Abstract
Strains of Streptococcus mutans lacking DnaK or GroEL appear not to be isolable. To better distinguish the roles played by these chaperones/chaperonins in the physiology of S. mutans, we created a knockdown strategy to lower the levels of DnaK by over 95% in strain SM12 and the level of GroEL about 80% in strain SM13. Interestingly, GroEL levels were approximately twofold higher in SM12 than in the parent strain, but the levels of DnaK were not altered in the GroEL knockdown strain. Both SM12 and SM13 grew slower than the parent strain, had a strong tendency to aggregate in broth culture, and showed major changes in their proteomes. Compared with the wild-type strain, SM12 and SM13 had impaired biofilm-forming capacities when grown in the presence of glucose. The SM12 strain was impaired in its capacity to grow at 44°C or at pH 5.0 and was more susceptible to H2O2, whereas SM13 behaved like the wild-type strain under these conditions. Phenotypical reversions were noted for both mutants when cells were grown in continuous culture at a low pH, suggesting the occurrence of compensatory mutations. These results demonstrate that DnaK and GroEL differentially affect the expression of key virulence traits, including biofilm formation and acid tolerance, and support that these chaperones have evolved to accommodate unique roles in the context of this organism and its niche.
Streptococcus mutans, a bacterial pathogen associated with human dental caries, thrives in multispecies biofilms on tooth surfaces, where it is subjected to a continuous assault by host defenses and to rapid and dramatic fluctuations in nutrient availability, carbohydrate source, and pH. The organism has evolved multiple physiologic and genetic adaptations to optimize growth under these dynamic conditions, including the capacity to scavenge and metabolize a wide variety of carbohydrates and to adapt to a wide range of stresses, especially low pH (15). The rapid and efficient responses to environmental stimuli are considered to be critical to the persistence and virulence of this organism, so efforts have been focused on dissecting the molecular control of responses to low pHs, other stresses, and nutrient flux. Through such studies, it has emerged that the expression of the class I heat shock proteins of S. mutans is highly responsive to intermittent and sustained exposure to relevant environmental stresses (13, 18).
The ubiquitously distributed class I stress proteins, the DnaK and GroEL molecular chaperones, are central to the tolerance to environmental stresses and participate in a variety of cellular processes including protein folding, protein translocation, and assembly and disassembly of protein complexes (8, 11, 26). The GroEL and DnaK complexes, which include GroES and DnaJ-GrpE, respectively, also regulate signal transduction pathways by controlling the stability and activities of transcriptional regulators and protein kinases (8, 11). In many gram-positive bacteria, transcription of the groE (groES-groEL) and dnaK (hrcA-grpE-dnaK-dnaJ) operons is negatively controlled by HrcA, which binds to a highly conserved cis-acting element (CIRCE) located in the regulatory regions of these operons (28, 30). In some cases, the groE and dnaK operons can also be under the negative control of CtsR, a repressor that binds to a conserved direct-repeat sequence and that was initially identified for its role in regulating clp gene expression (5, 7).
Previously, we demonstrated that the transcription of the dnaK operon in S. mutans is tightly controlled by HrcA (13, 18), which binds to two CIRCE elements located in the dnaK promoter region. The expression of groE in S. mutans is under the dual control of the HrcA and CtsR repressors, although the repression by CtsR is not as strong as the repression by HrcA (16). It was also demonstrated that the transcription of both operons is rapidly induced by acid shock and other stresses and that elevated levels of DnaK are maintained under acidic conditions (13, 18). In Escherichia coli, GroEL is essential for growth at all temperatures, while DnaK is essential only at temperatures above 37°C and below 15°C (9, 10, 12). In the gram-positive paradigm Bacillus subtilis, DnaK is essential only at temperatures above 52°C (27). In S. mutans, attempts to inactivate hrcA by inserting the strongly polar ΩKm cassette resulted in the isolation of only single-crossover insertions, even at lower temperatures or in buffered medium, indicating that the transcription of the downstream grpE-dnaK-dnaJ genes, coding for the DnaK machinery, was essential for cell viability. Similarly, strains lacking GroEL in S. mutans could not be isolated.
