Abstract
Genetic data suggest that the oligotrophic freshwater bacterium Caulobacter crescentus metabolizes d-xylose through a pathway yielding α-ketoglutarate, comparable to the recently described l-arabinose degradation pathway of Azospirillum brasilense. Enzymes of the C. crescentus pathway, including an NAD+-dependent xylose dehydrogenase, are encoded in the xylose-inducible xylXABCD operon (CC0823-CC0819).
d-Xylose (“wood sugar”) is the primary constituent of xylans that make up the bulk of hemicellulose in plant cell walls and is one of the more abundant carbohydrates in the biosphere. Two routes for d-xylose degradation in microorganisms have been described. Numerous bacteria, including Escherichia coli (15), Bacillus species (24, 25), and Lactobacillus species (16), use xylose isomerase to convert d-xylose to xylulose, which is then phosphorylated to enter the pentose phosphate pathway. Although some fungi have recently been shown to use this “bacterial” pathway (11), fungi more commonly transform d-xylose into xylitol by using xylose reductase and xylitol dehydrogenase (13). The freshwater bacterium Caulobacter crescentus, which readily uses d-xylose as a carbon and energy source, expresses an NAD-dependent xylose dehydrogenase (XDH) activity, suggesting that xylose metabolism occurs through a distinct pathway (21).
Prior to this work, the only known mutation affecting d-xylose utilization in C. crescentus was a Tn5-lacZ insertion that eliminated growth on xylose and exhibited strong xylose-dependent induction of β-galactosidase expression (18). The gene in which this insertion is located, designated “xylX” by Meisenzahl et al. (18) and later “CC0823” in the C. crescentus genome annotation (19), does not closely resemble any gene of known function. xylX is the first gene in a xylose-inducible operon (CC0823-CC0819) (12), referred to here as the xyl operon. We show that all of the genes in this operon are involved in xylose metabolism and propose a metabolic pathway employing these gene products.
Genetic analysis of d-xylose metabolism.
To identify genes required for d-xylose utilization, C. crescentus NA1000 was mutagenized with a kanamycin-resistant mini-Tn5 transposon (9). Insertion strains were selected on peptone-yeast extract (PYE) medium containing kanamycin (20 μg ml−1). Mutants in which xylose metabolism is defective were identified by patching Kanr colonies onto M2 minimal media (10) with glucose (M2G) or xylose as a carbon source. We also patched colonies on M2 medium containing both glucose and xylose to identify strains for which xylose had become toxic [xyl(Tox)]. Roughly 20,000 Kanr isolates were screened. Using chromosomal DNA as the template and primers derived from the Tn5 sequence, mutants with growth defects were analyzed by cycle sequencing to determine the location of the transposon insertion in comparison with that of the C. crescentus genome sequence (19). Strains unable to use glucose but unaffected in xylose utilization had mutations in genes previously implicated in glucose catabolism (12), including components of the Entner-Doudoroff (E-D) pathway (Fig. 1) (12, 22). The only gene identified here that was not previously associated with C. crescentus glucose catabolism is CC3065, which encodes a putative LacI superfamily transcription factor of unknown function.
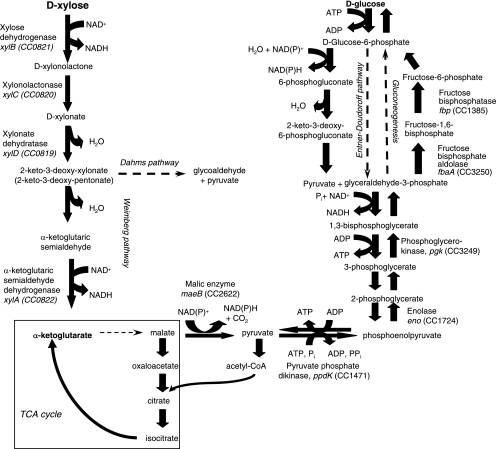
FIG. 1.
