their own expression (2, 5, 6, 9, 13, 16, 19, 26, 32, 33).
FIG. 6.
Roles of the various M. tuberculosis holoenzymes in the expression of σ factor genes. (A) Summary based on a threefold enrichment threshold. The dashed lines indicate in vitro transcription data for the sigB promoter obtained by Dainese et al. (9). (B) qPCR determination of the levels of various holoenzymes for the regions upstream of σ factor-encoding genes. Data for the Rv0449c intergenic region, putatively involved in expression of a sigK-containing operon, are also shown.
σA and σB appeared to recognize a promoter upstream of the σA-encoding gene. Moreover, additional intergenic regions were also bound by these two σ factors (Fig. 5), suggesting that they may recognize similar promoter sequences. In addition to the intergenic region upstream of its encoding gene, σF was also found to bind to the sigE upstream region and to a lesser extent to the region upstream of sigB (Fig. 6B). In vitro transcription experiments recently allowed identification of a σF-dependent promoter upstream of the sigB gene (9), and the relatively low level of enrichment observed by ChIP analysis could have resulted from the lower strength of this promoter compared to the σF consensus sequence. Our results also suggest that sigE can be expressed from a σH-dependent promoter, as proposed by other workers (33).
DISCUSSION
σ factors play a major role in the regulation of gene expression and hence contribute to several key process in the biology of mycobacteria. In spite of years of research, little is known about the conditions leading to activity of most σ factors in these pathogens. This is a major obstacle in determining the promoter specificities of the σ factors and their physiological roles. In an attempt to find direct target genes for M. tuberculosis σ factors, we expressed these proteins from an anhydrotetracycline-inducible promoter and performed ChIP assays. Genome-wide location assays for σC, σF, and σK were successful, yet relatively few binding sites were identified for the remaining σ factors. The ChIP localization of σ factors reported in this study is supported by in vitro transcription data and by additional previously described evidence, suggesting that they are real promoter sites.
A possible explanation for our inability to identify a greater number of target genes is that although the σ factors accumulated in the cells, the growth conditions were not suitable for proper activity of these proteins. Indeed, σ factors act in concert with other transcription regulators that are not necessarily expressed or active under the nonphysiological inducing conditions used. Since most promoters are regulated by multiple signals (1), it is likely that several target genes are not detected by this approach. Some posttranslational modifications may also be required for σ factor activity (25, 35). Furthermore, at least four M. tuberculosis σ factors are negatively regulated by anti-σ factors. While the activity of Myc-tagged σF was apparently not inhibited by the corresponding anti-σ factor under these conditions (there was 100% nucleotide identity between the functional M. tuberculosis gene and its M. bovis BCG-Russia equivalent [data not shown]), it is not clear whether other σ factors could have been maintained in an inactive state. For instance, the two other σ factors for which we successfully identified binding sites were apparently not regulated by anti-σ factors in the strain used in our study. We found no previously described evidence supporting the existence of a mycobacterial anti-σC factor, and a σK antagonist was recently shown to be mutated in M. bovis (36). This situation can probably be avoided by performing ChIP assays under physiological conditions for activity whenever this is possible.
Another factor to consider in the genome-wide location of σ factor binding sites is the possibility that only strong signals may have been detected, while the weaker signals may have been missed due to inherent experimental noise. Genome-wide location experiments were reported previously to have less dynamic range, greater noise, and reduced reproducibility compared to gene expression profiling (17). Moreover, it must be emphasized that the σ factor-promoter DNA interaction is dynamic and short-lived with respect to other DNA binding proteins. Consequently, efficient σ factor cross-linking may be harder to achieve and more susceptible to experimental variations.