To evaluate the role of HrcA as a repressor protein in chaperone expression, an HrcA-deficient strain, SM11, was constructed by allelic replacement of the 5′ portion of the gene with a polar kanamycin cassette (ΩKm) followed by the Streptococcus salivarius urease promoter (PureI) (6, 18). The HrcA mutant strain, which had constitutively elevated levels of GroES-GroEL, presumably due to a loss of the HrcA repressor, was more sensitive to acid killing and could not lower the pH as effectively as the parent (18). The acid-sensitive phenotype was, at least in part, attributable to lower F-ATPase activity (18). However, SM11 had only 50% of the DnaK protein found in the parent strain, probably because transcription from the S. salivarius PureI promoter was not as efficient as it was from the cognate promoter. Thus, the behavior of SM11 suggested that decreases in the levels of DnaK might be responsible for decreased acid resistance, although this strain lacked HrcA and overproduced GroES-GroEL. To distinguish the roles played by molecular chaperones in the physiology of S. mutans, we isolated strains that showed significant reductions in the levels of DnaK or GroEL. The data presented here demonstrate that the forced down-regulation of DnaK and GroEL had substantially different effects on S. mutans and confirmed that molecular chaperones play essential roles in core physiologic responses and virulence-associated attributes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. mutans UA159 and its derivatives were routinely grown in brain heart infusion (BHI) broth in a 5% CO2 aerobic atmosphere at 37°C. When needed, kanamycin (1 mg ml−1) was added to the medium. The ability to form stable biofilms was assessed by growing cells in wells of polystyrene microtiter plates using biofilm medium (BM) (19). For acid adaptation studies, cells were grown to an optical density at 600 nm (OD600) of 0.3 in BHI medium adjusted to pH 7.0 with KOH, harvested by centrifugation, resuspended in fresh BHI medium that was adjusted to pH 5.0 with HCl, and incubated for 2 h. Biofilms used for acid killing experiments and F-ATPase assays were grown in BM for 48 h in wells of 24-well polystyrene tissue culture plates (flat bottom). Cells were grown in continuous culture in a BioFloIII chemostat (New Brunswick Scientific, Edison, NJ) in TY base medium (3% tryptone, 0.5% yeast extract) containing 25 mM glucose, as previously described (13). The dilution rate (D) of the culture was 0.3 h−1, corresponding to a generation time of 2.3 h, and the pH of the vessel was maintained at either 7.0 or 5.0 by the addition of 1 N KOH. The steady state was considered to be achieved when the cultures were maintained under a particular growth condition for at least 10 generations.
DNA methods.
Chromosomal DNA was prepared from S. mutans as previously described (4). Restriction and DNA-modifying enzymes were obtained from Life Technologies Inc. (Gaithersburg, MD) or New England Biolabs (Beverly, MA). PCRs were carried out with 100 ng of S. mutans chromosomal DNA using Taq DNA polymerase, and PCR products were purified by using the QIAquick kit (QIAGEN). Plasmid DNA was introduced into E. coli by the calcium chloride method (25). Mutants of S. mutans were generated by natural transformation (23) with DNA from previously established strains or by using a PCR ligation mutagenesis approach (14). Southern blotting was carried out under high-stringency conditions as detailed elsewhere previously (25).
Construction of strains.
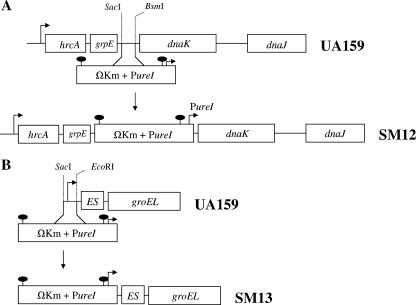
To down-regulate dnaK expression, a 1.0-kbp fragment containing the grpE-dnaK intergenic region and flanking portions of grpE and dnaK were amplified and cloned into pGEM-5Zf(+) (Promega, Madison, WI) to generate pJL71. In pJL71, a fragment containing 267 bp of the grpE-dnaK intergenic region was replaced by an antibiotic cassette that contains the polar ΩKm element (22) followed by the Streptococcus salivarius urease promoter (PureI) (6). The resulting plasmid was then isolated and used to transform S. mutans. By using this strategy, transcription of hrcA and grpE was still driven by the cognate promoter (PhrcA), whereas dnaK and dnaJ were transcribed through the weaker PureI promoter. To down-regulate groES-EL expression, a 40-bp region containing the −35 and −10 sequences of the groE operon was replaced by the ΩKm-PureI cassette. Briefly, two 0.5-kb fragments flanking the −35 and −10 sequences of the groE promoter were amplified by PCR, ligated into the ΩKm-PureI cassette, and used to transform S. mutans. Schematic diagrams depicting the construction of strains SM12 and SM13 are shown in Fig. 1.