Proposed pathway for d-xylose metabolism in C. crescentus. The reactions shown are based on biochemically confirmed degradation pathways for d-xylose metabolism in pseudomonads (7, 27). Both d-xylose and l-arabinose produce 2-keto-3-deoxy-pentonate. In the Dahms pathway (7), this compound is converted by an aldolase to pyruvate and glycoaldehyde. In an alternative reaction first demonstrated by Weimberg (27) and confirmed by Watanabe et al. (25, 26), for l-arabinose degradation in A. brasilense, a dehydratase produces α-ketoglutarate semialdehyde, which is then oxidized to α-ketoglutarate. The genes identified (through mutation) in this work as necessary for growth on d-xylose (Table 1) and the enzymes they encode are shown beside the appropriate reaction. The Entner-Doudoroff pathway and alternative reactions used in gluconeogenesis are shown at the upper right. TCA cycle reactions (in the box on the lower left, not shown in detail) are expected to be necessary for both d-xylose and d-glucose metabolism; genes encoding these enzymes were probably not found in this screen because they are also necessary for growth on PYE medium.
Twenty-two xyl mutants were isolated. Insertions were found in 11 genes, including 4 of the 5 genes of the xyl operon (CC0823-CC0819) (Table 1). No insertions were identified in the CC0820 coding region, but one was found upstream, between CC0821 and CC0820. Six genes yielding the xyl mutant phenotype were found in multiple independent isolates, suggesting that the mutagenesis was approaching saturation and that these represent most, if not all, of the genes required specifically for xylose metabolism. None of the xyl mutant strains had a mutation in a putative transcriptional activator, consistent with previous suggestions (12, 18) that C. crescentus xylose metabolism genes are controlled by an as-yet-unidentified repressor. In addition, no genes resembling transporters were identified in this screen. Perhaps there are multiple transport systems capable of importing xylose into C. crescentus, as there are in E. coli (1, 8), so that a single mutation cannot sufficiently impair xylose uptake to block growth.
TABLE 1.
Results of Tn5 mutant screen for C. crescentus xyl and xyl(Tox) mutants
| Phenotypea | Interrupted gene | No. of isolates | Annotation | Proposed function |
|---|---|---|---|---|
| Xyl− Gluc+ | xylX (CC0823) | 2 | Conserved hypothetical protein | Unknown |
| [Xyl(Tox)] | xylA (CC0822) | 2 | Aldehyde dehydrogenase | α-Ketoglutaric semialdehyde dehydrogenase |
| xylB (CC0821) | 1 | Oxidoreductase, short-chain dehydrogenase/reductase family | Xylose dehydrogenase | |
| xylC (CC0820) upstream regionb | 1 | CC0820: “SMP/Cgr family” | Xylonolactonase | |
| xylD (CC0819) | 1 | Dehydratase (IlvD/Edd family) | Xylonate dehydratase | |
| fbp (CC1385) | 1 | Fructose-1,6-bisphosphatase (EC 3.1.3.11) | Gluconeogenesis | |
| Xyl− Gluc+ | ppdK (CC1471) | 4 | Pyruvate phosphate dikinase (EC 2.7.9.1) | Gluconeogenesis |
| maeB (CC2622) | 1 | NADP-dependent malic enzyme (EC 1.1.1.40) | Gluconeogenesis | |
| fbaA (CC3250) | 2 | Fructose-bisphosphate aldolase (EC 4.1.2.13) | Gluconeogenesis | |
| CC3364 | 1 | Homoserine kinase (EC 2.1.7.13) | Unknown | |
| Xyl− Gluc− | eno (CC1724) | 2 | Enolase (EC 4.2.1.11) | Glycolysis and gluconeogenesis |
| pgk (CC3249) | 4 | Phosphoglycerate kinase (EC 2.7.2.3) | Glycolysis and gluconeogenesis | |
| Xyl+ Gluc− | zwf (CC2057) | 1 | Glucose-6-phosphate 1-dehydrogenase (EC 1.1.1.49) | Entner-Doudoroff pathway |
| CC2056 | 1 | 6-Phospho-glucono-lactonase (EC 3.1.1.31) | Entner-Doudoroff pathway | |
| ppc (CC1493) | 2 | Phosphoenolpyruvate carboxylase (EC 4.1.1.31) | Anaplerotic function | |
| CC3065 | 1 | Transcriptional regulator, LacI family | Unknown | |
| Xyl+ Gluc− [Gluc(Tox)] | eda (CC1495) | 1 | 4-Hydroxy-2-oxoglutarate aldolase (EC 4.1.2.14) | Entner-Doudoroff pathway |
The “Xyl−” phenotype refers to strains that were unable to grow on M2 medium containing 10 mM d-xylose as the sole carbon source. The “Gluc−” phenotype refers to strains that were unable to grow on M2 medium containing 10 mM d-glucose as the sole carbon source. The “Xyl(Tox)” phenotype refers to strains that were sensitive to the presence of d-xylose in the medium, i.e., strains that were able to grow on M2G but that did not form colonies when 10 mM d-xylose was added to M2G.