In previous studies workers proposed consensus promoter sequences for σF and σC on the basis of a comparison of the transcriptomes of the corresponding M. tuberculosis mutant strains to their wild-type counterparts and a search for conserved DNA motifs upstream of downregulated genes (13, 40). However, little similarity was observed between our results and the promoter sequences suggested in these reports. It is not clear whether the proper conditions for activity of σF and σC were used in the previous experiments. RNA was extracted at various times during growth, but no particular growth conditions were used to ensure that the direct effect of σF or σC on transcription could be monitored. Furthermore, no transcriptional start sites were identified to support the conclusions. In contrast, our results are consistent with those of Charlet and colleagues (6) in that we detected binding of Myc-tagged σK to mpt83, mpt70, and Rv2876, as well as upstream of the M. bovis BCG-Russia homolog of Rv0449c (Fig. 6B).
We also investigated the roles of various holoenzymes in the expression of every predicted M. tuberculosis σ factor gene. Although not all σ factor binding sites may have been detected by our approach, the data in Fig. 6 indicate that the transcription of many σ factor genes depends on autoregulated promoters. Moreover, by combining recent in vitro transcription data (9) with ChIP σ factor binding sites (Fig. 6A), putative regulatory cascades can be hypothesized that lead to up-regulation of sigB. Interestingly, σA and σB were found to be capable of binding to the same intergenic regions (Fig. 5 and 6B). Thus, it is tempting to speculate that the two proteins may use the same promoter at some of these genes. The latter hypothesis is supported by the high level of homology between σA and σB and by the fact that the corresponding holoenzymes allowed transcription initiation at the same nucleotide in vitro (19).
In spite of the unexpectedly low number of σ factor binding sites, our approach defined new target genes for many M. tuberculosis σ factors. Moreover, our results suggest that expression of a σ factor does not necessarily result in detectable activity since gene expression is likely to be regulated by numerous effectors. Novel insights into the biology of M. tuberculosis and into the molecular mechanisms used to infect a host should be obtained as the roles of σ factors and other transcription regulators are discovered.
Acknowledgments
We thank Sabine Ehrt for the kind gift of pUV15tetORm and related plasmids. We are also grateful to Marcel Behr for providing the M. bovis BCG-Russia strain used in this study and to Liette Laflamme for sequencing of the M. bovis BCG-Russia usfX gene. We thank Amy Svotelis for her comments on the manuscript.
This work was supported by a grant from NSERC genomics projects. L.G. holds a Canada Research Chair on mechanisms of gene transcription. S.R. is a recipient of fellowships from NSERC, and FRSQ. P.-É.J. was supported by FQRNT.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 2.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control sigmaF activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. [DOI] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Buck, M. J., and J. D. Lieb. 2004. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83:349-360. [DOI] [PubMed] [Google Scholar]
- 5.Calamita, H., C. Ko, S. Tyagi, T. Yoshimatsu, N. E. Morrison, and W. R. Bishai. 2005. The Mycobacterium tuberculosis SigD sigma factor controls the expression of ribosome-associated gene products in stationary phase and is required for full virulence. Cell. Microbiol. 7:233-244. [DOI] [PubMed] [Google Scholar]
- 6.Charlet, D., S. Mostowy, D. Alexander, L. Sit, H. G. Wiker, and M. A. Behr. 2005. Reduced expression of antigenic proteins MPB70 and MPB83 in Mycobacterium bovis BCG strains due to a start codon mutation in sigK. Mol. Microbiol. 56:1302-1313. [DOI] [PubMed] [Google Scholar]