FIG. 1.
(A) Construction of dnaK (SM12) and groE (SM13) knockdown strains. To down-regulate dnaK and dnaJ, a fragment containing 267 bp of the grpE-dnaK intergenic region was replaced by a cassette containing the ΩKm element and the PureI promoter. (B) To knock down groE expression, a 40-bp region containing the native groE promoter was replaced by the ΩKm-PureI cassette. For more details, see Materials and Methods.
RNA methods.
RNA was extracted by the hot acid-phenol method as described elsewhere previously (1). Levels of dnaK, groEL, and clpP mRNAs were quantified by real-time quantitative reverse transcriptase PCR (RT-PCR). Primer design, RT-PCRs, real-time RT-PCR cycling conditions, data analysis, and quality control were performed as described elsewhere previously (1). A Student's t test was employed to analyze the significance of the real-time RT-PCR quantifications.
Acid killing experiments.
For acid killing of planktonic cells, strains were grown in BHI medium to the desired growth phase, washed once with 0.1 M glycine buffer (pH 7.0), and resuspended in one-half of the original volume of 0.1 M glycine buffer (pH 2.8) for up to 60 min. For acid killing of biofilm-grown cells, cultures were grown in polystyrene plates in BM. Cells were incubated at 37°C in a 5% CO2 atmosphere for 48 h and then subjected to acid killing. Briefly, planktonic cells were discarded, and the plates were blotted onto absorbent paper. Biofilms were then incubated in 0.1 M glycine (pH 2.8) for up to 90 min. For each time point, biofilms from two wells were independently resuspended in the glycine solution by repeated pipetting, transferred into a 1.5-ml Eppendorf tube, and dispersed by vortexing at high speed for 30 s. Dispersed cells were serially diluted, plated in duplicates onto BHI plates, and incubated for 3 days before colonies were counted. Cell viability at each time point was expressed as the percentage of viable cells (CFU ml−1) at time zero.
ATPase assays.
For F-ATPase assays, cells were permeabilized with toluene and incubated with 5 mM ATP in ATPase buffer as previously described (2). Samples were removed at various intervals and assayed for inorganic phosphate release from ATP with the Fiske-Subbarow reagent (Sigma, St. Louis, MO). ATPase activity was expressed as nmol of PO4 min−1 mg protein−1. The protein concentration was determined using the BCA assay (Sigma) with bovine serum albumin as the standard.
Protein electrophoresis and Western blotting.
Whole-cell lysates for protein analysis were obtained by homogenization in the presence of glass beads with a Bead Beater (Biospec, Bartlesville, OK), as previously described (6). Protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto Immobilon-P membranes (Millipore, Bedford, MA), and subjected to Western blot analysis by using standard techniques. Membranes were incubated with antibodies raised against purified, recombinant Streptococcus pyogenes DnaK and GroEL proteins (17). Immune reactivity was detected by incubation with peroxidase-conjugated goat anti-rabbit immunoglobulin G followed by detection with 4-chloro-1-naphthol. Two-dimensional (2D) gel electrophoresis was performed by Kendrick Labs, Inc. (Madison, WI), according to a method described previously by O'Farrell (21). The protein concentration of samples was determined using the bicinchoninic assay (Sigma).
RESULTS AND DISCUSSION
Forced down-regulation of groES-groEL and dnaKJ.