Tn5 insertion was between the CC0821 and CC0820 coding regions.
The previously unnamed genes of the xyl operon are hereafter designated xylA (CC0822), xylB (CC0821), xylC (CC0820), and xylD (CC0819). Because transposon insertions in upstream genes of the operon (which is transcribed in the order xylX-xylA-xylB-xylC-xylD) could have polar effects, the role of each gene was assessed independently by constructing nonpolar in-frame deletions, using a PCR-based strategy (29). Deletion of any of the five genes rendered strains incapable of growth with d-xylose as the sole carbon source, confirming that all five genes are necessary for xylose utilization.
All strains with an insertion in one of the genes of the xyl operon exhibited a xyl(Tox) phenotype on M2G agar plates, with colony formation blocked by inclusion of 10 mM d-xylose in the medium. In logarithmically growing M2G broth cultures, all the mutant strains exhibited reduced growth rates following the addition of d-xylose (data not shown), but only the ΔxylD and ΔxylX strains suffered a loss of viability. Xylose toxicity was generally reduced on complex PYE medium, with the effects on growth rate and colony appearance being less pronounced. The exception was the ΔxylD strain, which generated no colonies on PYE plus xylose agar medium and still lost viability after the addition of xylose to PYE broth culture.
Analysis of d-xylose dehydrogenase activity.
Poindexter (21) observed d-xylose dehydrogenase activity in some C. crescentus strains grown in the presence of xylose. To determine whether any of the genes of the xyl operon encode this enzyme, XDH activity was assayed in extracts from wild-type and mutant strains. Cultures were grown in PYE broth at 30°C with constant shaking to an optical density at 600 nm of approximately 0.5, at which time xylose was added to a final concentration of 1 mM. After 2 h, cells were harvested by centrifugation and disrupted by sonication. XDH activity in cell extracts was measured by following the xylose-dependent reduction of NAD+, as indicated by an increase in absorption at 340 nm (21). Assays were carried out in a 1-ml quartz cuvette containing 50 mM phosphate buffer (pH 8), 5 mM d-xylose, and 4 mM NAD+. If present, xylose-independent NADH production (“background activity,” measured in control assays without xylose) was subtracted out. XDH activity was easily detectable in the wild-type strain induced with xylose (31.7 nmol NADH generated min−1 mg protein−1) but was not observable above background in cultures grown without xylose. The enzyme was unable to use NADP+ as the electron acceptor, as found by Poindexter (21). XDH activity was observed in extracts from the ΔxylX, ΔxylC, and ΔxylD mutant strains but was conspicuously absent from the ΔxylA and ΔxylB strains.
The xylA gene product was annotated by the C. crescentus genome project as a “short-chain aldehyde dehydrogenase,” while the xylB product was annotated as an “oxidoreductase” (19). To determine whether XDH activity is attributable to one of these gene products, the PCR-amplified coding regions were cloned separately into the pCR-CT-T7-Topo expression vector to allow production of C-terminal His-tagged proteins in E. coli strain BL21(λDE3) pLysS (Invitrogen). Cloning was carried out and protein expression was measured according to the manufacturer's protocols. Extracts from the E. coli strain expressing the cloned C. crescentus xylB gene displayed XDH activity (48.1 nmol NADH min−1 mg protein−1), which was absent from both the E. coli host strain and the strain expressing xylA. A 30-kDa polypeptide with XDH activity was purified from the xylB-expressing strain by Ni-affinity chromatography (Pharmacia nickel-nitrilotriacetic acid [nickel-NTA] column, developed with a 0 to 300 mM imidazole gradient in 50 mM sodium phosphate-50 mM NaCl-1 mM EDTA buffer on a Pharmacia fast protein liquid chromatography system). This polypeptide was confirmed as the XylB-His6 fusion protein by liquid chromatography-mass spectrometry analysis (Midwest Bio Services, Overland Park, KS). Affinity-purified XylB-His6 was at least 95% pure, based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. XylB is thus responsible for XDH catalytic activity. It is not clear why the ΔxylA strain lacked XDH activity; one possibility is that the xylA deletion may have somehow affected xylB expression, even though it was designed to be nonpolar.