- 7.Clark-Curtiss, J. E., and S. E. Haydel. 2003. Molecular genetics of Mycobacterium tuberculosis pathogenesis. Annu. Rev. Microbiol. 57:517-549. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Dainese, E., S. Rodrigue, G. Delogu, R. Provvedi, L. Laflamme, R. Brzezinski, G. Fadda, I. Smith, L. Gaudreau, G. Palu, and R. Manganelli. 2006. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor σL and roles in virulence and in global regulation of gene expression. Infect. Immun. 74:2457-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 11.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiman, D. E., D. Kaushal, C. Ko, S. Tyagi, Y. C. Manabe, B. G. Schroeder, R. D. Fleischmann, N. E. Morrison, P. J. Converse, P. Chen, and W. R. Bishai. 2004. Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect. Immun. 72:1733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerhart, E., H. Wagner, and J. Vogel. 2005. Approaches to identify novel non-messenger RNAs in bacteria and to investigate their biological functions: functional analysis of identified non-mRNAs, p. 614-642. In R. K. Hartmann, A. Bindereif, A. Schön, and E. Westhof (ed.), Handbook of RNA biochemistry, vol. 2. Wiley-VCH Verlag GmbH & Co, Weinheim, Germany. [Google Scholar]
- 15.Gonzalez-y-Merchand, J. A., M. J. Colston, and R. A. Cox. 1996. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology 142:667-674. [DOI] [PubMed] [Google Scholar]
- 16.Hahn, M. Y., S. Raman, M. Anaya, and R. N. Husson. 2005. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J. Bacteriol. 187:7062-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herring, C. D., M. Raffaelle, T. E. Allen, E. I. Kanin, R. Landick, A. Z. Ansari, and B. O. Palsson. 2005. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 187:6166-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, Y., and A. R. Coates. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacques, J. F., S. Rodrigue, R. Brzezinski, and L. Gaudreau. 2006. A recombinant Mycobacterium tuberculosis in vitro transcription system. FEMS Microbiol. Lett. 255:140-147. [DOI] [PubMed] [Google Scholar]
- 20.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendall, S. L., S. C. Rison, F. Movahedzadeh, R. Frita, and N. G. Stoker. 2004. What do microarrays really tell us about M. tuberculosis? Trends Microbiol. 12:537-544. [DOI] [PubMed] [Google Scholar]
- 22.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 23.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 24.Manganelli, R., L. Fattorini, D. Tan, E. Iona, G. Orefici, G. Altavilla, P. Cusatelli, and I. Smith. 2004. The extracytoplasmic function sigma factor σE is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 72:3038-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manganelli, R., R. Provvedi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 27.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 28.Matsuo, T., S. Matsumoto, N. Ohara, H. Kitaura, A. Mizuno, and T. Yamada. 1995. Differential transcription of the MPB70 genes in two major groups of Mycobacterium bovis BCG substrains. Microbiology 141:1601-1607. [DOI] [PubMed] [Google Scholar]
- 29.Mostowy, S., A. G. Tsolaki, P. M. Small, and M. A. Behr. 2003. The in vitro evolution of BCG vaccines. Vaccine 21:4270-4274. [DOI] [PubMed] [Google Scholar]
- 30.Mulder, M. A., H. Zappe, and L. M. Steyn. 1997. Mycobacterial promoters. Tuber. Lung Dis. 78:211-223. [DOI] [PubMed] [Google Scholar]
- 31.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 32.Raman, S., R. Hazra, C. C. Dascher, and R. N. Husson. 2004. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J. Bacteriol. 186:6605-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigue, S., R. Provvedi, P. E. Jacques, L. Gaudreau, and R. Manganelli. 2006. The σ factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30:926-941. [DOI] [PubMed] [Google Scholar]
- 36.Said-Salim, B., S. Mostowy, A. S. Kristof, and M. A. Behr. 2006. Mutations in Mycobacterium tuberculosis Rv0444c, the gene encoding anti-SigK, explain high level expression of MPB70 and MPB83 in Mycobacterium bovis. Mol. Microbiol. 62:1251-1263. [DOI] [PubMed] [Google Scholar]
- 37.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, I. 2003. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16:463-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 40.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 41.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unniraman, S., R. Prakash, and V. Nagaraja. 2002. Conserved economics of transcription termination in eubacteria. Nucleic Acids Res. 30:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma, A., A. K. Kinger, and J. S. Tyagi. 1994. Functional analysis of transcription of the Mycobacterium tuberculosis 16S rDNA-encoding gene. Gene 148:113-118. [DOI] [PubMed] [Google Scholar]