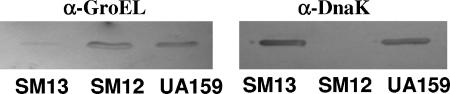
Previous unsuccessful attempts to isolate strains completely lacking GroEL or DnaK support that both proteins are essential for the viability of S. mutans. To gain further insight into the roles of DnaK and GroEL in the physiology of S. mutans, we created a knockdown strategy to lower the levels of DnaK and GroEL, resulting in strains SM12 and SM13, respectively (Fig. 1). Western blotting with polyclonal antisera against purified, recombinant GroEL or DnaK from S. pyogenes was used to confirm that the SM12 and SM13 strains had diminished levels of DnaK and GroEL, respectively (Fig. 2). Densitometric analysis revealed that the levels of DnaK in SM12 were <5% of that found in S. mutans UA159 grown under the same conditions, and GroEL levels in SM13 were about 20% of those found in the wild-type strain. Interestingly, when we used the same ΩKm-PureI cassette in a previous study (18) to inactivate the hrcA gene, the first gene in the dnaK operon, we observed only a twofold reduction in DnaK levels. Thus, as we posited previously (18), the grpE-dnaK intergenic region may play a significant role in dnaK mRNA stability.
FIG. 2.
Western blot analysis of DnaK and GroEL levels with polyclonal antibodies against S. pyogenes GroEL (1:500) (α-GroEL) and DnaK (1:1,000) (α-DnaK). Total cell lysates (10 μg per lane) were obtained from mid-exponential-phase cultures grown in BHI medium.
The strain producing low levels of DnaK, SM12, also showed enhanced expression of GroEL (Fig. 2). In B. subtilis, the GroES-GroEL complex has been shown to interact directly with HrcA (20), increasing the binding affinity of the HrcA repressor for the CIRCE element located in the dnaK and groE promoter regions. In S. mutans, the groE operon is under the control of HrcA, which binds to a CIRCE element in the promoter region but also is subject to regulation by CtsR, which binds a consensus heptad near the HrcA binding site (16). It does not appear that the reason that GroEL levels are increased in SM12 is due to effects on CtsR expression or activity. In particular, we demonstrated previously that clpP is under negative control by CtsR in S. mutans (16). Comparisons of the levels of clpP mRNA by real-time PCR did not reveal any differences between UA159 and SM12. Moreover, ctsR levels were not altered in either SM12 or SM13 (data not shown). In contrast, there is supporting evidence that S. mutans DnaK has a significant role in modulating the activity of HrcA. In particular, hrcA expression, which is autoregulated by HrcA, was found to be increased approximately 150-fold in SM12 but not in SM13 (data not shown). Western blot analysis using a polyclonal antibody raised against S. mutans HrcA (18) revealed higher levels of HrcA in the SM12 strain (data not shown), suggesting that the DNA binding activity of HrcA may be compromised, perhaps because DnaK is required for maintaining HrcA in an active state.
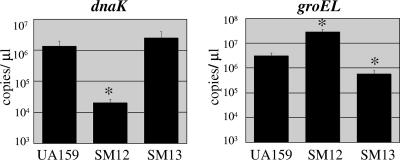
To confirm that the differences in chaperone expression observed at the protein level were due to changes in the transcription of the genes, Real-time RT-PCR was used to quantify groEL and dnaK mRNA in SM12 and SM13. Compared to the parent stain, the levels of dnaK mRNA were diminished by about 100-fold in SM12, and groEL mRNA was reduced about eightfold in SM13. The results also indicated that groEL was up-regulated in SM12, whereas dnaK expression levels were not altered in SM13 (Fig. 3). Of note, real-time PCR quantification also indicated that dnaJ levels were reduced by approximately 100-fold in SM12, consistent with our observation that dnaJ is cotranscribed with dnaK. The mRNA measurements are generally consistent with the protein measurements, although differences in the magnitudes of the changes in protein and mRNA in the mutants may reflect roles for the posttranscriptional control of the levels of DnaK and GroEL.
FIG. 3.
Real-time PCR quantification of dnaK and groEL mRNA. Strains UA159 (wild type), SM12 (dnaK knockdown), and SM13 (groE knockdown) were grown in BHI medium to the mid-exponential phase. The data represent the means ± standard deviations from three independent experiments. The asterisk indicates that the result was statistically significant (P < 0.01; Student's t test).
Characteristics of dnaK and groE knockdown strains.
The physiological characteristics of strains SM12 (dnaK knockdown) and SM13 (groE knockdown) are shown in Table 1. Both mutant strains grew slower than the wild type, formed long chains, and had a strong tendency to aggregate in broth culture. SM12 and SM13 had doubling times of 76 ± 11.5 min and 90 ± 17.3 min, respectively, while the doubling time for UA159 was 44 ± 3.6 min. Despite the slow-growth phenotype, both SM12 and SM13 were able to reach the same final optical density as the parent strain in approximately 22 and 28 h, respectively.