Purified recombinant XDH has a strong preference for d-xylose as a substrate. At sugar concentrations of up to 50 mM, d-arabinose, l-xylose, d-ribose, d-galactose, d-glucose, or d-glucose-6-phosphate produced little or no NADH. l-Arabinose was active as a substrate, but analysis of XDH activity over a range of substrate concentrations (0.1 to 500 mM d-xylose or l-arabinose) showed that the enzyme strongly prefers d-xylose as a substrate (for d-xylose, Km = 0.76 mM, Vmax = 27.5 μmol NADH min−1 mg−1; for l-arabinose, Km = 166 mM, Vmax = 20.5 μmol NADH min−1 mg−1). Preliminary analysis of partially purified native C. crescentus XDH (to be described elsewhere) showed an even lower Km for d-xylose of 70 μM, suggesting that the recombinant XylB-His6 is not completely native in structure when produced in E. coli, perhaps due to additional amino acids at the N and C termini introduced for expression and purification.
A few bacterial species have been shown to express XDH activity (3, 6, 28, 30), but only one dehydrogenase with high specificity for d-xylose has been identified genetically (14), in the halophilic archeon Haloarcula marismortui. A pairwise BLAST comparison identified no significant similarity between the H. marismortui XDH and the C. crescentus XylB polypeptide sequences.
Pathway for d-xylose degradation.
The two proposed pathways for xylose metabolism initiated by xylose dehydrogenase are identical through the production of 2-keto-3-deoxyxylonate (Fig. 1) (5, 7, 28). The initial series of reactions is analogous to the Entner-Doudoroff pathway, particularly the archaeal version of the E-D pathway in which glucose is not phosphorylated (23). One component of the Entner-Doudoroff and xylose degradation pathways appears to be evolutionarily related, since C. crescentus XylD (GenBank accession no. AAK22804) is notably similar in sequence to bacterial 6-phosphogluconate dehydratases (e.g., E. coli Edd; GenBank accession no. AAA23722; 31% identity over 446 amino acids with XylD) (4). Based on this, we hypothesize that XylD catalyzes the dehydration of d-xylonate to 2-keto-3-deoxyxylonate.
Watanabe et al. (26, 27) have recently shown that l-arabinose degradation in Azospirillum brasilense follows the pathway Weimberg proposed for l-arabinose and d-xylose (28). l-Arabinose and d-xylose are structurally related pentoses, and the l-arabinose in arabinogalactan polymers also contributes substantially to hemicellulose. Although the l-arabinose dehydrogenase cloned by Watanabe et al. (26) is unrelated by amino acid sequence to the C. crescentus XylB d-xylose dehydrogenase, other potential pathway components are related. Caulobacter crescentus XylC (GenBank accession no. AAK22805) aligns well with A. brasilense arabinolactonase (GenBank accession no. AB241136.1; 34% identity over 285 amino acids with XylC) and is thus a good candidate to catalyze the conversion of d-xylono-γ-lactone to d-xylonate. 2-Keto-deoxypentonate is produced by the subsequent dehydration reaction, which as noted above is predicted to be catalyzed by XylD. In the Weimberg pathway (28), 2-keto-deoxypentonate is dehydrated to α-ketoglutarate semialdehyde and oxidized to α-ketoglutarate by α-ketoglutarate semialdehyde dehydrogenase. The C. crescentus XylA sequence (GenBank accession no. AAK22807) aligns well with the A. brasilense α-ketoglutarate semialdehyde dehydrogenase (GenBank accession no. AB241137; 32% identity over 475 amino acids with XylA), suggesting that it executes this reaction (Fig. 1).