TABLE 1.
Characteristics of S. mutans dnaK knockdown (SM12) and groE knockdown (SM13) strainsa
| Strain | Doubling time (min) at 37°C | Growth characteristic(s) at 37°C | Growth at 43°C | Growth at pH 5.0 | H2O2 inhibition zone (cm) |
|---|---|---|---|---|---|
| UA159 | 44 ± 3.6 | Confluent growth | +++/extensive clumping | +++ | 1.22 ± 0.05 |
| SM12 | 76 ± 11.5 | Extensive clumping/long chains | NG | NG | Sensitive (1.52 ± 0.09) |
| SM13 | 90 ± 17.3 | Extensive clumping/long chains | +/extensive clumping | +++ | Like UA159 (1.12 ± 0.08) |
NG, no growth; +, poor growth; +++, wild-type-like growth.
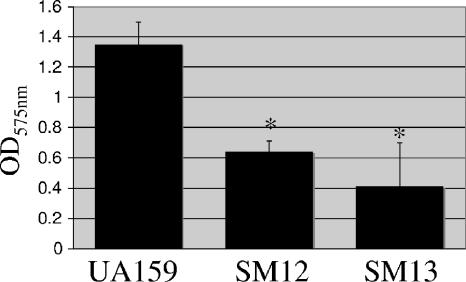
Biofilm formation assays were performed with BM supplemented with glucose at a final concentration of 20 mM. To exclude the possibility that any observed differences were due to variations in the growth capacities of the strains, cells were incubated for 48 h, and the final OD600 of the cultures in the microtiter plates was assessed prior to the assay. The data obtained indicated that although both mutants were capable of reaching the same final OD600 as the parent strain in BM, both SM12 and SM13 strains had a reduced capacity to form biofilms (Fig. 4).
FIG. 4.
Biofilm formation by S. mutans UA159 (wild type) and its derivatives. Cultures were grown in a microtiter plate containing BM supplemented with glucose at 37°C for 48 h. The graph shows the averages and standard deviations for five independent experiments. The asterisks indicate that the results were statistically significant in comparison to UA159 (P < 0.05; Student's t test).
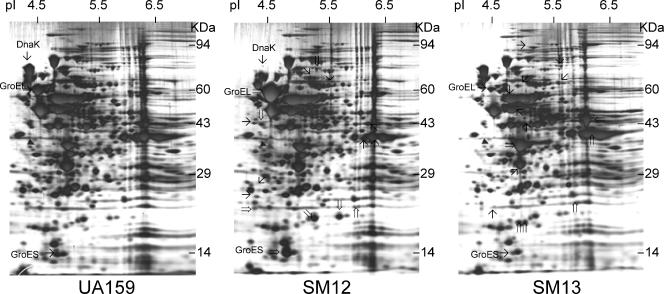
2D gels revealed that the forced down-regulation of DnaK and GroEL caused profound changes in the protein expression patterns of S. mutans (Fig. 5). Compared to UA159, at least 9 proteins were up-regulated and 10 were down-regulated in the dnaK knockdown strain (SM12). Consistent with the Western blot results, DnaK was down-regulated and GroES and GroEL were among the up-regulated proteins in SM12. The 2D protein pattern of the groEL knockdown strain (SM13) indicated that at least 10 proteins were up-regulated and 12 proteins were down-regulated in comparison to the parent strain. As expected, the expression of GroES and GroEL was markedly reduced in SM13, whereas DnaK levels were not affected. In S. mutans, the inactivation of trigger factor (TF), a ribosome-associated molecular chaperone, resulted in the up-regulation of both GroES-GroEL and DnaK (29). The 2D protein profile indicated that the expression of TF was not altered in the dnaK and groEL knockdown strains of S. mutans. It was previously demonstrated that TF has overlapping functions and cooperates with the DnaK machinery (10). In E. coli, a deficiency of both DnaK and TF results in a massive aggregation of cytosolic proteins and growth in only a very narrow temperature range, and these phenotypes were partially suppressed by the overexpression of GroES-GroEL (10).
FIG. 5.