This strategy for d-xylose metabolism in C. crescentus could explain the requirement for malic enzyme (maeB; CC2622) for growth on xylose (Table 1; Fig. 1). This enzyme would divert some malate (produced ultimately from α-ketoglutarate) to generate pyruvate, which is necessary for a variety of anabolic functions, including gluconeogenesis. The requirement for malic enzyme would be difficult to rationalize if C. crescentus metabolized d-xylose via the Dahms pathway (Fig. 1), because pyruvate would be generated by aldolase cleavage of 2-keto-deoxyxylonate (7). Gluconeogenesis presumably continues from pyruvate to phosphoenolpyruvate via pyruvate phosphate dikinase (ppdK; CC1471) (Table 1). Sinorhizobium meliloti, a close relative of C. crescentus, can use malic enzyme and PPDK to support gluconeogenesis during growth on tricarboxylic acid (TCA) cycle intermediates (20), which is comparable to what C. crescentus would experience if xylose metabolism proceeded via α-ketoglutarate. Other gene products required for growth on xylose, and likely identified in our screen because of gluconeogenic function, include enolase (eno; CC1724) and phosphoglycerate kinase (pgk; CC3249), which catalyze reversible reactions also required for glucose catabolism (Table 1). Fructose bisphosphate aldolase (fbaA; CC3250) also catalyzes a reversible reaction but is not necessary for growth on glucose because the Entner-Doudoroff pathway bypasses the fructose bisphosphate intermediate of glycolysis.
Given the similarity of the proposed C. crescentus d-xylose degradation pathway to the A. brasilense l-arabinose pathway and the fact that the xylB-encoded XDH can utilize l-arabinose as a substrate (albeit poorly), we examined whether this pathway has a role in l-arabinose metabolism. Wild-type C. crescentus strain CB15 grows very poorly in liquid M2 medium with l-arabinose as the sole carbon source but forms colonies on M2 agar containing l-arabinose. Growth levels of CB15 and the ΔxylA, ΔxylB, ΔxylC, and ΔxylD mutants were compared on M2 agar plates with either d-glucose, d-xylose, or l-arabinose (all at 10 mM) as the sole carbon sources. The strains grew similarly on glucose (i.e., 1-mm colonies within 3 days), and none of the mutants grew with xylose. CB15 produced 1-mm colonies within 3 to 4 days on xylose, and after 5 to 6 days, had formed 1-mm colonies on l-arabinose. The ΔxylA, ΔxylB, and ΔxylC strains formed smaller “microcolonies” (≤0.5 mm) on l-arabinose after 5 to 6 days and thus appear to be defective for growth on this substrate. Curiously, growth of the ΔxylD strain was similar to that of the parental strain CB15 on l-arabinose, indicating that the xylD product is dispensable for growth on l-arabinose. Deficiencies in growth on l-arabinose among the other xyl mutant strains were confirmed using Biolog phenotype microarray plates PM1 and PM2 (2) to examine carbon source utilization (data not shown). Thus, with the exception of the XylD-catalyzed step, the C. crescentus d-xylose degradation pathway probably contributes to l-arabinose degradation in vivo, but there may be an additional route for l-arabinose utilization.
Genes necessary for growth on d-xylose to which we cannot assign a role include xylX (CC0823) and CC3364. The xylX product falls into COG3970, the fumarylacetoacetate hydrolase family. CC3364 is annotated as a “homoserine kinase” due to weak similarity to the Pseudomonas aeruginosa thrB gene product. Functional characterization of these genes is an important future goal for understanding d-xylose metabolism in C. crescentus.
The basis for growth inhibition by xylose in strains with mutations in the xyl operon is not known. Interruption of a metabolic pathway can lead to toxicity if harmful intermediates accumulate. The ΔxylD mutant suffers the most severe effects in the presence of d-xylose, which could conceivably be due to the accumulation of d-xylonate, but we have no direct evidence at present to support that hypothesis. Excessive uptake of a nonmetabolized sugar, or the effects of xylose on gene expression, could also result in metabolic alterations that are harmful in the absence of metabolite flux through the xylose catabolic pathway. For example, xylose increases isocitrate lyase expression in C. crescentus (12). During growth on glucose in the absence of productive xylose metabolism, an increase in isocitrate lyase activity could excessively channel isocitrate into the glyoxylate bypass at the expense of critical TCA cycle intermediates, such as α-ketoglutarate, that are no longer being generated (directly or indirectly) from d-xylose. Potential explanations of the xyl(Tox) phenotype must also take into account the observation that growth inhibition is less severe in the complex PYE medium than in the defined M2 medium. If a metabolic imbalance is leading to growth inhibition, the diversity of organic metabolites present in PYE may alleviate some of the problems.
This route of d-xylose metabolism is not unique to C. crescentus, having been identified originally in a Pseudomonas strain (28), but a preliminary survey of other sequenced genomes suggests that this pathway is not common. Using BLAST, we were able to identify only three other bacteria containing possible operons with component genes closely related to most or all of the C. crescentus xyl operon genes: Caulobacter strain K31 (a freshwater α-proteobacterium isolated from chlorophenol-contaminated groundwater) (17), Burkholderia xenovorans strain LB400 (a PCB-degrading β-proteobacterium isolated from a landfill), and Chromohalobacter salexigens strain DSM 3043 (a halophilic γ-proteobacterium). These three genome sequences have not been described in publications but are available through the U.S. Department of Energy's Joint Genome Institute website (http://genome.jgi-psf.org/mic_home.html). Caulobacter strain K31 expresses d-xylose-inducible XDH activity (data not shown), but to our knowledge, xylose metabolism has not been further examined in these diverse species. We speculate that they share with C. crescentus a common pathway for d-xylose degradation, encoded in a gene cluster that may have been horizontally transferred in aquatic and/or soil habitats.
Acknowledgments
This work was supported by National Science Foundation grant MCB-0317037 to C.S. and Swiss National Science Foundation fellowship 3100A0-108186 to U.J.
We gratefully acknowledge the students of the 2003 and 2004 advanced bacterial genetics courses at Cold Spring Harbor Laboratory and the 2003 microbiology block course at the University of Basel who isolated the transposon mutants, as well as several Santa Clara University undergraduates who generated knockout strains in the recombinant DNA technology course (Desiree Yang, Naomi Arana, Nicole Robledo, Sarah Arriola, and Dennie Magcase). We also thank Angel Islas for advice on protein expression and purification and Minna Manisto (Finnish Forest Research Institute, Rovaniemi, Finland) for providing Caulobacter strain K31.
Footnotes
Published ahead of print on 15 December 2006.
REFERENCES
- 1.Ahlem, C., W. Huisman, G. Neslund, and A. S. Dahms. 1982. Purification and properties of a periplasmic d-xylose-binding protein from Escherichia coli K-12. J. Biol. Chem. 257:2926-2931. [PubMed] [Google Scholar]
- 2.Bochner, B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4:309-314. [DOI] [PubMed] [Google Scholar]
- 3.Buchert, J., L. Viikari, M. Linko, and P. Markkanen. 1986. Production of xylonic acid by Pseudomonas fragi. Biotechnol. Lett. 8:541-546. [Google Scholar]
- 4.Carter, A. T., B. M. Pearson, J. R. Dickinson, and W. E. Lancashire. 1993. Sequence of the Escherichia coli K-12 edd and eda genes of the Entner-Doudoroff pathway. Gene 130:155-156. [DOI] [PubMed] [Google Scholar]