2D protein pattern of S. mutans UA159 (wild type), SM12 (dnaK knockdown), and SM13 (groE knockdown). Samples were grown in BHI medium and harvested in the exponential growth phase (OD600 ≅ 0.5). Protein extracts (50 μg per gel) were separated by isoelectric focusing in the pI range of 4 to 8 in the first dimension and by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis in the second dimension. The silver-stained proteins that exhibited more obvious differences in the SM12 and SM13 strains in comparison to UA159 are indicated. Proteins with enhanced expression are indicated with double arrows, and proteins with reduced synthesis are indicated with black arrows. The filled triangle indicates tropomyosin protein loaded as an internal control.
Effects of DnaK and GroEL down-regulation on stress tolerance.
The capacity of the mutant strains to grow on BHI agar plates that had been adjusted to pH 5.0 with HCl was tested. Both UA159 and SM13 could form normal-sized colonies on acidified BHI after 48 h, whereas no growth of the DnaK down-regulated strain was observed, even when the plates were incubated for up to 7 days (Table 1). The capacity of the strains to grow at temperatures above 37°C was tested. At 43°C, growth rates of the parent strain were significantly reduced, and extensive cell clumping was observed. SM13 showed very poor growth yields at 43°C, whereas no growth was observed when SM12 was incubated at this temperature (Table 1). Next, we evaluated the capacity of the strains to form colonies in the presence of H2O2. For that purpose, a uniform layer of cells was spread onto BHI agar plates, and paper filter discs saturated with 0.5% H2O2 were placed onto the agar. The zone of inhibition caused by H2O2 diffusion was similar for the parent and SM13 strains (approximately 1.22 ± 0.05 cm and 1.2 ± 0.08 cm, respectively), whereas SM12 was more sensitive, with an inhibition halo of 1.52 ± 0.09 cm (Table 1). Collectively, these results indicate a stress-sensitive phenotype of the dnaK knockdown strain but reveal that the down-regulation of GroEL has only a minor effect on the stress tolerance properties of S. mutans, although it should be taken into consideration that there was a more dramatic reduction in the levels of DnaK in SM12 than in the levels of GroEL in SM13.
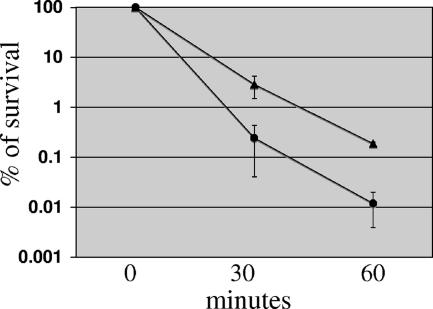
To better assess the acid tolerance properties of SM12, acid killing experiments were performed with biofilm-grown cells that were exposed to pH 2.8. Unexpectedly, biofilm cultures of SM12 were more resistant to acid killing (approximately 1 log) than the parent strain (Fig. 6). Similar results were obtained when mid-exponential- or stationary-phase planktonic cultures were used (data not shown). At first, the impaired capacity to grow at pH 5.0 and enhanced tolerance to acid killing of SM12 appeared to be contradictory. However, acid killing experiments test the capacity of the strain to survive a lethal pH for a limited period of time (up to 90 min) and then resume growth when transferred to medium at a neutral pH. Because the growth of SM12 was considerably slower than that of the parent strain, one possible explanation for this unexpected phenotype is that cells with a slower metabolism become less susceptible to the damage caused by acidification. In fact, using chemostat-grown cells, it was demonstrated that fast-growing cells of E. coli were more sensitive to stresses than slow-growing cells (3). To investigate this possibility, steady-state cultures of the wild-type strain were grown in continuous culture under two different dilution rates and subjected to acid killing. The results obtained clearly indicated that cells grown at a lower D of 0.1 h−1 (generation time, 6.9 h) were more resistant to acid killing than cells grown at a higher dilution rate (D of 0.3 and a generation time of 2.3 h) (J. A. Lemos and R. A. Burne, unpublished data).
FIG. 6.
Survival of S. mutans strains UA159 (wild type) (circles) and SM12 (dnaK knockdown) (triangles) after acid challenge. Cells from 48-h biofilms grown on the surface of polystyrene plates were subjected to acid killing in 0.1 M glycine (pH 2.8). Cell viability at each time point is expressed as the percentage of viable cells (CFU ml of culture−1) at time zero. A Student's t test indicated that the differences observed at 60 min were statistically significant (P < 0.001).