- 5.Dagley, S., and P. W. Trudgill. 1965. The metabolism of galactarate, d-glucarate and various pentoses by species of Pseudomonas. Biochem. J. 95:48-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahms, A. S., and J. Russo. 1982. D-xylose dehydrogenase. Methods Enzymol. 89D:226-228. [DOI] [PubMed] [Google Scholar]
- 7.Dahms, A. S. 1974. 3-Deoxy-d-pentulosonic acid aldolase and its role in a new pathway of d-xylose degradation. Biochem. Biophys. Res. Commun. 60:1433-1439. [DOI] [PubMed] [Google Scholar]
- 8.Davis, E. O., and P. J. F. Henderson. 1987. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J. Biol. Chem. 262:13928-13932. [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 11.Harhangi, H. R., A. S. Akhmanova, R. Emmens, C. van der Drift, W. T. de Laat, J. P. van Dijken, M. S. Jetten, J. T. Pronk, and H. J. Op den Camp. 2003. Xylose metabolism in the anaerobic fungus Piromyces sp. strain E2 follows the bacterial pathway. Arch. Microbiol. 180:134-141. [DOI] [PubMed] [Google Scholar]
- 12.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 186:1448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffries, T. W. 1983. Utilization of xylose by bacteria, yeasts, and fungi. Adv. Biochem. Eng. Biotechnol. 27:1-32. [DOI] [PubMed] [Google Scholar]
- 14.Johnsen, U., and P. Schönheit. 2004. Novel xylose dehydrogenase in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 186:6198-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawlis, V. B., M. S. Dennis, E. Y. Chen, D. H. Smith, and D. J. Henner. 1984. Cloning and sequencing of the xylose isomerase and xylulose kinase genes of Escherichia coli. Appl. Environ. Microbiol. 47:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lokman, B. C., P. Van Santen, J. C. Verdoes, J. Kruse, R. J. Leer, M. Posno, and P. H. Pouwels. 1991. Organization and characterization of three genes involved in d-xylose catabolism in Lactobacillus pentosus. Mol. Gen. Genet. 230:161-169. [DOI] [PubMed] [Google Scholar]
- 17.Mannisto, M. K., M. A. Tiirola, M. S. Salkinoja-Salonen, M. S. Kulomaa, and J. A. Puhakka. 1999. Diversity of chlorophenol-degrading bacteria isolated from contaminated boreal groundwater. Arch. Microbiol. 171:189-197. [DOI] [PubMed] [Google Scholar]
- 18.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osteras, M., B. T. Driscoll, and T. M. Finan. 1997. Increased pyruvate orthophosphate dikinase activity results in an alternative gluconeogenic pathway in Rhizobium (Sinorhizobium) meliloti. Microbiology 143:1639-1648. [DOI] [PubMed] [Google Scholar]
- 21.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riley, R. G., and B. J. Kolodziej. 1976. Pathway of glucose catabolism in Caulobacter crescentus. Microbios 16:219-226. [PubMed] [Google Scholar]
- 23.Romano, A. H., and T. Conway. 1996. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 147:448-455. [DOI] [PubMed] [Google Scholar]
- 24.Rygus, T., A. Scheler, R. Allmansberger, and W. Hillen. 1991. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus megaterium encoded regulon for xylose utilization. Arch. Microbiol. 155:535-542. [DOI] [PubMed] [Google Scholar]
- 25.Scheler, A., T. Rygus, R. Allmansberger, and W. Hillen. 1991. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus licheniformis-encoded regulon for xylose utilization. Arch. Microbiol. 155:526-534. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe, S., T. Kodaki, and M. Keisuke. 2006. Cloning, expression and characterization of bacterial l-arabinose 1-dehydrogenase involved in an alternative pathway of l-arabinose metabolism. J. Biol. Chem. 281:2612-2623. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe, S., T. Kodaki, and K. Makino. 2006. A novel alpha-ketoglutaric semialdehyde dehydrogenase: evolutionary insight into an alternative pathway of bacterial l-arabinose metabolism. J. Biol. Chem. 281:28876-28888. [DOI] [PubMed] [Google Scholar]
- 28.Weimberg, R. 1961. Pentose oxidation by Pseudomonas fragi. J. Biol. Chem. 236:629-635. [PubMed] [Google Scholar]
- 29.West, L., D. Yang, and C. Stephens. 2002. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J. Bacteriol. 184:2155-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamanaka, K., M. Gino, and R. Kaneda. 1977. A specific NAD-d-xylose dehydrogenase from Arthrobacter sp. Agric. Biol. Chem. 41:1493-1499. [Google Scholar]