Effects of DnaK down-regulation on F-ATPase activity.
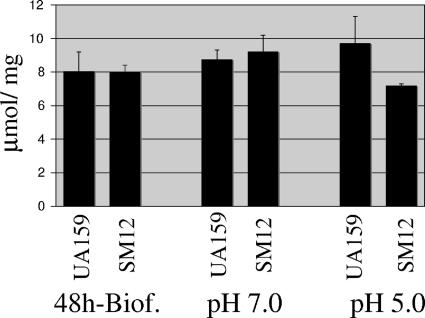
In S. mutans, the proton-translocating ATPase (F-ATPase) is considered to be the primary mechanism of proton extrusion to maintain ΔpH during growth in acidic conditions (24). Previously, a strain lacking the HrcA regulator (SM11), which displayed constitutively high levels of GroEL and lower levels of DnaK, showed diminished F-ATPase activity when grown at pH 5.0 in a continuous chemostat culture (18), although no differences were noted at a neutral pH. It was speculated that DnaK participates in the biogenesis of the F-ATPase complex or in stabilizing this complex at a low pH. Although the dnaK knockdown strain (SM12) isolated in this study also displayed high levels of GroEL, the levels of DnaK were drastically reduced in comparison to the SM11 strain. F-ATPase activity of the wild-type and SM12 strains was assayed in permeabilized cells from 48-h biofilms and in cells growing exponentially at pH 7.0 or following acid adaptation at pH. 5.0. No differences in the F-ATPase activities of the parent and SM12 strains were observed when cells were grown in biofilms or in batch culture at pH 7.0. In contrast, acid-adapted cells of SM12 showed consistent reductions in F-ATPase activity compared to the wild-type strain grown under the same conditions, although the differences observed were not statistically significant (Fig. 7). This finding coupled with our previous results with the SM11 strain (18) clearly support that the DnaK chaperone complex is important for the biogenesis or stabilization of the F-ATPase complex at a low pH, when the expression of the ATPase is known to be elevated. The manifestation of the difference only during growth at low pH values may be attributable to the titration of DnaK to damaged proteins in acidic conditions or perhaps because the enzyme requires DnaK, directly or indirectly, for stability at a low pH. An alternative explanation may be related to a role for DnaK in stabilizing a transcriptional activator of the atp operon that is required for efficient induction at a low pH. Real-time PCR did not indicate differences in the mRNA levels of atpF (Fo domain, b subunit) between the UA159 and SM12 strains of cells growing exponentially in rich medium (data not shown), but further studies will be needed to assess the way in which DnaK impacts ATPase activity.
FIG. 7.
F-ATPase activity of S. mutans strains UA159 (wild-type) and SM12 (dnaK knockdown). Extracts were prepared from exponentially growing cells at pH 7.0, batch-cultured acid-adapted cells at pH 5.0, and 48-h biofilms. The results are the averages and standard deviations of three independent cultures. The differences observed in cells grown at pH 5.0 were not statistically significant (Student's t test).
Instability of dnaK and groEL knockdown strains in culture.
The initial concern for growing SM12 and SM13 in the chemostat was that the strains would wash out because of their slow-growth phenotype. After allowing cells to establish batchwise in the chemostat without pH control, both strains were able to maintain constant and adequate cell densities at a dilution rate of 0.3 h−1 (generation time, 2.3 h) when the pH of the vessel was kept at 7.0. After sampling steady-state cultures growing at pH 7.0, cultures were acid shocked to pH 5.0 by titration with HCl over the course of about 1 min. Neither SM12 nor SM13 was capable of tolerating such rapid acidification of the vessel, and washout was evident shortly after the acid challenge. To obtain steady-state cells of SM12 and SM13 growing at pH 5.0, the pH control of the vessel was adjusted to 5.0, and the culture was allowed to slowly lower the pH through glycolysis. The wild-type strain was capable of lowering the pH from 7.0 to 5.0 in a matter of hours, whereas SM12 and SM13 took as long as 48 h to drop the pH to 5.0.
Interestingly, when aliquots were collected from the chemostat at pH 5.0 and plated onto BHI agar plates, both mutant strains were able to form visible colonies after overnight incubation, suggesting that the slow-growth phenotype was lost. A closer examination of these cultures indicated that the strains no longer aggregated in broth, although resistance to kanamycin associated with the insertion of foreign DNA was retained. The levels of DnaK and GroEL in cells obtained from the chemostat kept at pH 7.0 or 5.0 were assessed by Western blot analysis. SM12 cells grown at pH 5.0 demonstrated that the reversion of the growth phenotypes was accompanied by the restoration of DnaK levels (data not shown). In contrast, phenotypic reversion of SM13 was not associated with restored levels of GroEL. PCR and sequence analysis of the mutated regions did not reveal changes in the site where the foreign DNA and promoter had inserted in either strain. The instability of both mutants indicates that the suppression of the phenotype occurs rapidly when cells are cultivated under stressed conditions and that reversion is associated with increases in the levels of chaperones in the case of DnaK but that extragenic suppression may be a factor for the reversion of both SM12 and SM13.
Concluding remarks.
In this study, the down-regulation of dnaK and groEL generated pleiotropic effects and confirmed the essential nature of the two major molecular chaperones in S. mutans physiology. Moreover, the results presented here provide further evidence of molecular linkages between stress responses and biofilm formation in S. mutans (15). Thus, it is becoming clear that an appropriately regulated response by the microorganism to the environmental stresses encountered during the development and maturation of a biofilm have a profound influence on the biofilm structure or whether biofilms will form at all. Studies to dissect how chaperones affect the expression or biogenesis of known virulence attributes involved in biofilm formation are ongoing.
Acknowledgments
This study was supported by NIH-NIDCR award DE13239.
Footnotes
Published ahead of print on 15 December 2006.
REFERENCES
- 1.Ahn, S. J., J. A. Lemos, and R. A. Burne. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J. Bacteriol. 187:3028-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belli, W. A., and R. E. Marquis. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57:1134-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berney, M., H. U. Weilenmann, J. Ihssen, C. Bassin, and T. Egli. 2006. Specific growth rate determines the sensitivity of Escherichia coli to thermal, UVA, and solar disinfection. Appl. Environ. Microbiol. 72:2586-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burne, R. A., K. Schilling, W. H. Bowen, and R. E. Yasbin. 1987. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J. Bacteriol. 169:4507-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y. Y., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 8.Erbse, A., M. P. Mayer, and B. Bukau. 2004. Mechanism of substrate recognition by Hsp70 chaperones. Biochem. Soc. Trans. 32:617-621. [DOI] [PubMed] [Google Scholar]
- 9.Fayet, O., T. Ziegelhoffer, and C. Georgopoulos. 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 171:1379-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genevaux, P., F. Keppel, F. Schwager, P. S. Langendijk-Genevaux, F. U. Hartl, and C. Georgopoulos. 2004. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 5:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 12.Itikawa, H., and J.-I. Ryu. 1979. Isolation and characterization of a temperature-sensitive dnaK mutant of Escherichia coli B. J. Bacteriol. 138:339-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 14.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 15.Lemos, J. A., J. Abranches, and R. A. Burne. 2005. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 7:95-107. [PubMed] [Google Scholar]
- 16.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemos, J. A., R. A. Burne, and A. C. Castro. 2000. Molecular cloning, purification and immunological responses of recombinants GroEL and DnaK from Streptococcus pyogenes. FEMS Immunol. Med. Microbiol. 28:121-128. [DOI] [PubMed] [Google Scholar]
- 18.Lemos, J. A., Y. Y. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry, D., and H. K. Kuramitsu. 1981. Genetic transformation of Streptococcus mutans. Infect. Immun. 32:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quivey, R. G., Jr., W. L. Kuhnert, and K. Hahn. 2000. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42:239-274. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cols Spring Harbor, NY.
- 26.Schlieker, C., B. Bukau, and A. Mogk. 2002. Prevention and reversion of protein aggregation by molecular chaperones in the E. coli cytosol: implications for their applicability in biotechnology. J. Biotechnol. 96:13-21. [DOI] [PubMed] [Google Scholar]
- 27.Schulz, A., B. Tzschaschel, and W. Schumann. 1995. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol. Microbiol. 15:421-429. [DOI] [PubMed] [Google Scholar]
- 28.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen, Z. T., P. Suntharaligham, D. G. Cvitkovitch, and R. A. Burne. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73:219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]